The Roles of RUNX Proteins in Lymphocyte Function and Anti-Tumor Immunity
Abstract
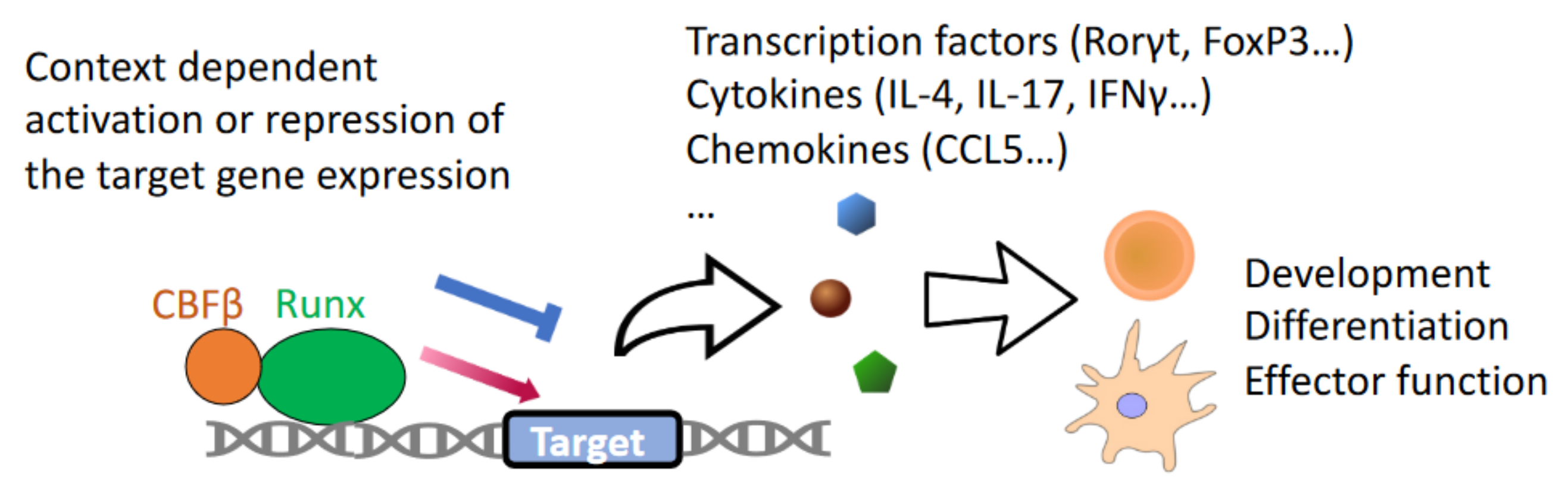
1. Introduction
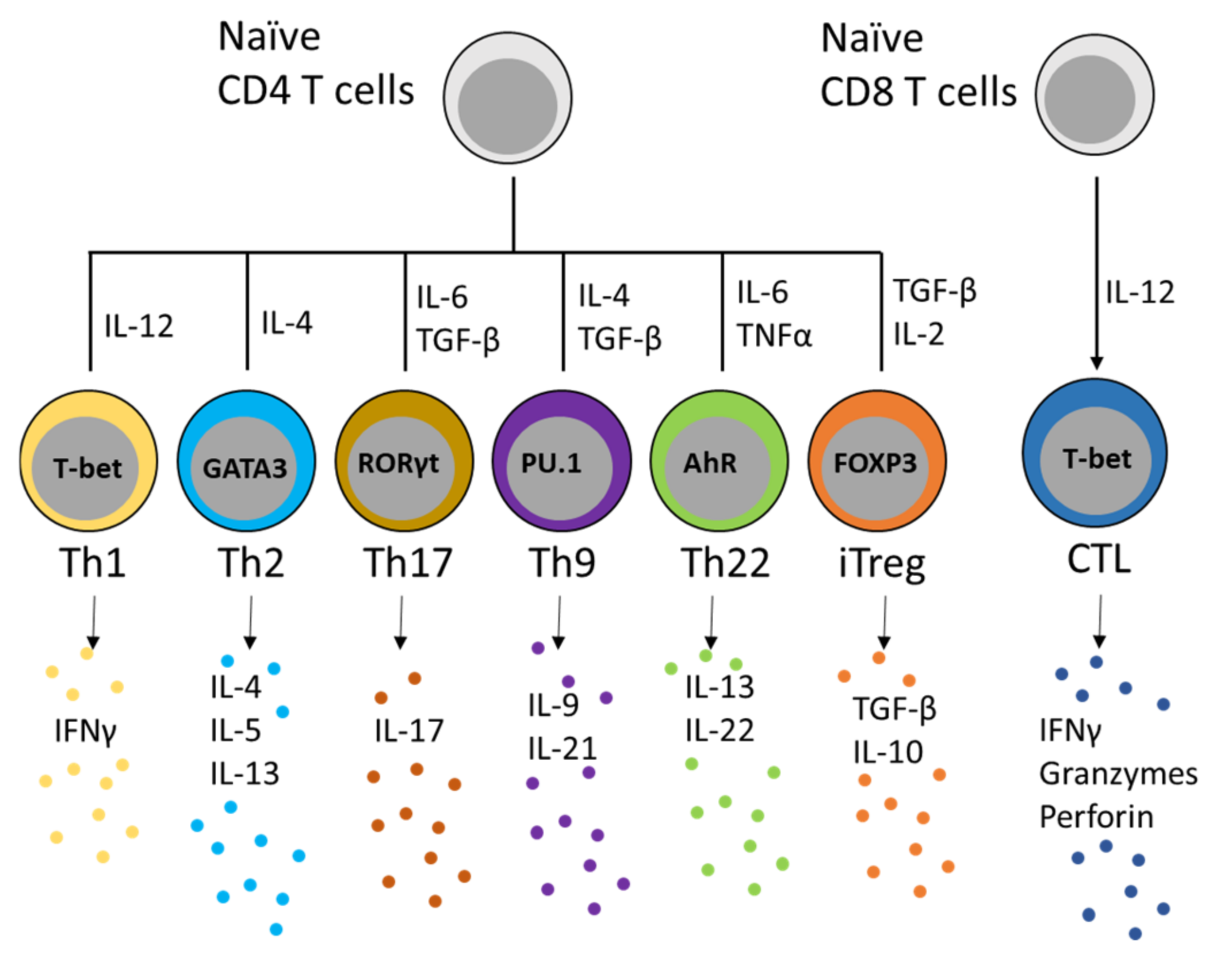
2. RUNX in Th1 and Th-1 like Th17 Function
3. RUNX in CD4 and CD8 CTL
4. RUNX in Tissue Resident memory T Cells (TRM)
5. RUNX in Regulatory T Cell Function
6. RUNX in ILC Function
7. RUNX in NK Cell Function
8. Conclusive Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Okuda, T.; van Deursen, J.; Hiebert, S.W.; Grosveld, G.; Downing, J.R. AML1, the Target of Multiple Chromosomal Translocations in Human Leukemia, Is Essential for Normal Fetal Liver Hematopoiesis. Cell 1996, 84, 321–330. [Google Scholar] [CrossRef]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted Disruption of Cbfa1 Results in a Complete Lack of Bone Formation owing to Maturational Arrest of Osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.H.; Beddington, R.S.P.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a Candidate Gene for Cleidocranial Dysplasia Syndrome, Is Essential for Osteoblast Differentiation and Bone Development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Inoue, K.-i.; Ozaki, S.; Shiga, T.; Ito, K.; Masuda, T.; Okado, N.; Iseda, T.; Kawaguchi, S.; Ogawa, M.; Bae, S.-C.; et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 2002, 5, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.-C.; Komori, T.; Ito, Y.; Littman, D.R. Differential Requirements for Runx Proteins in CD4 Repression and Epigenetic Silencing during T Lymphocyte Development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Huang, G.; Shigesada, K.; Ito, K.; Wee, H.-J.; Yokomizo, T.; Ito, Y. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin–proteasome-mediated degradation. EMBO J. 2001, 20, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Tahirov, T.H.; Inoue-Bungo, T.; Morii, H.; Fujikawa, A.; Sasaki, M.; Kimura, K.; Shiina, M.; Sato, K.; Kumasaka, T.; Yamamoto, M.; et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell 2001, 104, 755–767. [Google Scholar] [CrossRef]
- Seo, W.; Taniuchi, I. The Roles of RUNX Family Proteins in Development of Immune Cells. Mol. Cells 2020, 43, 107–113. [Google Scholar] [CrossRef]
- Levanon, D.; Groner, Y. Structure and regulated expression of mammalian RUNX genes. Oncogene 2004, 23, 4211–4219. [Google Scholar] [CrossRef]
- Egawa, T.; Tillman, R.E.; Naoe, Y.; Taniuchi, I.; Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007, 204, 1945–1957. [Google Scholar] [CrossRef]
- Fortunel, N.O.; Hatzfeld, A.; Hatzfeld, J.A. Transforming growth factor-β: Pleiotropic role in the regulation of hematopoiesis. Blood 2000, 96, 2022–2036. [Google Scholar] [CrossRef]
- Ito, Y.; Miyazono, K. RUNX transcription factors as key targets of TGF-β superfamily signaling. Curr. Opin. Genet. Dev. 2003, 13, 43–47. [Google Scholar] [CrossRef]
- Russ, B.E.; Prier, J.E.; Rao, S.; Turner, S.J. T cell immunity as a tool for studying epigenetic regulation of cellular differentiation. Front. Genet. 2013, 4, 218. [Google Scholar] [CrossRef]
- Jiang, S.; Dong, C. A complex issue on CD4(+) T-cell subsets. Immunol. Rev. 2013, 252, 5–11. [Google Scholar] [CrossRef]
