Abstract
We have previously reported specific swine leukocyte antigen (SLA) haplotype associations with significant effects on several reproduction performance traits in a highly inbred miniature pig population of Microminipigs (MMPs). In this study, to clarify the effects on farrowing rates of SLA similarity between mating partners in the MMP population, we compared the farrowing rates as a measure of reproductive success after 1063-cumulative matings among the following three groups of mating partners: (1) completely sharing SLA class I or class II haplotypes or alleles between partners (CS), (2) only one sharing the haplotypes or alleles (OS), and (3) non-sharing the haplotypes or alleles (NS). Average farrowing rates in CS groups consisting of completely sharing SLA class II haplotypes or DRBI and DQB1 alleles were lowest in the three groups. Moreover, lower farrowing rates were indicated in mating pairs with smaller amino acid pairwise genetic distances of SLA-1, SLA-3, DRB1 and DQB1 alleles between the pairs. These results suggested that the dissimilarity of SLA class I and class II alleles between mating partners markedly improved reproductive performance; therefore, SLA alleles or haplotypes are potentially useful genetic markers for the selection of mating pairs in breeding programs and epistatic studies of reproductive traits of MMPs.
1. Introduction
Many genetic and environmental factors play an important role for successful fertilization and conceptus growth through the gestation period, culminating in the birth of healthy offspring. Successful programs for breeding pigs from conception to birth depend on the outcomes of mating selected sows and sires, fertilization, implantation, gestation, and farrowing achievements. [1,2].
In pigs, as well as in other mammals such as mice, cattle, and humans, genes within the major histocompatibility complex (MHC) genomic region are one of the genetic factors involved in fertilization, selective abortions, and modulating mate choice [3,4,5]. Many studies showed that the sharing of certain maternal–fetal or paternal human leukocyte antigens (HLA) and bovine leukocyte antigens (BoLA) may influence fetal development and survival [5,6]. In pigs, the swine leukocyte antigen (SLA) class I and class II genes within the SLA genomic region are highly polymorphic and have important roles in the regulation of immune recognition and immune responses to foreign antigens and allo- or xeno-grafts in transplantation [7,8,9,10,11,12,13]. The polymorphisms of SLA genes enabled the analysis of associations between genotypes, alleles, haplotypes, and various immune responses and reproductive performance [7,14]. The influence of SLA-encoded genes on reproductive performances in domestic pigs has been reported as the association between specific SLA haplotypes and genital tract development in males, ovulation rates, litter sizes, and piglet weights at birth and weaning [7,15,16,17,18]. In selectively bred Duroc pigs with two low resolution SLA class II haplotypes (Lr), Lr-0.13 and Lr-0.30, assigned by a PCR-SSP method, Lr-0.30 was associated with higher weaning and rearing rates [16]. Furthermore, in a Landrace pig line with eleven SLA class II haplotypes, pigs with Lr-0.23 or Lr-0.13 had less severe pathological lesions of mycoplasmal pneumonia, increased leukocyte phagocytic activity, and higher white blood cell counts in comparison to pigs with other haplotypes [14].
A Microminipig (MMP) was developed by Fuji Micra Inc. (Fujinomiya, Shizuoka, Japan) as a novel miniature pig with an extremely small body size for use in laboratory biological and medical research. The body sizes, such as body weight, height, chest width, and chest circumference at 4–6 months of age, were much smaller than those of young mature beagles at 10 months old [17,18,19,20]. In the population of MMPs, 11 SLA class I and II haplotypes, including three recombinant haplotypes, were identified in the 14 parents or the progenitors of the highly inbred MMP herd. A total of 25 class I alleles (nine alleles of the SLA-1, eight alleles of the SLA-2 and eight alleles of the SLA-3 genes) were identified previously in MMPs with 8 SLA class I haplotypes. Homozygous MMPs with high resolution haplotype (Hp-) 35.0 had two different alleles at the SLA-1 gene, SLA-1*12:01 and SLA-1*13:01, suggesting duplicated SLA-1 genes [21]. Recently, in the MMP population, we analyzed associations between the SLA class II haplotypes and reproductive performances, such as the fertility index that is expressed as the farrowing rate (reproductive success), gestation periods, litter sizes, and stillbirth rates in MMPs [18,22]. Two SLA class II haplotypes, Lr-0.13 and Lr-0.18, in both dams and sires showed lower and higher fertility indices, respectively, in comparison to the other six haplotypes. A significant effect of Lr-0.23 in dams also showed smaller litter size in comparison to seven other haplotypes [18]. Moreover, the SLA complex also appears to influence stillbirth rates in the MMP population [22].
In the present study, to evaluate the contribution of SLA class I and class II genes on reproductive traits in MMPs, we investigated the involvement of SLA sharing between mating pairs on their farrowing rates among SLA class I and class II haplotypes, including SLA-homozygous individuals. Furthermore, relationships among farrowing rates and amino acid distances of SLA alleles between mating pairs were also analyzed.
2. Materials and Methods
2.1. Animals
In this study, we used an MMP herd bred at Fuji Micra Inc. (Fujinomiya, Japan) from June 2008 to February 2017. In the herd, the records of 1063 cumulative matings of MMPs consisting of 114 sows and 44 boars assigned to 11 different SLA class I and class II haplotypes including 3 recombinant ones were utilized for the measurement of farrowing rates (reproductive success). The matings of MMPs were basically random. However, during some generations, especially with initial matings, mating pairs with relatively small body sizes were preferentially selected to establish the characteristics of the MMP breed [19].
This study was approved by the Animal Care and Use Committee of Gifu University (#17042, 26 May 2017). The care and use of the laboratory animals were in compliance with the guidelines of Good Laboratory Practice of Gifu University and Fuji Micra Inc.
