Transcriptome Changes in Glioma Cells Cultivated under Conditions of Neurosphere Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Patient-Derived Cell Cultures
2.3. Cell Culture for Neurosphere Formation
2.4. RNA Isolation
2.5. RNA Sequencing
2.6. Transcriptome Analysis
2.7. Real Time RT-PCR Analysis of RNA
2.8. Statistical Analysis
3. Results
3.1. Neurosphere Formation from Primary Brain Tumors and Immortalized Cell Lines
3.2. The Formation of Neurospheres Occurs in a Common Way in Patient-Derived Glioma Cell Cultures and Immortalized Cell Lines
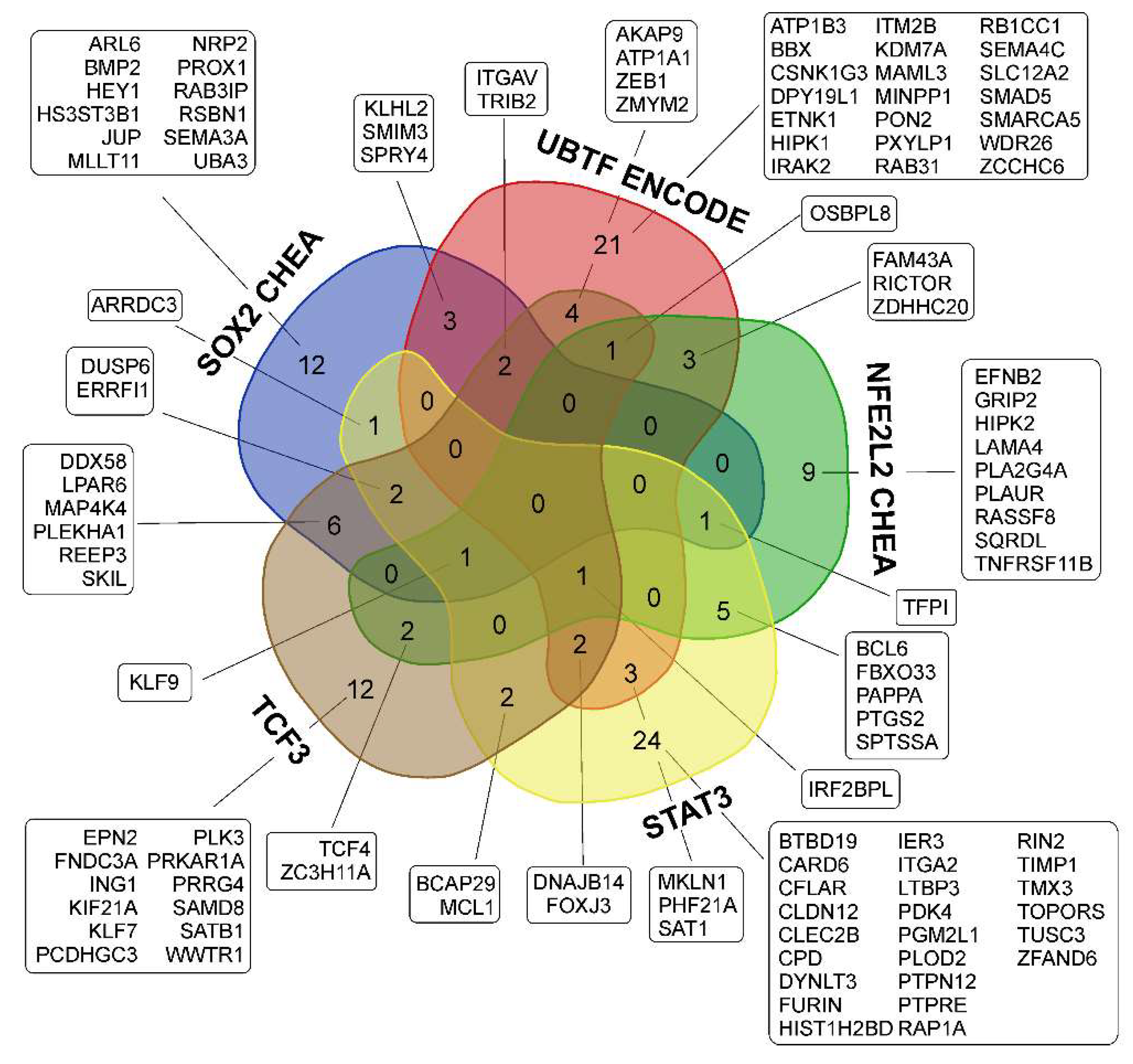
3.3. Common Gene Expression Changes in Both Patient-Derived and Immortalized Glioma Cell Cultures
3.3.1. SOX2-Dependent Gene Activation under Conditions of Neurosphere Formation by Glioma Cells
3.3.2. Activation of UBTF-Dependent Genes
3.3.3. ZEB1 Transcription Factor mRNA- and ZEB1-Controlled Genes
3.3.4. Activation of NFE2L2-Dependent Genes
3.3.5. Activation of STAT3 TF- and STAT3-Dependent Genes
3.3.6. Transcription Factor TCF3 and TCF3-Dependent Genes
3.3.7. Repression of MYC- and MAX-Controlled Genes
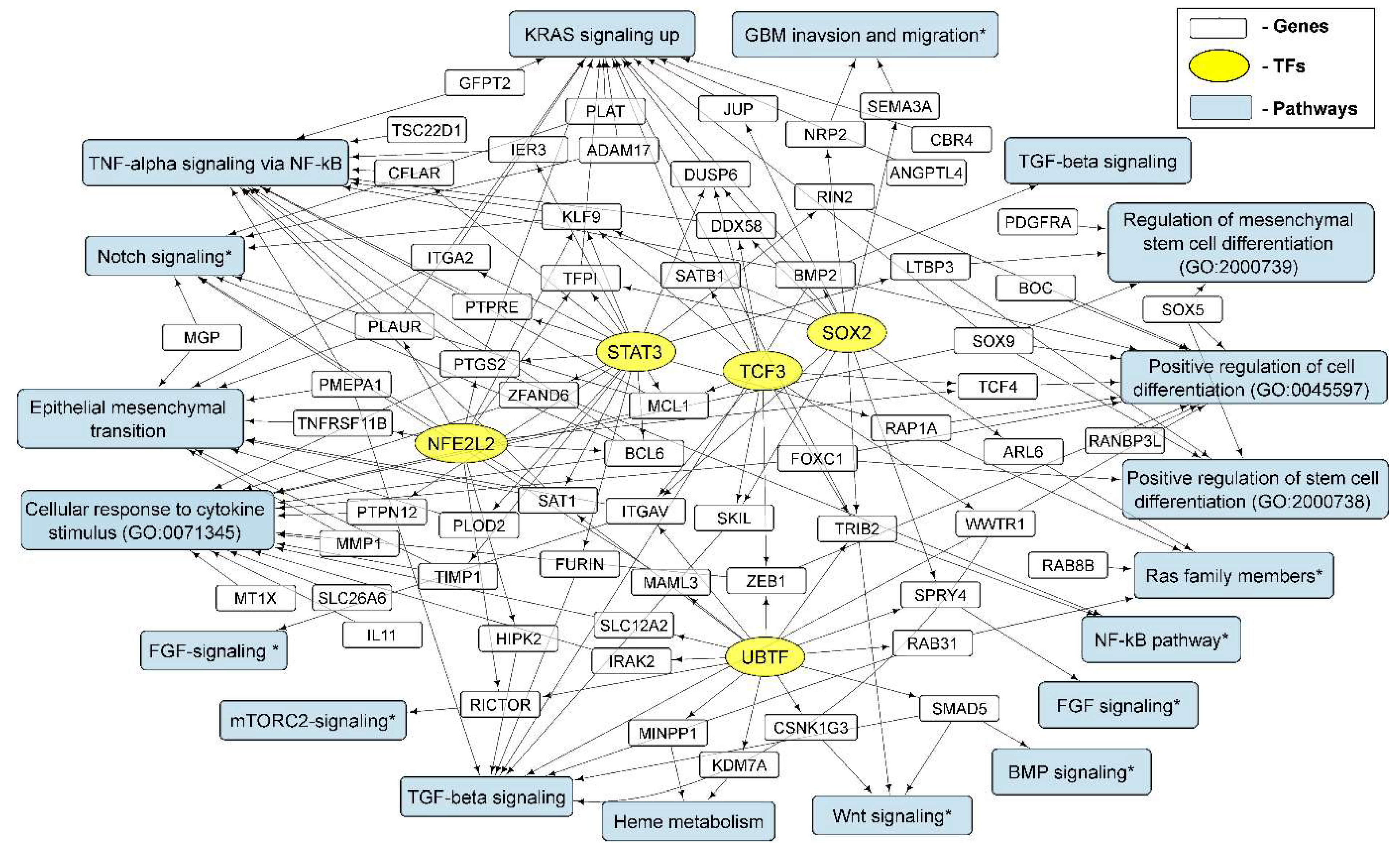
3.4. Cellular Processes and Signaling Pathways Underlying the Formation of Glioma Neurospheres
3.4.1. KRAS Signaling
3.4.2. TGF-Beta Signaling and TNF-Alpha Signaling via NF-kB
3.4.3. Epithelial to Mesenchymal Transition
3.4.4. GO Annotations and KEGG Pathways
3.5. Validation of NGS Data with qRT-PCR
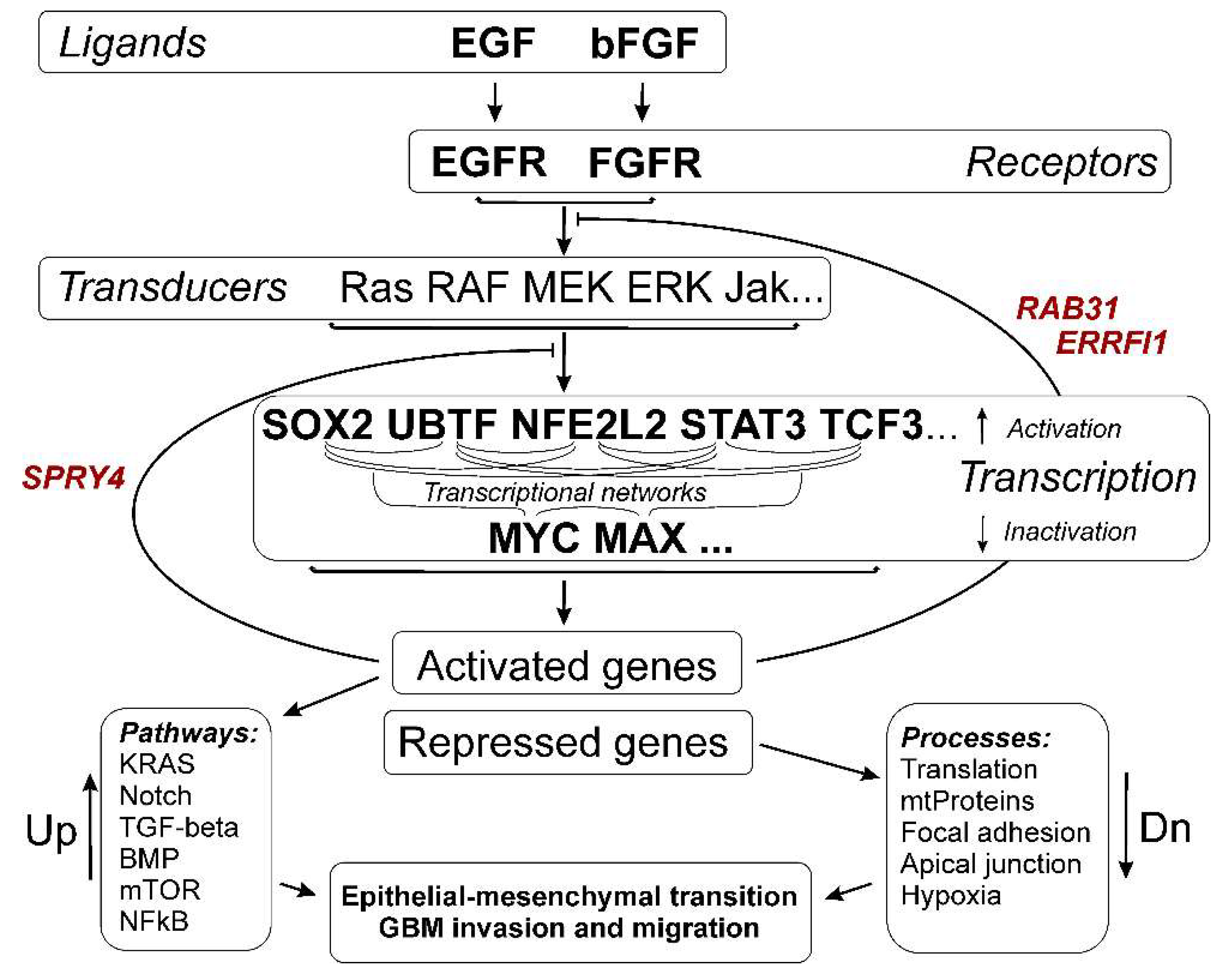
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| bFGF | basic fibroblast growth factor |
| CNS | central nervous system |
| CSCs | cancer stem cells |
| EGF | epidermal growth factor |
| EMT | epithelial-mesenchymal transition |
| GBM | glioblastoma |
| HC | hierarchical clustering |
| MN | monolayer |
| NGS | next generation sequencing |
| NS | neurosphere |
| PMT | pro-neural to mesenchymal transition |
| TFs | transcription factors |
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro. Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Miller, K.D.; Ostrom, Q.T.; Kruchko, C.; Patil, N.; Tihan, T.; Cioffi, G.; Fuchs, H.E.; Waite, K.A.; Jemal, A.; Siegel, R.L.; et al. Brain and other central nervous system tumor statistics, 2021. CA. Cancer J. Clin. 2021, 71, 381–406. [Google Scholar] [CrossRef]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef]
- Wang, D.; Wang, C.; Wang, L.; Chen, Y. A comprehensive review in improving delivery of small-molecule chemotherapeutic agents overcoming the blood-brain/brain tumor barriers for glioblastoma treatment. Drug Deliv. 2019, 26, 551–565. [Google Scholar] [CrossRef]