- Taniuchi, I. CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annu. Rev. Immunol. 2018, 36, 579–601. [Google Scholar] [CrossRef]
- Erickson, P.; Gao, J.; Chang, K.S.; Look, T.; Whisenant, E.; Raimondi, S.; Lasher, R.; Trujillo, J.; Rowley, J.; Drabkin, H. Identification of Breakpoints in t(8;21) Acute Myelogenous Leukemia and Isolation of a Fusion Transcript, AML1/ETO, With Similarity to Drosophila Segmentation Gene, runt. Blood 1992, 80, 1825–1831. [Google Scholar] [CrossRef]
- Liu, P.; Tarlé, S.A.; Hajra, A.; Claxton, D.F.; Marlton, P.; Freedman, M.; Siciliano, M.J.; Collins, F.S. Fusion Between Transcription Factor CBFβ/PEBP2β and a Myosin Heavy Chain in Acute Myeloid Leukemia. Science 1993, 261, 1041–1044. [Google Scholar] [CrossRef]
- Nucifora, G.; Begy, C.R.; Erickson, P.; Drabkin, H.A.; Rowley, J.D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER 1. Proc. Natl. Acad. Sci. 1993, 90, 7784–7788. [Google Scholar] [CrossRef]
- Chi, X.-Z.; Yang, J.-O.; Lee, K.-Y.; Ito, K.; Sakakura, C.; Li, Q.-L.; Kim, H.-R.; Cha, E.-J.; Lee, Y.-H.; Kaneda, A.; et al. RUNX3 Suppresses Gastric Epithelial Cell Growth by Inducing p21WAF1/Cip1 Expression in Cooperation with Transforming Growth Factor β-Activated SMAD. Mol. Cell. Biol. 2005, 25, 8097–8107. [Google Scholar] [CrossRef]
- Szabo, S.J.; Kim, S.T.; Costa, G.L.; Zhang, X.; Fathman, C.G.; Glimcher, L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000, 100, 655–669. [Google Scholar] [CrossRef]
- Naoe, Y.; Setoguchi, R.; Akiyama, K.; Muroi, S.; Kuroda, M.; Hatam, F.; Littman, D.R.; Taniuchi, I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 2007, 204, 1749–1755. [Google Scholar] [CrossRef]
- Djuretic, I.M.; Levanon, D.; Negreanu, V.; Groner, Y.; Rao, A.; Ansel, K.M. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 2007, 8, 145–153. [Google Scholar] [CrossRef]
- Brenner, O.; Levanon, D.; Negreanu, V.; Golubkov, O.; Fainaru, O.; Woolf, E.; Groner, Y. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc. Natl. Acad. Sci. USA 2004, 101, 16016–16021. [Google Scholar] [CrossRef]
- Fainaru, O.; Woolf, E.; Lotem, J.; Yarmus, M.; Brenner, O.; Goldenberg, D.; Negreanu, V.; Bernstein, Y.; Levanon, D.; Jung, S.; et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004, 23, 969–979. [Google Scholar] [CrossRef]
- Fang, D.; Cui, K.; Cao, Y.; Zheng, M.; Kawabe, T.; Hu, G.; Khillan, J.S.; Li, D.; Zhong, C.; Jankovic, D.; et al. Differential regulation of transcription factor T-bet induction during NK cell development and T helper-1 cell differentiation. Immunity 2022, 55, 639–655.e637. [Google Scholar] [CrossRef]
- Ivanov, I.I.; McKenzie, B.S.; Zhou, L.; Tadokoro, C.E.; Lepelley, A.; Lafaille, J.J.; Cua, D.J.; Littman, D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006, 126, 1121–1133. [Google Scholar] [CrossRef]
- Manel, N.; Unutmaz, D.; Littman, D.R. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 2008, 9, 641–649. [Google Scholar] [CrossRef]
- Lazarevic, V.; Chen, X.; Shim, J.H.; Hwang, E.S.; Jang, E.; Bolm, A.N.; Oukka, M.; Kuchroo, V.K.; Glimcher, L.H. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat. Immunol. 2011, 12, 96–104. [Google Scholar] [CrossRef]
- Miossec, P.; Kolls, J.K. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 2012, 11, 763–776. [Google Scholar] [CrossRef]