2.2. SLA Class I and Class II Typing
Polymorphic SLA alleles for three class I (SLA-1, SLA-2, and SLA-3) and two class II (DRB1 and DQB1) genes were assigned according to low-resolution SLA genotyping in 158 MMPs (114 sows and 44 boars), using a PCR-sequence-specific primers (SSP) method, as described previously [21]. Eleven types of high-resolution SLA class I and class II haplotypes (Hp-6.7, Hp-10.11, Hp-16.16, Hp-17.17, Hp-20.18, Hp-31.13, Hp-35.23 and Hp-43.37) including three class I and class II recombinant haplotypes (Hp-10.23, Hp-35.17 and Hp-43.17) were deduced from the preliminary results in the herd that were determined by performing an analysis of the inheritance and segregation of alleles of the three class I (SLA-1, SLA-2, and SLA-3), and two class II genes (DRB1 and DQB1), respectively, in descendants of the MMP population (Table 1). To analyze the association between SLA class I, class II alleles, haplotypes and farrowing rates, and evolutionary amino acid divergences among SLA class I and class II alleles, the SLA alleles in four-digit genotypes of the 158 MMPs were deduced from the two-digit genotypes for the three class I (SLA-1, SLA-2, and SLA-3), and two class II (DRB1 and DQB1) genes and the eleven types of low-resolution SLA class I and class II haplotypes (Table 1).

Table 1.
SLA-class I and II genotypes and number of SLA haplotypes in Microminipigs.
2.3. Measurement of Farrowing Rates
To clarify the influence of the SLA allele sharing on farrowing rates, each mating pair was classified into one of three mating groups, CS, OS, and NS, that may or may not have shared SLA class I or class II alleles and haplotypes between sows and boars. Firstly, the completely shared (CS) alleles and haplotypes group consisted of mating pairs that completely shared SLA class I or class II alleles and haplotypes encoding the SLA-1, SLA-2, SLA-3, DRB1 and DQB1 genes. Secondly, the only-one-shared (OS) group, consisted of mating pairs that shared only one SLA class I or class II allele or haplotype. Thirdly, the non-shared (NS) SLA alleles and haplotypes group consisted of mating pairs that had a completely different SLA class I or class II alleles and haplotypes.
Average farrowing rates in the three mating groups with different shared SLA haplotypes or alleles between sows and boars were calculated as the ratios of the number of deliveries including both live and still births compared to the number of matings. The farrowing rate in each mating group was calculated as the ratio of the number of deliveries to that of matings. The farrowing rates were compared among the three mating groups with the different shared SLA alleles or haplotypes.
2.4. Influence of Amino Acid Distance of SLA Class I and Class II Genotypes between Mating Partners on Farrowing Rates
To evaluate the relationship between farrowing rates and amino acid divergences of SLA class I (SLA-1, SLA-2, and SLA-3) or class II (DRB1 and DQB1) alleles in each mating pair, amino acid pairwise distances among the alleles were analyzed with MEGA X software using the JTT matrix-based model with a gamma distribution (Supplementary Tables S1–S5). The pairwise distances were calculated based on the number of amino acid substitutions per site between their allele sequences [23,24]. The sum of all the pairwise amino-acid distances (D) of the four possible alleles (A, a, B, and b) in each mating pair was calculated as follows:
Alleles with sire: A, a;
Alleles with dam: B, b.
The sum of all the pairwise amino-acid distances = (DAB + DAb + DaB + Dab). If the mating pair has exactly the same alleles, the sum of all the pairwise amino-acid distances is ‘0’.
Farrowing rates were calculated as the ratios of the number of deliveries to those of matings in range groups that were divided according to the sum of all the amino acid pairwise distances in each SLA locus among the possible four alleles between mating partners shown in Table 2. The sums of the amino acid pairwise distances among four alleles in each of the SLA class I genes and class II-DRB1 genes carried by each mating pair were classified into nine to eleven groups, which increased in distance from each other in intervals of 0.1. On the other hand, the sum of the amino acid pairwise distances among the SLA four alleles of the class II- DQB1 gene carried by each mating pair was classified into eight groups that increased in distance from each other by 0.05 ranges. Correlation coefficients were calculated between the central value of each range group and farrowing rate.

Table 2.
Number of matings in each range classified by the sum of the amino acid pairwise distances of SLA class I and class II alleles between mating pairs for correlation analyses among farrowing rates and the amino acid pairwise distances.
2.5. Statistical Analyses
Farrowing rates indicated as absolute numbers were evaluated by the Chi-square for independence test, using an m×n contingency table (BellCurve in Excel, Social Survey Research Information Co., Ltd. Tokyo, Japan). In Figure 1, Figure 2, Figure 3 and Figure 4, the farrowing rates were indicated as percentages for simpler comparisons. Correlations among farrowing rates and amino acid pairwise distances of SLA-1, SLA-2, SLA-3, DRB1 and DQB1 alleles between mating partners were evaluated by Spearman’s rank correlation coefficient. p-values of less than 0.05 were considered significant.
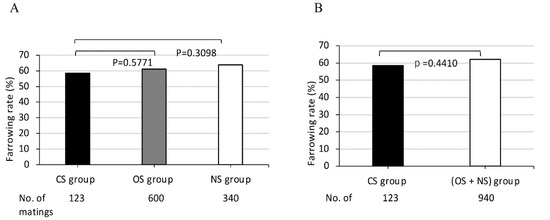
Figure 1.
Comparison of farrowing rates in MMPs among (A) three groups CS, OS, NS, and (B) between two groups (B), CS and (OS + NS). CS, completely sharing SLA class I haplotypes between partners; OS, sharing only one SLA class I haplotype between partners; NS, non-sharing of SLA class I haplotypes between partners. X-axis shows CS, OS, and NS groups (A) and CS and (OS + NS) groups (B) and the number of matings for each group. Y-axis shows farrowing rate as indicated by the ratio (%) of the number of deliveries to the number of matings, expressed as the mean value (bar). Black and white bars represent lower and higher farrowing rate, respectively, of the mean values. Gray bars represent intermediate farrowing rate of the mean values between those of black and white bars.