- Baskaran, S.; Mayrhofer, M.; Kultima, H.G.; Bergström, T.; Elfineh, L.; Cavelier, L.; Isaksson, A.; Nelander, S. Primary glioblastoma cells for precision medicine: A quantitative portrait of genomic (in)stability during the first 30 passages. Neuro. Oncol. 2018, 20, 1080–1091. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, T.N.; Kukekov, V.G.; Laywell, E.D.; Suslov, O.N.; Vrionis, F.D.; Steindler, D.A. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002, 39, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Wislet-Gendebien, S.; Leprince, P.; Moonen, G.; Rogister, B. Regulation of neural markers nestin and GFAP expression by cultivated bone marrow stromal cells. J. Cell Sci. 2003, 116, 3295–3302. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, B.A.; Weiss, S. Generation of Neurons and Astrocytes from Isolated Cells of the Adult Mammalian Central Nervous System. Science 1992, 255, 1707–1710. [Google Scholar] [CrossRef]
- Yuan, X.; Curtin, J.; Xiong, Y.; Liu, G.; Waschsmann-Hogiu, S.; Farkas, D.L.; Black, K.L.; Yu, J.S. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 2004, 23, 9392–9400. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- Marzagalli, M.; Fontana, F.; Raimondi, M.; Limonta, P. Cancer Stem Cells—Key Players in Tumor Relapse. Cancers 2021, 13, 376. [Google Scholar] [CrossRef]
- Joseph, J.V.; van Roosmalen, I.A.M.; Busschers, E.; Tomar, T.; Conroy, S.; Eggens-Meijer, E.; Peñaranda Fajardo, N.; Pore, M.M.; Balasubramanyian, V.; Wagemakers, M.; et al. Serum-Induced Differentiation of Glioblastoma Neurospheres Leads to Enhanced Migration/Invasion Capacity That Is Associated with Increased MMP9. PLoS ONE 2015, 10, e0145393. [Google Scholar] [CrossRef]
- Peñaranda-Fajardo, N.M.; Meijer, C.; Liang, Y.; Dijkstra, B.M.; Aguirre-Gamboa, R.; den Dunnen, W.F.A.; Kruyt, F.A.E. ER stress and UPR activation in glioblastoma: Identification of a noncanonical PERK mechanism regulating GBM stem cells through SOX2 modulation. Cell Death Dis. 2019, 10, 690. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, J.; Schiapparelli, P.; Lal, B.; Ying, M.; Quinones-Hinojosa, A.; Xia, S.; Laterra, J. Regulation of Glioblastoma Tumor-Propagating Cells by the Integrin Partner Tetraspanin CD151. Neoplasia 2016, 18, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Meleis, A.M.; Mahtabfar, A.; Danish, S.; Foty, R.A. Dexamethasone-mediated inhibition of Glioblastoma neurosphere dispersal in an ex vivo organotypic neural assay. PLoS ONE 2017, 12, e0186483. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.L.V.; Gomes, I.N.F.; Carloni, A.C.; Rosa, M.N.; da Silva, L.S.; Evangelista, A.F.; Reis, R.M.; Silva, V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 2021, 12, 206. [Google Scholar] [CrossRef]