- Zou, W.; Restifo, N.P. T(H)17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 2010, 10, 248–256. [Google Scholar] [CrossRef]
- Vitiello, G.A.; Miller, G. Targeting the interleukin-17 immune axis for cancer immunotherapy. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.R.; Nelson, M.H.; Himes, R.A.; Li, Z.; Mehrotra, S.; Paulos, C.M. Th17 cells in cancer: The ultimate identity crisis. Front. Immunol. 2014, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Duarte, J.H.; Veldhoen, M.; Hornsby, E.; Li, Y.; Cua, D.J.; Ahlfors, H.; Wilhelm, C.; Tolaini, M.; Menzel, U.; et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 2011, 12, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Godec, J.; Ben-Aissa, K.; Cui, K.; Zhao, K.; Pucsek, A.B.; Lee, Y.K.; Weaver, C.T.; Yagi, R.; Lazarevic, V. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 2014, 40, 355–366. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Cruz-Guilloty, F.; Pipkin, M.E.; Djuretic, I.M.; Levanon, D.; Lotem, J.; Lichtenheld, M.G.; Groner, Y.; Rao, A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 2009, 206, 51–59. [Google Scholar] [CrossRef]
- Borst, J.; Ahrends, T.; Babala, N.; Melief, C.J.M.; Kastenmuller, W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- Oh, D.Y.; Fong, L. Cytotoxic CD4(+) T cells in cancer: Expanding the immune effector toolbox. Immunity 2021, 54, 2701–2711. [Google Scholar] [CrossRef]
- Cheroutre, H.; Husain, M.M. CD4 CTL: Living up to the challenge. Semin. Immunol. 2013, 25, 273–281. [Google Scholar] [CrossRef]
- Swain, S.L.; McKinstry, K.K.; Strutt, T.M. Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 2012, 12, 136–148. [Google Scholar] [CrossRef]
- Kido, K.; Yamanaka, S.; Nakano, S.; Motani, K.; Shinohara, S.; Nozawa, A.; Kosako, H.; Ito, S.; Sawasaki, T. AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. eLife 2020, 9, e54983. [Google Scholar] [CrossRef]
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front. Immunol. 2017, 8, 194. [Google Scholar] [CrossRef]
- Patil, V.S.; Madrigal, A.; Schmiedel, B.J.; Clarke, J.; O’Rourke, P.; de Silva, A.D.; Harris, E.; Peters, B.; Seumois, G.; Weiskopf, D.; et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef]
- Takeuchi, A.; Badr Mel, S.; Miyauchi, K.; Ishihara, C.; Onishi, R.; Guo, Z.; Sasaki, Y.; Ike, H.; Takumi, A.; Tsuji, N.M.; et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 2016, 213, 123–138. [Google Scholar] [CrossRef]
- Serroukh, Y.; Gu-Trantien, C.; Hooshiar Kashani, B.; Defrance, M.; Vu Manh, T.P.; Azouz, A.; Detavernier, A.; Hoyois, A.; Das, J.; Bizet, M.; et al. The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. Elife 2018, 7. [Google Scholar] [CrossRef]
- Gulich, A.F.; Preglej, T.; Hamminger, P.; Alteneder, M.; Tizian, C.; Orola, M.J.; Muroi, S.; Taniuchi, I.; Ellmeier, W.; Sakaguchi, S. Differential Requirement of Cd8 Enhancers E8I and E8VI in Cytotoxic Lineage T Cells and in Intestinal Intraepithelial Lymphocytes. Front. Immunol. 2019, 10, 409. [Google Scholar] [CrossRef]
- Reis, B.S.; Rogoz, A.; Costa-Pinto, F.A.; Taniuchi, I.; Mucida, D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat. Immunol. 2013, 14, 271–280. [Google Scholar] [CrossRef]