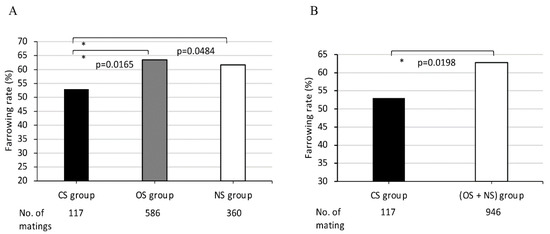
Figure 2.
Comparison of farrowing rates in MMPs among (A) three groups CS, OS, NS, and (B) between two groups, CS and (OS + NS). CS, completely sharing SLA class II haplotypes or DRB1 alleles between partners; OS, sharing only one SLA class II haplotype or DRB1 alleles between partners; NS, non-sharing, that is, SLA class II haplotypes or DRB1 alleles were completely different between partners. X-axis shows CS, OS, and NS groups (A), and CS and (OS + NS) groups (B), and the number of matings for each group. Y-axis shows farrowing rate as indicated by the ratio (%) of the number of deliveries to the number of matings, expressed as the mean value (bar). Black and white bars represent lower and higher farrowing rate, respectively, of the mean values. Gray bars represent intermediate farrowing rate of the mean values between those of black and white bars. Probabilities of significant differences among groups are indicated by single (p < 0.05) asterisks.
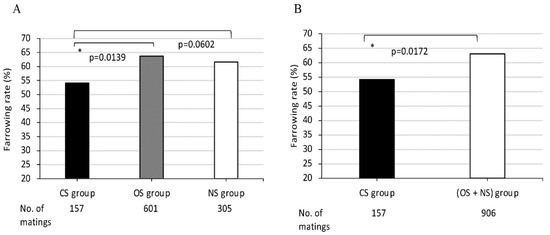
Figure 3.
Comparison of farrowing rates in MMPs among (A) three groups CS, OS, NS, and (B) between two groups CS and (OS + NS). CS, completely sharing SLA class II-DQB1 alleles between partners; OS, sharing only one DQB1 allele between partners; NS, non-sharing, that is, completely different DQB1 alleles between partners. X-axis shows CS, OS, and NS groups (A) and CS and (OS + NS) groups (B) and the number of matings for each group. Y-axis shows farrowing rate as indicated by the ratio (%) of the number of deliveries to the number of matings, expressed as the mean value (bar). Black and white bars represent lower and higher farrowing rate, respectively, of the mean values. Gray bars represent intermediate farrowing rate of the mean values between those of black and white bars. Probabilities of significant differences among groups are indicated by single (p < 0.05) asterisks.
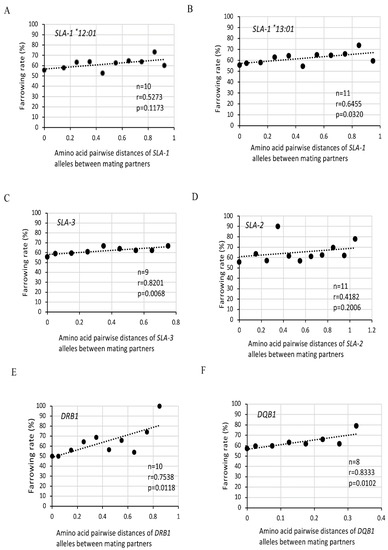
Figure 4.
Effects of amino acid pairwise distances of SLA class I and class II alleles between partners on farrowing rates in MMPs. Dots are indicated as farrowing rates (%) in the center value of each group of amino acid pairwise distances of SLA-1 (A,B), SLA-3 (C), SLA-2 (D), DRB1 (E), and DQB1 (F) alleles between partners as shown in Table 2. Dotted line shows the approximated line. n: number of groups on the sum of pairwise amino acid differences of SLA-1 (A,B), SLA-3 (C), SLA-2 (D), DRB1 (E), and DQB1 (F) alleles between partners as shown in Table 2. r: correlation coefficient, and p: p value evaluated by Spearman’s rank correlation coefficient.
3. Results
3.1. Association between Farrowing Rates and Sharing of SLA Class I Haplotypes or Alleles
A total of 656 pregnancies were obtained as the result of 1063 matings of 158 MMPs consisting of 114 sows and 44 boars, representing a farrowing rate of 61.7%. The farrowing rates in MMPs was considerably lower than 88.4 ± 4.6 (standard deviation (SD)) in mixed breed domestic pigs in Japan [25].
In the present study, 25 SLA class I alleles including two SLA-1 alleles (SLA-1*12:01 and SLA-1*13:01) encoded by duplicated SLA-1 genes in 114 dams and 44 sires were inherited as eight class I haplotypes without cross over among the three class I loci, SLA-1, SLA-2, and SLA-3. Therefore, the eight SLA class I haplotypes in MMPs consist of nine, eight, and eight classical SLA class I alleles in the SLA-1, SLA-2, and SLA-3 loci, respectively. Since associations among farrowing rates and the sharing of SLA class I haplotypes amount to the same results as association among farrowing rates and the sharing of SLA class I alleles, the following farrowing rates were found mostly as the sharing of SLA class I haplotypes.
The average farrowing rates of 114 dams of MMPs in the three mating groups, CS, OS, and NS, as classified according to the sharing of SLA class I haplotypes between partners, were 58.5%, 61.2% and 63.8%, respectively.
The farrowing rate of the CS group was lowest in comparison to those of the other two groups, the OS, and NS. However, no significant differences of the farrowing rates were obtained among the three groups consisting of different shared SLA class I haplotypes (Figure 1A). Furthermore, in the comparison of the farrowing rates between the CS group and other mating groups, no significant differences of the farrowing rates were obtained between the two groups, the CS and (OS + NS), consisting of only one sharing or non-sharing SLA class I haplotype between partners (Figure 1B).
3.2. Association between Farrowing Rates and Sharing of SLA Class II Haplotypes or Alleles
In addition to the three SLA class I recombinant haplotypes, there were only eight distinct SLA class II haplotypes in this MMP population (Table 1). These were Hp-0.7, Hp-0.11, Hp-0.13, Hp-0.16, Hp-0.17, Hp-0.18, Hp-0.23 and Hp-0.37, as determined by an analysis of the inheritance and segregation of eight and four genotypes of the DRB1 and DQB1 genes, respectively. Thus, we used the DRB1 and DQB1 alleles individually as well as their haplotypic linkages to assess their effects on farrowing rates. Since each of the DRB1 alleles corresponds specifically to only one of the eight class II haplotypes in the MMP population (Table 1), we used the DRB1 alleles as genetic markers of the class II haplotypes.