- Rajalingam, K.; Schreck, R.; Rapp, U.R.; Albert, Š. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 1177–1195. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.T.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 1263–1284. [Google Scholar] [CrossRef]
- Gimple, R.C.; Wang, X. RAS: Striking at the Core of the Oncogenic Circuitry. Front. Oncol. 2019, 9, 965. [Google Scholar] [CrossRef]
- Gonzalez, D.M.; Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014, 7, re8. [Google Scholar] [CrossRef]
- Liang, Q.; Guan, G.; Li, X.; Wei, C.; Wu, J.; Cheng, P.; Wu, A.; Cheng, W. Profiling pro-neural to mesenchymal transition identifies a lncRNA signature in glioma. J. Transl. Med. 2020, 18, 378. [Google Scholar] [CrossRef]
- Zhu, Q.-C.; Gao, R.-Y.; Wu, W.; Qin, H.-L. Epithelial-mesenchymal Transition and Its Role in the Pathogenesis of Colorectal Cancer. Asian Pacific J. Cancer Prev. 2013, 14, 2689–2698. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Hartley, S.W.; Mullikin, J.C. QoRTs: A comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics 2015, 16, 224. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Iacopino, F.; Angelucci, C.; Piacentini, R.; Biamonte, F.; Mangiola, A.; Maira, G.; Grassi, C.; Sica, G. Isolation of Cancer Stem Cells from Three Human Glioblastoma Cell Lines: Characterization of Two Selected Clones. PLoS ONE 2014, 9, e105166. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.K. Neuronal Cell Culture; Methods in Molecular Biology; Amini, S., White, M.K., Eds.; Humana Press: Totowa, NJ, NJ, 2013; Volume 1078, ISBN 978-1-62703-639-9. [Google Scholar]
- Shimizu, M.; Shibuya, H.; Tanaka, N. Enhanced O-GlcNAc modification induced by the RAS/MAPK/CDK1 pathway is required for SOX2 protein expression and generation of cancer stem cells. Sci. Rep. 2022, 12, 2910. [Google Scholar] [CrossRef]
- Novak, D.; Hüser, L.; Elton, J.J.; Umansky, V.; Altevogt, P.; Utikal, J. SOX2 in development and cancer biology. Semin. Cancer Biol. 2020, 67, 74–82. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- David, C.J.; Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018, 19, 419–435. [Google Scholar] [CrossRef]