- Reis, B.S.; Hoytema van Konijnenburg, D.P.; Grivennikov, S.I.; Mucida, D. Transcription factor T-bet regulates intraepithelial lymphocyte functional maturation. Immunity 2014, 41, 244–256. [Google Scholar] [CrossRef]
- Sujino, T.; London, M.; Hoytema van Konijnenburg, D.P.; Rendon, T.; Buch, T.; Silva, H.M.; Lafaille, J.J.; Reis, B.S.; Mucida, D. Tissue adaptation of regulatory and intraepithelial CD4(+) T cells controls gut inflammation. Science 2016, 352, 1581–1586. [Google Scholar] [CrossRef]
- Oliveira, G.; Stromhaug, K.; Cieri, N.; Iorgulescu, J.B.; Klaeger, S.; Wolff, J.O.; Rachimi, S.; Chea, V.; Krause, K.; Freeman, S.S.; et al. Landscape of helper and regulatory antitumour CD4(+) T cells in melanoma. Nature 2022, 605, 532–538. [Google Scholar] [CrossRef]
- Veatch, J.R.; Lee, S.M.; Shasha, C.; Singhi, N.; Szeto, J.L.; Moshiri, A.S.; Kim, T.S.; Smythe, K.; Kong, P.; Fitzgibbon, M.; et al. Neoantigen-specific CD4(+) T cells in human melanoma have diverse differentiation states and correlate with CD8(+) T cell, macrophage, and B cell function. Cancer Cell 2022, 40, 393–409.e399. [Google Scholar] [CrossRef] [PubMed]
- Lowery, F.J.; Krishna, S.; Yossef, R.; Parikh, N.B.; Chatani, P.D.; Zacharakis, N.; Parkhurst, M.R.; Levin, N.; Sindiri, S.; Sachs, A.; et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science 2022, 375, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Cachot, A.; Bilous, M.; Liu, Y.C.; Li, X.; Saillard, M.; Cenerenti, M.; Rockinger, G.A.; Wyss, T.; Guillaume, P.; Schmidt, J.; et al. Tumor-specific cytolytic CD4 T cells mediate immunity against human cancer. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Abblett, R.; Ryan, J.M.; Hagymasi, A.T.; Agyekum-Yamoah, A.; Svedova, J.; Reiner, S.L.; St Rose, M.C.; Hanley, M.P.; Vella, A.T.; et al. An Immunotherapeutic CD137 Agonist Releases Eomesodermin from ThPOK Repression in CD4 T Cells. J. Immunol. 2018, 200, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Schenkel, J.M.; Fraser, K.A.; Beura, L.K.; Pauken, K.E.; Vezys, V.; Masopust, D. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 2014, 346, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Sathaliyawala, T.; Kubota, M.; Yudanin, N.; Turner, D.; Camp, P.; Thome, J.J.; Bickham, K.L.; Lerner, H.; Goldstein, M.; Sykes, M.; et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 2013, 38, 187–197. [Google Scholar] [CrossRef]
- Szabo, P.A.; Miron, M.; Farber, D.L. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef]
- Park, S.L.; Buzzai, A.; Rautela, J.; Hor, J.L.; Hochheiser, K.; Effern, M.; McBain, N.; Wagner, T.; Edwards, J.; McConville, R.; et al. Tissue-resident memory CD8(+) T cells promote melanoma-immune equilibrium in skin. Nature 2019, 565, 366–371. [Google Scholar] [CrossRef]
- Mackay, L.K.; Rahimpour, A.; Ma, J.Z.; Collins, N.; Stock, A.T.; Hafon, M.-L.; Vega-Ramos, J.; Lauzurica, P.; Mueller, S.N.; Stefanovic, T.; et al. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 2013, 14, 1294–1301. [Google Scholar] [CrossRef]
- Mackay, L.K.; Minnich, M.; Kragten, N.A.; Liao, Y.; Nota, B.; Seillet, C.; Zaid, A.; Man, K.; Preston, S.; Freestone, D.; et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352, 459–463. [Google Scholar] [CrossRef]