The 1063 matings of 158 MMPs were classified into the following three groups: the CS, OS, and NS groups, according to the sharing of SLA class II-DRB1 or -DQB1 alleles between each mating pair. The average farrowing rates in the three mating groups, CS OS, and NS, classified according to the sharing of DRB1 alleles between dams and sires in each mating pair, were 53.0%, 63.5% and 61.7%, respectively. The average farrowing rate of the CS group was lowest when compared to those of the other two groups, the OS and NS groups. The average farrowing rate of the CS group was significantly lower than those of OS and NS groups (p = 0.0165 and p = 0.0484, respectively (Figure 2A)). Furthermore, the average farrowing rate of the CS group was also significantly lower than that of the (OS + NS) group consisting of only one sharing and non-sharing DRB1 with completely different alleles between partners (p = 0.0198 (Figure 2B)).
The average farrowing rates in the three mating groups that were classified according to the sharing of DQB1 alleles between dams and sires in each mating pair were 54.1%, 63.7% and 61.6%, respectively. The average farrowing rate of the CS group was lowest in comparison to those of the other two groups, the OS and NS groups. A significant difference was observed between average farrowing rates of CS group and OS group (p = 0.0139 (Figure 3A)). When the average farrowing rates were compared between the CS and (OS + NS) groups, a significant difference was also observed (p = 0.0172 (Figure 3B)).
In comparing the sharing of SLA class II haplotypes and DRB1 alleles between dams and sires, the mating pairs were classified also into the CS, OS, and NS groups. The average farrowing rates of dams in CS groups with completely shared DRB1 or DQB1 genotypes between dams and sires were lowest when compared with the other mating groups with different class II alleles (Figure 2A and Figure 3A).
3.3. Effects of Amino Acid Pairwise Distances between SLA Class I Alleles of Mating Pairs on Farrowing Rates in MMPs
Amino acid distances among nine SLA-1, eight SLA-3 and eight SLA-2 alleles of mating pairs showed various values depending on the class I loci (Supplementary Tables S1–S3). The overall mean distances among the SLA-1, SLA-3 and SLA-2 alleles were 0.1972, 0.1178, and 0.2330, respectively. Since MMPs with Hp-35.0 duplicated SLA-1 genes that encode two alleles named as SLA-1*12:01 and SLA-1*13:01, the sum of pairwise amino acid distances of SLA-1 alleles of mating pairs was calculated separately for SLA-1*12:01 and SLA-1*13:01 (Table 2). The sum of the pairwise distances in 1063 matings among four SLA-1 alleles between partners increased from 0 to 0.910, and 0 to 0.994 using the pairwise distances with SLA-1*12:01 and SLA-1*13:01, respectively, when either or both of the partners had Hp-35.0. These pairwise distances among four SLA-1 alleles between partners were classified into ten and eleven groups that were divided across 0.1 ranges using the pairwise distances with SLA-1*12:01 and SLA-1*13:01, respectively. As shown in Figure 4A,B, lower farrowing rates were indicated in mating pairs with relatively smaller pairwise genetic distances of SLA-1 alleles. A significant correlation was observed by Spearman’s correlation coefficient analysis (p = 0.0320) in Figure 4B among farrowing rates and amino acid pairwise distances of SLA-1*13:01 between the mating pairs.
The sum of the pairwise distances in 1063 matings among SLA-3 alleles between partners increased from 0 to 0.718, and was classified into nine groups that were divided across 0.1 intervals (Table 2). As shown in Figure 4C, lower farrowing rates were indicated in mating pairs with smaller pairwise genetic distances of SLA-3 alleles. A significant correlation among farrowing rates and amino acid distances of SLA-3 alleles between partners was observed (p = 0.0501 (Figure 4C)). The sum of the pairwise distances among SLA-2 alleles between partners increased from 0 to 1.154, and was classified into eleven groups that were divided across 0.1 ranges (Table 2). There was no significant differences among the farrowing rates and amino acid distances of SLA-2 alleles between the mating pairs (Figure 4D).
3.4. Effects of Amino Acid Pairwise Distances between SLA Class II Alleles of Mating Pairs on Farrowing Rates in MMPs
As shown in Supplementary Table S4, amino acid distances among eight DRB1 alleles of mating pairs showed relatively high values, and the overall mean distance among the DRB1 alleles was 0.1695. The sum of the amino acid pairwise distances in each of the 1063 matings among DRB1 alleles was classified into ten groups from 0 to 0.899 across 0.1 ranges (Table 2). The ranges of the sum of the pairwise distances from 0 to 0.831 among DRB1 alleles between partners were comparable with those of the class I SLA-1, SLA-2, and SLA-3 alleles between partners (Table 2). In contrast, the differences of the amino acid pairwise distances among six kinds of SLA class II-DQB1 alleles between mating partners ranging from 0.0235 to 0.0867 (overall mean distance: 0.0651) were at a relatively low level when compared with those differences of the SLA-1, SLA-2, SLA-3, and DRB1 alleles with the mating partners (Supplementary Tables S1–S5). Due to the narrow distribution of pairwise distances and small number of different DQB1 alleles, the sum of the amino acid pairwise distances in each of the 1063 matings among DQB1 alleles was classified into only eight groups that ranged from 0 to 0.390 with ranges of 0.05 (Table 2). As shown in Figure 3A,B, lower farrowing rates were indicated in mating pairs with smaller pairwise genetic distances of DRB1 or DQB1 alleles. Significant correlations among farrowing rates and amino acid distances of DRB1 and DQB1 alleles between partners were observed by Spearman’s correlation coefficient analysis (p = 0.0118 (Figure 4E) and p = 0.0102 (Figure 4F)) for DRB1 and DQB1 alleles, respectively.