- Higgins, D.M.O.; Caliva, M.; Schroeder, M.; Carlson, B.; Upadhyayula, P.S.; Milligan, B.D.; Cheshier, S.H.; Weissman, I.L.; Sarkaria, J.N.; Meyer, F.B.; et al. Semaphorin 3A mediated brain tumor stem cell proliferation and invasion in EGFRviii mutant gliomas. BMC Cancer 2020, 20, 1213. [Google Scholar] [CrossRef]
- Sasaki, A.; Taketomi, T.; Kato, R.; Saeki, K.; Nonami, A.; Sasaki, M.; Kuriyama, M.; Saito, N.; Shibuya, M.; Yoshimura, A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol. 2003, 5, 427–432. [Google Scholar] [CrossRef]
- Sanij, E.; Diesch, J.; Lesmana, A.; Poortinga, G.; Hein, N.; Lidgerwood, G.; Cameron, D.P.; Ellul, J.; Goodall, G.J.; Wong, L.H.; et al. A novel role for the pol I transcription factor ubtf in maintaining genome stability through the regulation of highly transcribed pol II genes. Genome Res. 2015, 25, 201–212. [Google Scholar] [CrossRef]
- Kitagawa, M. Notch signalling in the nucleus: Roles of Mastermind-like (MAML) transcriptional coactivators. J. Biochem. 2016, 159, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, S.V.; O’Keeffe, G.W.; Sullivan, A.M. BMP-Smad 1/5/8 signalling in the development of the nervous system. Prog. Neurobiol. 2013, 109, 28–41. [Google Scholar] [CrossRef]
- Mayoral-Varo, V.; Jiménez, L.; Link, W. The Critical Role of TRIB2 in Cancer and Therapy Resistance. Cancers 2021, 13, 2701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, J.; Liu, W.; Ge, R.; Gao, T.; Tian, Q.; Mu, X.; Zhao, L.; Li, X. UBTF facilitates melanoma progression via modulating MEK1/2-ERK1/2 signalling pathways by promoting GIT1 transcription. Cancer Cell Int. 2021, 21, 543. [Google Scholar] [CrossRef] [PubMed]
- Vizoso-Vázquez, A.; Barreiro-Alonso, A.; González-Siso, M.I.; Rodríguez-Belmonte, E.; Lamas-Maceiras, M.; Cerdán, M.E. HMGB proteins involved in TOR signaling as general regulators of cell growth by controlling ribosome biogenesis. Curr. Genet. 2018, 64, 1205–1213. [Google Scholar] [CrossRef]
- Ramaiah, M.J.; Kumar, K.R. mTOR-Rictor-EGFR axis in oncogenesis and diagnosis of glioblastoma multiforme. Mol. Biol. Rep. 2021, 48, 4813–4835. [Google Scholar] [CrossRef]
- Caramel, J.; Ligier, M.; Puisieux, A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018, 78, 30–35. [Google Scholar] [CrossRef]
- Siebzehnrubl, F.A.; Silver, D.J.; Tugertimur, B.; Deleyrolle, L.P.; Siebzehnrubl, D.; Sarkisian, M.R.; Devers, K.G.; Yachnis, A.T.; Kupper, M.D.; Neal, D.; et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol. Med. 2013, 5, 1196–1212. [Google Scholar] [CrossRef]
- Anandhan, A.; Dodson, M.; Schmidlin, C.J.; Liu, P.; Zhang, D.D. Breakdown of an Ironclad Defense System: The Critical Role of NRF2 in Mediating Ferroptosis. Cell Chem. Biol. 2020, 27, 436–447. [Google Scholar] [CrossRef]