- Hombrink, P.; Helbig, C.; Backer, R.A.; Piet, B.; Oja, A.E.; Stark, R.; Brasser, G.; Jongejan, A.; Jonkers, R.E.; Nota, B.; et al. Programs for the persistence, vigilance and control of human CD8(+) lung-resident memory T cells. Nat. Immunol. 2016, 17, 1467–1478. [Google Scholar] [CrossRef]
- Parga-Vidal, L.; Behr, F.M.; Kragten, N.A.M.; Nota, B.; Wesselink, T.H.; Kavazovic, I.; Covill, L.E.; Schuller, M.B.P.; Bryceson, Y.T.; Wensveen, F.M.; et al. Hobit identifies tissue-resident memory T cell precursors that are regulated by Eomes. Sci. Immunol. 2021, 6. [Google Scholar] [CrossRef]
- Frizzell, H.; Fonseca, R.; Christo, S.N.; Evrard, M.; Cruz-Gomez, S.; Zanluqui, N.G.; von Scheidt, B.; Freestone, D.; Park, S.L.; McWilliam, H.E.G.; et al. Organ-specific isoform selection of fatty acid-binding proteins in tissue-resident lymphocytes. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef]
- Crowl, J.T.; Heeg, M.; Ferry, A.; Milner, J.J.; Omilusik, K.D.; Toma, C.; He, Z.; Chang, J.T.; Goldrath, A.W. Tissue-resident memory CD8(+) T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat. Immunol. 2022, 23, 1121–1131. [Google Scholar] [CrossRef]
- Caushi, J.X.; Zhang, J.; Ji, Z.; Vaghasia, A.; Zhang, B.; Hsiue, E.H.; Mog, B.J.; Hou, W.; Justesen, S.; Blosser, R.; et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 2021, 596, 126–132. [Google Scholar] [CrossRef]
- Wu, T.D.; Madireddi, S.; de Almeida, P.E.; Banchereau, R.; Chen, Y.J.; Chitre, A.S.; Chiang, E.Y.; Iftikhar, H.; O’Gorman, W.E.; Au-Yeung, A.; et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020, 579, 274–278. [Google Scholar] [CrossRef]
- Casanova-Acebes, M.; Dalla, E.; Leader, A.M.; LeBerichel, J.; Nikolic, J.; Morales, B.M.; Brown, M.; Chang, C.; Troncoso, L.; Chen, S.T.; et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 2021, 595, 578–584. [Google Scholar] [CrossRef]
- Luoma, A.M.; Suo, S.; Wang, Y.; Gunasti, L.; Porter, C.B.M.; Nabilsi, N.; Tadros, J.; Ferretti, A.P.; Liao, S.; Gurer, C.; et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 2022, 185, 2918–2935.e2929. [Google Scholar] [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 2021, 374, abe6474. [Google Scholar] [CrossRef]
- Milner, J.J.; Toma, C.; He, Z.; Kurd, N.S.; Nguyen, Q.P.; McDonald, B.; Quezada, L.; Widjaja, C.E.; Witherden, D.A.; Crowl, J.T.; et al. Heterogenous Populations of Tissue-Resident CD8(+) T Cells Are Generated in Response to Infection and Malignancy. Immunity 2020, 52, 808–824.e807. [Google Scholar] [CrossRef]
- Hirai, T.; Zenke, Y.; Yang, Y.; Bartholin, L.; Beura, L.K.; Masopust, D.; Kaplan, D.H. Keratinocyte-Mediated Activation of the Cytokine TGF-β Maintains Skin Recirculating Memory CD8+ T Cells. Immunity 2019, 50, 1249–1261.e1245. [Google Scholar] [CrossRef]
- Zhang, N.; Bevan, M.J. Transforming Growth Factor-β Signaling Controls the Formation and Maintenance of Gut-Resident Memory T Cells by Regulating Migration and Retention. Immunity 2013, 39, 687–696. [Google Scholar] [CrossRef]