4. Discussion
The farrowing rate in MMPs of 61.7% was considerably lower than in mixed breed domestic pigs in Japan [25]. The reason why the farrowing rate in MMPs was lower than in the mixed breed domestic pigs is not known. However, other reproductive performances such as gestation periods, litter sizes at birth, and weaning in MMPs [18] were similar to those of other breeds of domestic pigs [15,25,26] including Göttingen and NIBS minipigs [27,28]. Thus, apart from the farrowing rates, the MMP population used in the present study appears to have relatively normal porcine reproductive traits.
In this study, eleven SLA class I and class II haplotypes including three recombinant haplotypes, Hp-10.23, Hp-35.17 and Hp-43.17, were found in 228 dams and 88 sires of MMPs (Table 1), which is consistent with our previous MHC haplotyping results of the MMP population. In the three recombinant haplotypes, all of the DNA crossover regions in the haplotypes were found within the class III region [21]. Frequencies of dams and sires for the mating performance with three recombinant haplotypes, Lr-10.23, Lr-35.17, and Lr-43.17, were found to be between 0% and 8% (Table 1). Due to the mating pairs with the recombinant haplotypes, the numbers of three mating groups on SLA class I haplotypes, CS, OS, and NS, were different from those on SLA class II haplotypes. Therefore, we separately analyzed the average farrowing rates of mating partners sharing SLA class I and class II haplotypes. The average farrowing rates of mating partners sharing SLA class II haplotypes DRB1 and/or DQB1 alleles were relatively low compared to those of non-sharing haplotypes. Since the two p values (p = 0.0198 and p = 0.0172) that were obtained from the association analyses between farrowing rates and the sharing of DRB1 and DQB1 showed almost the same level, it is difficult to determine which gene, DRB1 or DQB1, has higher significant effects for better farrowing rates. In contrast to the DRB1 and the DQB1 results, the degree of sharing SLA class I haplotypes or alleles between mating partners had no effect on the average farrowing rates. Taken together, these results suggested that the dissimilarities of SLA class II haplotypes or alleles between partners could provide more benefits on farrowing rates than that of SLA class I haplotypes or alleles. Nevertheless, we analyzed the genetic association between farrowing rates and SLA alleles or haplotypes using a relatively small number of MMPs in this study. Therefore, it might be difficult to exclude the possibilities of biased results in the comparison of farrowing rates between sharing or non-sharing SLA alleles or haplotypes. The reliability of the present results could be confirmed in future studies by using a larger number of MMPs and other pig breeds.
Moreover, higher farrowing rates were observed in mating pairs with bigger amino acid pairwise genetic distances of SLA-1 alleles using those of SLA-1*13:01, SLA-3, DRB1 or DQB1 alleles between the pairs. Relatively higher farrowing rates were also observed in mating pairs with bigger amino acid pairwise genetic distances of SLA-1 alleles using those of SLA-1*12:01 or SLA-2 alleles between the pairs, although no obvious significant correlations among farrowing rates and amino acid distances of SLA-1 alleles using those of SLA-1*12:01 or SLA-2 alleles between partners were observed by Spearman’s correlation coefficient analysis. A high farrowing rate, 90%, was obtained for the sum of the amino acid pairwise distances of SLA-2 between 0.3 and 0.399, although this may be due to a potential bias using only 20 matings which suggests that the number of matings might have an influence on the correlation coefficient analysis (Table 2, Figure 4D). Nevertheless, these results revealed that amino acid dissimilarities of SLA-1, SLA-3, DRB1 and DQB1 alleles between mating pairs had significant effects on the farrowing rates in MMPs.
Relationships between farrowing rates and MHC-dissimilarity or MHC heterozygosity have been reported in mate choice studies on many kinds of animals including rodents [3,4,29], humans [30,31], non-human primates [31], horses [32], birds [33] and fish [34]. Namely, in the theory of mate choice based on heterozygosity, partners with MHC-dissimilarity have a greater tendency to choose each other than those with MHC-similarity. The preference for MHC-dissimilarity between potential partners may function to avoid inbreeding and promote offspring heterozygosity for greater resistance to pathogens [35,36]. Our present results also showed that the dissimilarities of SLA class I and class II alleles between the pairs might be involved in farrowing rates in the MMP population. Thus, our findings in this and previous studies of reproduction in the MMP population [16] support the overall hypothesis that MHC heterozygosity enhances reproductive success. However, we have not analyzed whether the offspring heterozygosity would be promoted by MHC-dissimilarity between potential partners in the MMPs. Recently, a tendency for humans to prefer MHC-dissimilar mates was also demonstrated by using HLA types and 9,010 single nucleotide polymorphisms (SNPs) densely distributed across the MHC in 30 European American couples [37]. Furthermore, the same research group reported that the MHC influences mate choice, although social constraints in some populations also affect the mate choice [38]. On the other hand, meta-analyses in humans focusing on genomic mate selection, relationship satisfaction, odor preference and relationship satisfaction revealed that there was no association between MHC-dissimilarity and mate choice in actual couples [39]. Nevertheless, to clarify the mechanism of MHC-based mating preference, further genome wide association studies (GWAS) will be necessary to identify candidate genes correlated with mate choice and the effects of epistasis.
In the MMP population, SLA haplotype sharing and amino acid dissimilarities of SLA class I or class II alleles between partners had a weak influence on their farrowing rates. In general, the overall effects of MHC-based mating preferences are relatively weak. For instance, in the overall effect sizes of primates, Fisher’s Z correlation coefficient for dissimilarity showed Zr = 0.044 [31]. Furthermore, small effect sizes on mate choice for MHC-dissimilarity in non-human vertebrates were also found using formal phylogenetic meta-analysis and meta-regression techniques.
Additionally, the effect of MHC dissimilarity in mating preference was found when the dissimilarity was characterized at multiple loci including MHC class I and II loci [40]. Therefore, the effect of MHC dissimilarity by multiple loci is consistent with our results that dissimilarities of SLA-1, SLA-3, DRB1 and DQB1 alleles between mating pairs had significant effects on the farrowing rates in MMPs.