- Gilder, A.S.; Natali, L.; Van Dyk, D.M.; Zalfa, C.; Banki, M.A.; Pizzo, D.P.; Wang, H.; Klemke, R.L.; Mantuano, E.; Gonias, S.L. The Urokinase Receptor Induces a Mesenchymal Gene Expression Signature in Glioblastoma Cells and Promotes Tumor Cell Survival in Neurospheres. Sci. Rep. 2018, 8, 2982. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhou, L.; Sun, X.; Li, Q. Homeodomain-interacting protein kinase 2 (HIPK2): A promising target for anti-cancer therapies. Oncotarget 2017, 8, 20452–20461. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, K.; Shi, Z.; Zou, J.; Wang, Y.; Jia, Z.; Zhang, A.; Han, L.; Yue, X.; Liu, N.; et al. High-catenin/Tcf-4 activity confers glioma progression via direct regulation of AKT2 gene expression. Neuro. Oncol. 2011, 13, 600–609. [Google Scholar] [CrossRef]
- Ju, Q.; Li, X.; Zhang, H.; Yan, S.; Li, Y.; Zhao, Y. NFE2L2 Is a Potential Prognostic Biomarker and Is Correlated with Immune Infiltration in Brain Lower Grade Glioma: A Pan-Cancer Analysis. Oxid. Med. Cell. Longev. 2020, 2020, 3580719. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, Y.; Biswas, A.; Kumar, U.; Das, S.; Banerjee, I.; Banik, P.; Bharti, R.; Nayak, S.; Ghosh, S.K.; Mandal, M. Targeting NFE2L2, a transcription factor upstream of MMP-2: A potential therapeutic strategy for temozolomide resistant glioblastoma. Biochem. Pharmacol. 2019, 164, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Vilchez Mercedes, S.A.; Bocci, F.; Ahmed, M.; Eder, I.; Zhu, N.; Levine, H.; Onuchic, J.N.; Jolly, M.K.; Wong, P.K. Nrf2 Modulates the Hybrid Epithelial/Mesenchymal Phenotype and Notch Signaling During Collective Cancer Migration. Front. Mol. Biosci. 2022, 9, 807324. [Google Scholar] [CrossRef]
- Giannopoulou, A.-I.; Kanakoglou, D.S.; Piperi, C. Transcription Factors with Targeting Potential in Gliomas. Int. J. Mol. Sci. 2022, 23, 3720. [Google Scholar] [CrossRef]
- Zhang, H.-F.; Lai, R. STAT3 in Cancer—Friend or Foe? Cancers 2014, 6, 1408–1440. [Google Scholar] [CrossRef]
- Ying, M.; Sang, Y.; Li, Y.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Vescovi, A.L.; Eberhart, C.G.; Xia, S.; Laterra, J. Krüppel-Like Family of Transcription Factor 9, a Differentiation-Associated Transcription Factor, Suppresses Notch1 Signaling and Inhibits Glioblastoma-Initiating Stem Cells. Stem Cells 2011, 29, 20–31. [Google Scholar] [CrossRef]