- Fonseca, R.; Burn, T.N.; Gandolfo, L.C.; Devi, S.; Park, S.L.; Obers, A.; Evrard, M.; Christo, S.N.; Buquicchio, F.A.; Lareau, C.A.; et al. Runx3 drives a CD8(+) T cell tissue residency program that is absent in CD4(+) T cells. Nat. Immunol. 2022, 23, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Milner, J.J.; Toma, C.; Yu, B.; Zhang, K.; Omilusik, K.; Phan, A.T.; Wang, D.; Getzler, A.J.; Nguyen, T.; Crotty, S.; et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature 2017, 552, 253–257. [Google Scholar] [CrossRef]
- Chen, J.; Lopez-Moyado, I.F.; Seo, H.; Lio, C.J.; Hempleman, L.J.; Sekiya, T.; Yoshimura, A.; Scott-Browne, J.P.; Rao, A. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 2019, 567, 530–534. [Google Scholar] [CrossRef]
- Li, R.; Liu, H.; Cao, Y.; Wang, J.; Chen, Y.; Qi, Y.; Lv, K.; Liu, X.; Yu, K.; Lin, C.; et al. Identification and validation of an immunogenic subtype of gastric cancer with abundant intratumoural CD103(+)CD8(+) T cells conferring favourable prognosis. Br. J. Cancer 2020, 122, 1525–1534. [Google Scholar] [CrossRef]
- Anadon, C.M.; Yu, X.; Hanggi, K.; Biswas, S.; Chaurio, R.A.; Martin, A.; Payne, K.K.; Mandal, G.; Innamarato, P.; Harro, C.M.; et al. Ovarian cancer immunogenicity is governed by a narrow subset of progenitor tissue-resident memory T cells. Cancer Cell 2022, 40, 545–557.e513. [Google Scholar] [CrossRef]
- Wang, D.; Diao, H.; Getzler, A.J.; Rogal, W.; Frederick, M.A.; Milner, J.; Yu, B.; Crotty, S.; Goldrath, A.W.; Pipkin, M.E. The Transcription Factor Runx3 Establishes Chromatin Accessibility of cis-Regulatory Landscapes that Drive Memory Cytotoxic T Lymphocyte Formation. Immunity 2018, 48, 659–674.e656. [Google Scholar] [CrossRef]
- Nowyhed, H.N.; Huynh, T.R.; Blatchley, A.; Wu, R.; Thomas, G.D.; Hedrick, C.C. The nuclear receptor nr4a1 controls CD8 T cell development through transcriptional suppression of runx3. Sci. Rep. 2015, 5, 9059. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Josefowicz, S.Z.; Kas, A.; Chu, T.-T.; Gavin, M.A.; Rudensky, A.Y. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 2007, 445, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Sakaguchi, S. Targeting Treg cells in cancer immunotherapy. Eur. J. Immunol. 2019, 49, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Vignali, D.A.; Collison, L.W.; Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008, 8, 523–532. [Google Scholar] [CrossRef]
- Gavin, M.A.; Rasmussen, J.P.; Fontenot, J.D.; Vasta, V.; Manganiello, V.C.; Beavo, J.A.; Rudensky, A.Y. Foxp3-dependent programme of regulatory T-cell differentiation. Nature 2007, 445, 771–775. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Zhu, B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015, 6, e1792. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Bruno, L.; Mazzarella, L.; Hoogenkamp, M.; Hertweck, A.; Cobb, B.S.; Sauer, S.; Hadjur, S.; Leleu, M.; Naoe, Y.; Telfer, J.C.; et al. Runx proteins regulate Foxp3 expression. J. Exp. Med. 2009, 206, 2329–2337. [Google Scholar] [CrossRef]
- Klunker, S.; Chong, M.M.W.; Mantel, P.-Y.; Palomares, O.; Bassin, C.; Ziegler, M.; Rückert, B.; Meiler, F.; Akdis, M.; Littman, D.R.; et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 2009, 206, 2701–2715. [Google Scholar] [CrossRef]