In humans, non-classical MHC class I molecules, HLA-E and HLA-G, can interact to maintain immune homeostasis of the maternal–fetus interface with many kinds of receptors on maternal immune cells in the uterus. HLA-E is the ligand of inhibitory and activating receptors, CD94/NKG2A and CD94/NKG2C, respectively, and HLA-G is the ligand of inhibitory receptors, ILT-2 (CD85j/LILRB1) and ILT-4 (CD85d/LILRB2) expressed on natural killer (NK) cells [41,42]. The pig appears to have many novel LILR genes, and only one KIR gene [43], but the specific gene products that are expressed and which play a role in porcine reproduction is still not known. Regarding the achievement of porcine deliveries, there are many biological and genetic factors other than MHC-based mating preferences involved in the important processes from fertilization to successful deliveries. To facilitate porcine pregnancy success, many immune cells such as uterine NK cells, dendritic cells, and macrophages are involved in the regulation of placental development, homeostasis, and tolerance of the fetal allograft [2]. Uterine endometrial expression of SLA class I molecules in stromal cells and luminal epithelium cells has been analyzed at each pregnancy stage, and it is generally accepted that the expression of SLA class I and β-2 microglobulin (β2M) molecules decrease in the placenta to prevent conceptus as a semi-allograft from host-vs-graft immune rejection. Nevertheless, classical SLA class I (SLA-1, SLA-2, and SLA-3) and nonclassical SLA class I (SLA-6, SLA-7, and SLA-8) and β2M molecules increased in uterine stromal cells and luminal epithelium cells during the peri-attachment period; then, their abundant expression decreased after successful conceptus attachment.
Cell-type specific regulation of the SLA and B2M genes is controlled by progesterone and IFNs on uterine cells [44]. However, the correlation between differential fertilization success depending on SLA sharing and the pregnancy stage specific expression of SLA class I molecules remains unclear. Regarding SLA class II expression during pregnancy, DQ molecules increased in response to conceptus-derived interferon gamma (IFNG) and likely regulate immune response at the maternal–fetal interface to support the maintenance of pregnancy in pigs [45]. Moreover, HLA class I and class II molecules were expressed in ejaculated spermatozoa, thereby suggesting some undefined roles of the HLA molecules on spermatozoa function and immune activation in female reproductive tracts. The expression of the HLA class II molecules was significantly higher than those of HLA class I in human spermatozoa [46]. In mice, it was suggested that the class II molecules on the posterior region of the sperm head plays an adhesive role in the recognition between sperm and egg during fertilization [47]. These reports on the expression of SLA class I and class II molecules suggested that the SLA molecules might be involved in porcine pregnancy success with many other immune factors. Here, we described the relatively weak and variable effects of the dissimilarity of SLA class I and II alleles between mating pairs on farrowing rates. Therefore, the SLA class I and class II genes or haplotypes might not be directly responsible for porcine pregnancy success with high farrowing rates. Although the polymorphic features of SLA genes or haplotypes and SLA dissimilarity between mating pairs might correlate only indirectly with the pregnancy process of MMPs; it nevertheless is evident from the present study that SLA class I and class II alleles or haplotypes could be useful genetic markers for the selection of mating pairs in breeding programs for assisting with a more effective breeding management of MMPs.
Since the effects of SLA dissimilarity between mating partners on farrowing rates in our present study were analyzed using only a few SLA haplotypic class I and II loci in a limited pig population and breed, further studies are needed to confirm the effects of SLA dissimilarity on pregnancy and the achievement of deliveries using other pig breeds with other SLA haplotypes.
5. Conclusions
Farrowing rates of mating pairs with shared SLA class II haplotypes or alleles were relatively lower than those with non-sharing ones. Furthermore, analyses between amino acid distances of SLA alleles between mating pairs exhibited that the dissimilarities of SLA class I and class II alleles might be involved in farrowing rates in the MMP population. These relationships between the farrowing rates and SLA suggested that the SLA class I and class II alleles or haplotypes have the potential to be useful genetic markers for the selection of mating pairs in breeding programs and epistatic studies of reproductive traits of MMPs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11193138/s1, Table S1: Amino acid pairwise distances among nine SLA-1 alleles; Table S2: Amino acid pairwise distances among eight SLA-3 alleles; Table S3: Amino acid pairwise distances among eight SLA-2 alleles; Table S4: Amino acid pairwise distances among eight DRB1 alleles; Table S5: Amino acid pairwise distances among six DQB1 alleles.
Author Contributions
Conceptualization, A.A. and H.K.; methodology, A.A., Y.K. and H.K.; software, S.S. and H.K.; validation, A.A., T.M., N.I., A.S., A.M. and S.O.; formal analysis, S.S. and H.K.; investigation, T.M., N.I., M.T., A.S., A.M. and S.O.; resources, T.M., N.I. and M.T.; data curation, A.A. and T.M.; writing—original draft preparation, A.A. and H.K.; writing—review and editing, T.S., J.K.K. and H.K.; visualization, A.A. and H.K.; supervision, T.S. and H.K.; project administration, Y.K., T.S. and H.K.; funding acquisition, H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care and Use Committee of Gifu University (#17042, 26 May 2017). The care and use of the laboratory animals were conducted in compliance with the guidelines of Good Laboratory Practice of Gifu University and Fuji Micra Inc.