- De Santis, F.; Romero-Cordoba, S.L.; Castagnoli, L.; Volpari, T.; Faraci, S.; Fucà, G.; Tagliabue, E.; De Braud, F.; Pupa, S.M.; Di Nicola, M. BCL6 and the Notch pathway: A signaling axis leading to a novel druggable biotarget in triple negative breast cancer. Cell. Oncol. 2022, 45, 257–274. [Google Scholar] [CrossRef]
- Tan, C.; Liu, L.; Liu, X.; Qi, L.; Wang, W.; Zhao, G.; Wang, L.; Dai, Y. Activation of PTGS2 /NF-κB signaling pathway enhances radiation resistance of glioma. Cancer Med. 2019, 8, 1175–1185. [Google Scholar] [CrossRef]
- Widden, H.; Placzek, W.J. The multiple mechanisms of MCL1 in the regulation of cell fate. Commun. Biol. 2021, 4, 1029. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-M.; Hong, X.-W.; Wen, X.; Han, X.-R.; Wang, S.; Wang, Y.-J.; Shen, M.; Fan, S.-H.; Zhuang, J.; Zhang, Z.-F.; et al. MCL1 gene silencing promotes senescence and apoptosis of glioma cells via inhibition of the PI3K/Akt signaling pathway. IUBMB Life 2019, 71, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Adorno-Cruz, V.; Liu, H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Dis. 2019, 6, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, C. Gene 33/Mig6/ERRFI1, an Adapter Protein with Complex Functions in Cell Biology and Human Diseases. Cells 2021, 10, 1574. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Pereira, L.; Merrill, B.J. Tcf3 Functions as a Steady-State Limiter of Transcriptional Programs of Mouse Embryonic Stem Cell Self-Renewal. Stem Cells 2008, 26, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Yan, B.; Ma, J.; Jiang, H.; Li, L.; Tang, H.; Ji, F.; Yao, Z. Expression of TCF3 in Wilms’ tumor and its regulatory role in kidney tumor cell viability, migration and apoptosis in vitro. Mol. Med. Rep. 2021, 24, 642. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Hu, X.; Lian, H.; Wang, L.; Fu, H. Transcription factor 3 controls cell proliferation and migration in glioblastoma multiforme cell lines. Biochem. Cell Biol. 2016, 94, 247–255. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, C.; Huo, G.; Deng, J.; Zhao, H.; Xu, R.; Jiang, L.; Chen, S.; Wang, S. ATP1A1 Integrates AKT and ERK Signaling via Potential Interaction With Src to Promote Growth and Survival in Glioma Stem Cells. Front. Oncol. 2019, 9, 320. [Google Scholar] [CrossRef]
- Lee, D.; Apelt, K.; Lee, S.O.; Chan, H.R.; Luijsterburg, M.S.; Leung, J.W.C.; Miller, K.M. ZMYM2 restricts 53BP1 at DNA double-strand breaks to favor BRCA1 loading and homologous recombination. Nucleic Acids Res. 2022, 50, 160. [Google Scholar] [CrossRef]