- Kitoh, A.; Ono, M.; Naoe, Y.; Ohkura, N.; Yamaguchi, T.; Yaguchi, H.; Kitabayashi, I.; Tsukada, T.; Nomura, T.; Miyachi, Y.; et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 2009, 31, 609–620. [Google Scholar] [CrossRef]
- Rudra, D.; Egawa, T.; Chong, M.M.; Treuting, P.; Littman, D.R.; Rudensky, A.Y. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2009, 10, 1170–1177. [Google Scholar] [CrossRef]
- Ono, M.; Yaguchi, H.; Ohkura, N.; Kitabayashi, I.; Nagamura, Y.; Nomura, T.; Miyachi, Y.; Tsukada, T.; Sakaguchi, S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 2007, 446, 685–689. [Google Scholar] [CrossRef]
- Khosravi, M.; Bidmeshkipour, A.; Moravej, A.; Hojjat-Assari, S.; Naserian, S.; Karimi, M.H. Induction of CD4(+)CD25(+)Foxp3(+) regulatory T cells by mesenchymal stem cells is associated with RUNX complex factors. Immunol. Res. 2018, 66, 207–218. [Google Scholar] [CrossRef]
- Zhou, L.; Lopes, J.E.; Chong, M.M.; Ivanov, I.I.; Min, R.; Victora, G.D.; Shen, Y.; Du, J.; Rubtsov, Y.P.; Rudensky, A.Y.; et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 2008, 453, 236–240. [Google Scholar] [CrossRef]
- Zhang, F.; Meng, G.; Strober, W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 2008, 9, 1297–1306. [Google Scholar] [CrossRef]
- Eberl, G.; Di Santo, J.P.; Vivier, E. The brave new world of innate lymphoid cells. Nat. Immunol. 2015, 16, 1–5. [Google Scholar] [CrossRef]
- Ebihara, T.; Song, C.; Ryu, S.H.; Plougastel-Douglas, B.; Yang, L.; Levanon, D.; Groner, Y.; Bern, M.D.; Stappenbeck, T.S.; Colonna, M.; et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 2015, 16, 1124–1133. [Google Scholar] [CrossRef]
- Jacquelot, N.; Seillet, C.; Vivier, E.; Belz, G.T. Innate lymphoid cells and cancer. Nat. Immunol. 2022, 23, 371–379. [Google Scholar] [CrossRef]
- Miyamoto, C.; Kojo, S.; Yamashita, M.; Moro, K.; Lacaud, G.; Shiroguchi, K.; Taniuchi, I.; Ebihara, T. Runx/Cbfbeta complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat. Commun. 2019, 10, 447. [Google Scholar] [CrossRef]
- Mirchandani, A.S.; Besnard, A.G.; Yip, E.; Scott, C.; Bain, C.C.; Cerovic, V.; Salmond, R.J.; Liew, F.Y. Type 2 innate lymphoid cells drive CD4+ Th2 cell responses. J. Immunol. 2014, 192, 2442–2448. [Google Scholar] [CrossRef]
- Grisaru-Tal, S.; Itan, M.; Klion, A.D.; Munitz, A. A new dawn for eosinophils in the tumour microenvironment. Nat. Rev. Cancer 2020, 20, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Hollande, C.; Boussier, J.; Ziai, J.; Nozawa, T.; Bondet, V.; Phung, W.; Lu, B.; Duffy, D.; Paradis, V.; Mallet, V.; et al. Inhibition of the dipeptidyl peptidase DPP4 (CD26) reveals IL-33-dependent eosinophil-mediated control of tumor growth. Nat. Immunol. 2019, 20, 257–264. [Google Scholar] [CrossRef]
- Jacquelot, N.; Seillet, C.; Wang, M.; Pizzolla, A.; Liao, Y.; Hediyeh-Zadeh, S.; Grisaru-Tal, S.; Louis, C.; Huang, Q.; Schreuder, J.; et al. Blockade of the co-inhibitory molecule PD-1 unleashes ILC2-dependent antitumor immunity in melanoma. Nat. Immunol. 2021, 22, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Carretero, R.; Sektioglu, I.M.; Garbi, N.; Salgado, O.C.; Beckhove, P.; Hammerling, G.J. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8(+) T cells. Nat. Immunol. 2015, 16, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Seo, W.; Shimizu, K.; Kojo, S.; Okeke, A.; Kohwi-Shigematsu, T.; Fujii, S.I.; Taniuchi, I. Runx-mediated regulation of CCL5 via antagonizing two enhancers influences immune cell function and anti-tumor immunity. Nat. Commun. 2020, 11, 1562. [Google Scholar] [CrossRef] [PubMed]