Informed Consent Statement
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Takashi Nishimura and Toshiaki Nishimura (Fuji Micra Inc., Fujinomiya, Japan) for providing us with recorded information on reproduction performances for MMPs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Linton, N.F.; Wessels, J.M.; Cnossen, S.A.; Croy, B.A.; Tayade, C. Immunological mechanisms affecting angiogenesis and their relation to porcine pregnancy success. Immunol. Investig. 2008, 37, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Bidarimath, M.; Tayade, C. Pregnancy and spontaneous fetal loss: A pig perspective. Mol. Reprod. Dev. 2017, 84, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, K.; Beauchamp, G.K. Genetic basis for MHC-dependent mate choice. Adv. Genet. 2007, 59, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Thoss, M.; Ilmonen, P.; Musolf, K.; Penn, D.J. Major histocompatibility complex heterozygosity enhances reproductive success. Mol. Evol. 2011, 20, 1546–1557. [Google Scholar] [CrossRef]
- Agenor, A.; Bhattacharya, S. Infertility and miscarriage: Common pathways in manifestation and management. Womens Health 2015, 11, 527–541. [Google Scholar] [CrossRef]
- Rapacz-Leonard, A.; Dąbrowska, M.; Janowski, T. Major histocompatibility complex I mediates immunological tolerance of the trophoblast during pregnancy and may mediate rejection during parturition. Mediators Inflamm. 2014, 2014, 579279. [Google Scholar] [CrossRef]
- Conley, A.J.; Jung, Y.C.; Schwartz, N.K.; Warner, C.M.; Rothschild, M.F.; Ford, S.P. Influence of SLA haplotype on ovulation rate and litter size in miniature pigs. J. Reprod. Fertil. 1988, 82, 595–601. [Google Scholar] [CrossRef]
- Lunney, J.K.; Ho, C.S.; Wysocki, M.; Smith, D.M. Molecular genetics of the swine major histocompatibility complex, the SLA complex. Dev. Comp. Immunol. 2009, 33, 362–374. [Google Scholar] [CrossRef]
- Lassiter, R.; Wang, Y.; Fang, X.; Winn, M.; Ghaffari, A.; Ho, C.-S.; Helman, S.; Jajosky, R.; Kleven, D.; Nahman, N.S., Jr.; et al. A model of acute renal allograft rejection in outbred Yorkshire piglets. Transpl. Immunol. 2017, 42, 40–46. [Google Scholar] [CrossRef]
- Hammer, S.E.; Ho, C.S.; Ando, A.; Rogel-Gaillard, C.; Charles, M.; Tector, M.; Tector, A.J.; Lunney, J.K. Importance of the major histocompatibility complex (swine leukocyte antigen) in swine health and biomedical research. Annu. Rev. Anim. Biosci. 2020, 8, 171–198. [Google Scholar] [CrossRef]
- Duran-Struuck, R.; Huang, C.A.; Orf, K.; Bronson, R.T.; Sachs, D.H.; Spitzer, T.R. Miniature swine as a clinically relevant model of graft-versus-host disease. Comp. Med. 2015, 65, 429–443. [Google Scholar] [PubMed]
- Carvalho-Oliveira, M.; Valdivia, E.; Blasczyk, R.; Figueiredo, C. Immunogenetics of xenotransplantation. Int. J. Immunogenet. 2021, 48, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Abicht, J.M.; Sfriso, R.; Reichart, B.; Langin, M.; Gahle, K.; Yung, G.L.P.; Seebach, J.D.; Rieben, R.; Ayares, D.; Wolf, E.; et al. Multiple genetically modified GTKO/hCD46/HLA-E/hβ2-mg porcine hearts are protected from complement activation and natural killer cell infiltration during ex vivo perfusion with human blood. Xenotransplantation 2018, 25, e12390. [Google Scholar] [CrossRef]
- Ando, A.; Shigenari, A.; Kojima-Shibata, C.; Nakajoh, M.; Suzuki, K.; Kitagawa, H.; Shiina, T.; Inoko, H.; Uenishi, H. Association of swine leukocyte antigen class II haplotypes and immune-related traits in a swine line selected for resistance to mycoplasmal pneumonia. Comp. Immunol. Microbiol. Infect. Dis. 2016, 48, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gautschi, C.; Gaillard, C. Influence of major histocompatibility complex on reproduction and production traits in swine. Anim. Genet. 1989, 21, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Imaeda, N.; Ando, A.; Takasu, M.; Matsubara, T.; Nishii, N.; Takashima, S.; Shigenari, A.; Shiina, T.; Kitagawa, H. Influences of swine leukocyte antigen haplotypes on serum antigen titers against swine erysipelas vaccine and traits of reproductive ability and meat production in SLA-defined Duroc pigs. J. Vet. Med. Sci. 2018, 80, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Takasu, M.; Imaeda, N.; Nishii, N.; Takashima, S.; Nishimura, T.; Shiina, T.; Ando, A.; Kitagawa, H. Genetic association of swine leukocyte antigen class II haplotypes and body weight in Microminipigs. Asian-Aust. J. Animal Sci. 2018, 31, 163–166. [Google Scholar] [CrossRef]
- Ando, A.; Imaeda, N.; Matsubara, T.; Takasu, M.; Miyamoto, A.; Ohshima, S.; Nishii, N.; Kametani, Y.; Shiina, T.; Kulski, J.K.; et al. Genetic association between swine leukocyte antigen class II haplotypes and reproduction traits in Microminipigs. Cells 2019, 8, 783. [Google Scholar] [CrossRef]
- Kaneko, N.; Itoh, K.; Sugiyama, A.; Izumi, Y. Microminipig, a non-rodent experimental animal optimized for life science research: Preface. J. Pharmacl. Sci. 2011, 115, 112–114. [Google Scholar] [CrossRef]
- Takasu, M.; Tsuji, E.; Imaeda, N.; Kaneko, N.; Matsubara, T.; Maeda, M.; Ito, Y.; Shibata, S.; Ando, A.; Nishii, N.; et al. Body and, major organ sizes of young mature Microminipigs determined by computed tomography. Lab. Anim. 2015, 49, 65–70. [Google Scholar] [CrossRef]