- Higashimori, A.; Dong, Y.; Zhang, Y.; Kang, W.; Nakatsu, G.; Ng, S.S.M.; Arakawa, T.; Sung, J.J.Y.; Chan, F.K.L.; Yu, J. Forkhead Box F2 Suppresses Gastric Cancer through a Novel FOXF2–IRF2BPL–β-Catenin Signaling Axis. Cancer Res. 2018, 78, 1643–1656. [Google Scholar] [CrossRef] [PubMed]
- Escoll, M.; Lastra, D.; Pajares, M.; Robledinos-Antón, N.; Rojo, A.I.; Fernández-Ginés, R.; Mendiola, M.; Martínez-Marín, V.; Esteban, I.; López-Larrubia, P.; et al. Transcription factor NRF2 uses the Hippo pathway effector TAZ to induce tumorigenesis in glioblastomas. Redox Biol. 2020, 30, 101425. [Google Scholar] [CrossRef]
- Frömberg, A.; Engeland, K.; Aigner, A. The Special AT-rich Sequence Binding Protein 1 (SATB1) and its role in solid tumors. Cancer Lett. 2018, 417, 96–111. [Google Scholar] [CrossRef]
- Poole, C.J.; van Riggelen, J. MYC—Master Regulator of the Cancer Epigenome and Transcriptome. Genes 2017, 8, 142. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Chua, C.E.L.; Tang, B.L. The role of the small GTPase Rab31 in cancer. J. Cell. Mol. Med. 2015, 19, 1–10. [Google Scholar] [CrossRef]
- Zunke, F.; Rose-John, S. The shedding protease ADAM17: Physiology and pathophysiology. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2059–2070. [Google Scholar] [CrossRef]
- Mirzaei, S.; Saghari, S.; Bassiri, F.; Raesi, R.; Zarrabi, A.; Hushmandi, K.; Sethi, G.; Tergaonkar, V. NF-κB as a regulator of cancer metastasis and therapy response: A focus on epithelial–mesenchymal transition. J. Cell. Physiol. 2022. [Google Scholar] [CrossRef]
- Iser, I.C.; Pereira, M.B.; Lenz, G.; Wink, M.R. The Epithelial-to-Mesenchymal Transition-Like Process in Glioblastoma: An Updated Systematic Review and In Silico Investigation. Med. Res. Rev. 2017, 37, 271–313. [Google Scholar] [CrossRef] [PubMed]
- Arlt, A.; Schäfer, H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur. J. Cell Biol. 2011, 90, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Merk, B.C.; Owens, J.L.; Lopes, M.-B.S.; Silva, C.M.; Hussaini, I.M. STAT6 expression in glioblastoma promotes invasive growth. BMC Cancer 2011, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Kreße, N.; Schröder, H.; Stein, K.-P.; Wilkens, L.; Mawrin, C.; Sandalcioglu, I.E.; Dumitru, C.A. PLOD2 Is a Prognostic Marker in Glioblastoma That Modulates the Immune Microenvironment and Tumor Progression. Int. J. Mol. Sci. 2022, 23, 6037. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, P.; Arozarena, I. Tyrosine Kinase Inhibitors in Adult Glioblastoma: An (Un)Closed Chapter? Cancers 2021, 13, 5799. [Google Scholar] [CrossRef] [PubMed]