- Mayer, J.U.; Hilligan, K.L.; Chandler, J.S.; Eccles, D.A.; Old, S.I.; Domingues, R.G.; Yang, J.; Webb, G.R.; Munoz-Erazo, L.; Hyde, E.J.; et al. Homeostatic IL-13 in healthy skin directs dendritic cell differentiation to promote TH2 and inhibit TH17 cell polarization. Nat. Immunol. 2021, 22, 1538–1550. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Sato, T.; Kohu, K.; Takeda, K.; Okumura, K.; Satake, M.; Habu, S. Runx proteins are involved in regulation of CD122, Ly49 family and IFN-gamma expression during NK cell differentiation. Int. Immunol. 2008, 20, 71–79. [Google Scholar] [CrossRef]
- Levanon, D.; Negreanu, V.; Lotem, J.; Bone, K.R.; Brenner, O.; Leshkowitz, D.; Groner, Y. Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol. Cell Biol. 2014, 34, 1158–1169. [Google Scholar] [CrossRef]
- Lotem, J.; Levanon, D.; Negreanu, V.; Leshkowitz, D.; Friedlander, G.; Groner, Y. Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS ONE 2013, 8, e80467. [Google Scholar] [CrossRef]
- Wahlen, S.; Matthijssens, F.; Van Loocke, W.; Taveirne, S.; Kiekens, L.; Persyn, E.; Van Ammel, E.; De Vos, Z.; De Munter, S.; Matthys, P.; et al. The transcription factor RUNX2 drives the generation of human NK cells and promotes tissue residency. Elife 2022, 11. [Google Scholar] [CrossRef]
- Sun, J.C.; Beilke, J.N.; Lanier, L.L. Adaptive immune features of natural killer cells. Nature 2009, 457, 557–561. [Google Scholar] [CrossRef]
- Chiossone, L.; Dumas, P.Y.; Vienne, M.; Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018, 18, 671–688. [Google Scholar] [CrossRef]
- Laskowski, T.J.; Biederstadt, A.; Rezvani, K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat. Rev. Cancer 2022. [Google Scholar] [CrossRef]
- Rapp, M.; Lau, C.M.; Adams, N.M.; Weizman, O.E.; O’Sullivan, T.E.; Geary, C.D.; Sun, J.C. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef]
- Angkasekwinai, P.; Dong, C. IL-9-producing T cells: Potential players in allergy and cancer. Nat. Rev. Immunol. 2021, 21, 37–48. [Google Scholar] [CrossRef]
- Papotto, P.H.; Ribot, J.C.; Silva-Santos, B. IL-17(+) gammadelta T cells as kick-starters of inflammation. Nat. Immunol. 2017, 18, 604–611. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, W.; Nomura, A.; Taniuchi, I. The Roles of RUNX Proteins in Lymphocyte Function and Anti-Tumor Immunity. Cells 2022, 11, 3116. https://doi.org/10.3390/cells11193116
Seo W, Nomura A, Taniuchi I. The Roles of RUNX Proteins in Lymphocyte Function and Anti-Tumor Immunity. Cells. 2022; 11(19):3116. https://doi.org/10.3390/cells11193116
Chicago/Turabian StyleSeo, Wooseok, Aneela Nomura, and Ichiro Taniuchi. 2022. "The Roles of RUNX Proteins in Lymphocyte Function and Anti-Tumor Immunity" Cells 11, no. 19: 3116. https://doi.org/10.3390/cells11193116
APA StyleSeo, W., Nomura, A., & Taniuchi, I. (2022). The Roles of RUNX Proteins in Lymphocyte Function and Anti-Tumor Immunity. Cells, 11(19), 3116. https://doi.org/10.3390/cells11193116