- Ando, A.; Imaeda, N.; Ohshima, S.; Miyamoto, A.; Kaneko, N.; Takasu, M.; Shiina, T.; Kulski, J.K.; Inoko, H.; Kitagawa, H. Characterization of swine leucocyte antigen alleles and haplotypes on a novel miniature pig line, Microminipig. Anim. Gen. 2014, 45, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Imaeda, N.; Ando, A.; Matsubara, T.; Takasu, M.; Nishii, N.; Miyamoto, A.; Ohshima, S.; Kametani, Y.; Suzuki, S.; Shiina, T.; et al. Stillbirth rates and their association with swine leucocyte antigen class II haplotypes in Microminipigs. Anim. Biosci. 2021, 34, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molec. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Suzuki, K.; Somei, H. Present condition and opinions of artificial insemination in pig farming. Bull. Bull. Chiba Prefect. Livest. Res. Cent. 1982, 6, 39–43. (In Japanese) [Google Scholar]
- Rothkötter, H.J.; Sowa, E.; Pabst, R. The pig as a model of developmental immunology. Hum. Exp. Toxicol. 2002, 21, 533–536. [Google Scholar] [CrossRef]
- Schuleri, K.H.; Boyle, A.J.; Centola, M.; Amado, L.C.; Evers, R.; Zimmet, J.M.; Evers, K.S.; Ostbye, K.M.; Scorpio, D.G.; Hare, J.M.; et al. The adult Göttingen Minipig as a model for chronic heart failure after myocardial infarction: Focus on cardiovascular imaging and regenerative therapies. Comp. Med. 2008, 58, 568–579. [Google Scholar]
- Saitoh, T. Utilization of a swine other than food. All About Swine 2009, 35, 14–20. (In Japanese) [Google Scholar]
- Tregenza, T.; Wedell, N. Genetic compatibility, mate choice and patterns of parentage: Invited review. Mol. Ecol. 2000, 9, 1013–1027. [Google Scholar] [CrossRef]
- Havlicek, J.; Roberts, S.C. MHC-correlated mate choice in humans: A review. Psychoneuroendocrinol 2009, 34, 497–512. [Google Scholar] [CrossRef]
- Winternitz, J.; Abbate, J.L.; Huchard, E.; Havlicek, J.; Garamszeg, L.Z. Patterns of MHC-dependent mate selection in humans and nonhuman primates: A meta-analysis. Molec. Ecol. 2017, 26, 668–688. [Google Scholar] [CrossRef] [PubMed]
- Burger, D.; Thomas, S.; Aepli, H.; Dreyer, M.; Fabre, G.; Marti, E.; Sieme, H.; Robinson, M.R.; Wedekind, C. Major histocompatibility complex-linked social signaling affects female fertility. Proc. R. Soc. B 2017, 284, 1824–1829. [Google Scholar] [CrossRef] [PubMed]
- Bonneaud, C.; Chastel, O.; Federici, P.; Westerdahl, H.; Sorci, G. Complex MHC-based mate choice in a wild passerine. Proc. R. Soc. B Biol. Sci. 2006, 273, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Landry, C.; Grant, D.; Duchesne, P.; Bernatchez, L. Good genes as heterozygosity: The major histocompatibility complex and mate choice in Atlantic salmon. Proc. R. Soc. 2001, 268, 1279–1285. [Google Scholar] [CrossRef]
- Potts, W.K.; Manning, C.J.; Wakeland, E.K. The role of infectious disease inbreeding and mating preferences in maintaining MHC diversity: An experimental test. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994, 346, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L. A theory of mate choice based on heterozygosity. Behav. Ecol. 1997, 8, 60–65. [Google Scholar] [CrossRef]
- Chaix, R.; Cao, C.; Donnelly, P. Is mate choice in humans MHC-dependent? PLOS Genet. 2008, 4, e1000184. [Google Scholar] [CrossRef]
- Dandine-Roulland, C.; Laurent, R.; Dall’Ara, I.; Toupance, B.; Chiax, R. Genomic evidence for MHC disassortative mating in humans. Proc. Biol. Sci. 2019, 286, 20182664. [Google Scholar] [CrossRef]
- Havlicek, J.; Winternitz, J.; Roberts, S.C. Major histocompatibility complex associated odour preferences and human mate choice: Near and far horizons. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190260. [Google Scholar] [CrossRef]
- Kamiya, T.; O’Dwyer, H.; Westerdahl, H.; Senior, A.; Nakagawa, S. Quantitative review of MHC-based mating preference: The role of diversity and dissimilarity. Molec. Ecol. 2014, 23, 5151–5163. [Google Scholar] [CrossRef]
- Vales-Gomez, M.; Reyburn, H.T.; Erskine, R.A.; Lopez-Botet, M.; Strominger, J.L. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999, 18, 4250–4260. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, B.; Shang, J.; Yao, Y. HLA-G: An important mediator of maternal-fetal immune-tolerance. Front. Immunol. 2021, 12, 744324. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.C.; Hammond, J.A. The unique evolution of the pig LRC, a single KIR but expansion of LILR and a novel Ig receptor family. Immunogenetics 2018, 70, 661–669. [Google Scholar] [CrossRef]
- Joyce, M.M.; Burghardt, J.R.; Burghardt, R.C.; Hooper, R.N.; Bazer, F.W.; Johnson, G.A. Uterine MHC class I molecules and beta 2-microglobulin are regulated by progesterone and conceptus interferons during pig pregnancy. J. Immunol. 2008, 181, 2494–2505. [Google Scholar] [CrossRef]
- Kim, M.; Seo, H.; Choi, Y.; Shim, J.; Bazer, F.W.; Ka, H. Swine leukocyte antigen-DQ expression and its regulation by interferon gamma at the maternal-fetal interface in pigs. Biol. Reprod. 2012, 86, 43. [Google Scholar] [CrossRef] [PubMed]
- Sereshki, N.; Andalib, A.; Ghahiri, A.; Mehrabian, F.; Sherkat, R.; Rezaei, A.; Wilkinson, D. The expression of human leukocyte antigen by human ejaculated spermatozoa. Mol. Genet. Genomic. Med. 2019, 7, e1005. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Wu, G.M.; Mori, E.; Shindo, Y.; Mori, N.; Fukuda, A.; Mori, T. Expression of class II major histocompatibility complex antigen on mouse sperm and its roles in fertilization. Am. J. Reprod. Immunol. 1990, 24, 9–14. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).