- de la Rocha, A.M.A.; Sampron, N.; Alonso, M.M.; Matheu, A. Role of SOX family of transcription factors in central nervous system tumors. Am. J. Cancer Res. 2014, 4, 312–324. [Google Scholar]
- Zhang, L.; Yu, H.; Yuan, Y.; Yu, J.S.; Lou, Z.; Xue, Y.; Liu, Y. The necessity for standardization of glioma stem cell culture: A systematic review. Stem Cell Res. Ther. 2020, 11, 84. [Google Scholar] [CrossRef]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma Stem Cell Lines Expanded in Adherent Culture Have Tumor-Specific Phenotypes and Are Suitable for Chemical and Genetic Screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef]
- Vinel, C.; Rosser, G.; Guglielmi, L.; Constantinou, M.; Pomella, N.; Zhang, X.; Boot, J.R.; Jones, T.A.; Millner, T.O.; Dumas, A.A.; et al. Comparative epigenetic analysis of tumour initiating cells and syngeneic EPSC-derived neural stem cells in glioblastoma. Nat. Commun. 2021, 12, 6130. [Google Scholar] [CrossRef]
- LeBlanc, V.G.; Trinh, D.L.; Aslanpour, S.; Hughes, M.; Livingstone, D.; Jin, D.; Ahn, B.Y.; Blough, M.D.; Cairncross, J.G.; Chan, J.A.; et al. Single-cell landscapes of primary glioblastomas and matched explants and cell lines show variable retention of inter- and intratumor heterogeneity. Cancer Cell 2022, 40, 379–392. [Google Scholar] [CrossRef]
- Bastola, S.; Pavlyukov, M.S.; Yamashita, D.; Ghosh, S.; Cho, H.; Kagaya, N.; Zhang, Z.; Minata, M.; Lee, Y.; Sadahiro, H.; et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun. 2020, 11, 4660. [Google Scholar] [CrossRef]
- Collet, B.; Avril, T.; Aubry, M.; Hamlat, A.; Le Reste, P.-J.; Chiforeanu, D.; Vauleon, E.; Mosser, J.; Quillien, V. Proteomic analysis underlines the usefulness of both primary adherent and stem-like cell lines for studying proteins involved in human glioblastoma. J. Proteomics 2014, 110, 7–19. [Google Scholar] [CrossRef]
- Peixoto, J.; Janaki-Raman, S.; Schlicker, L.; Schmitz, W.; Walz, S.; Winkelkotte, A.M.; Herold-Mende, C.; Soares, P.; Schulze, A.; Lima, J. Integrated Metabolomics and Transcriptomics Analysis of Monolayer and Neurospheres from Established Glioblastoma Cell Lines. Cancers 2021, 13, 1327. [Google Scholar] [CrossRef] [PubMed]
- Celik-Selvi, B.E.; Stütz, A.; Mayer, C.-E.; Salhi, J.; Siegwart, G.; Sutterlüty, H. Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells. Cells 2019, 8, 808. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Yoo, C.; Roy, D. Reverse Engineering of Modified Genes by Bayesian Network Analysis Defines Molecular Determinants Critical to the Development of Glioblastoma. PLoS ONE 2013, 8, e64140. [Google Scholar] [CrossRef] [PubMed]
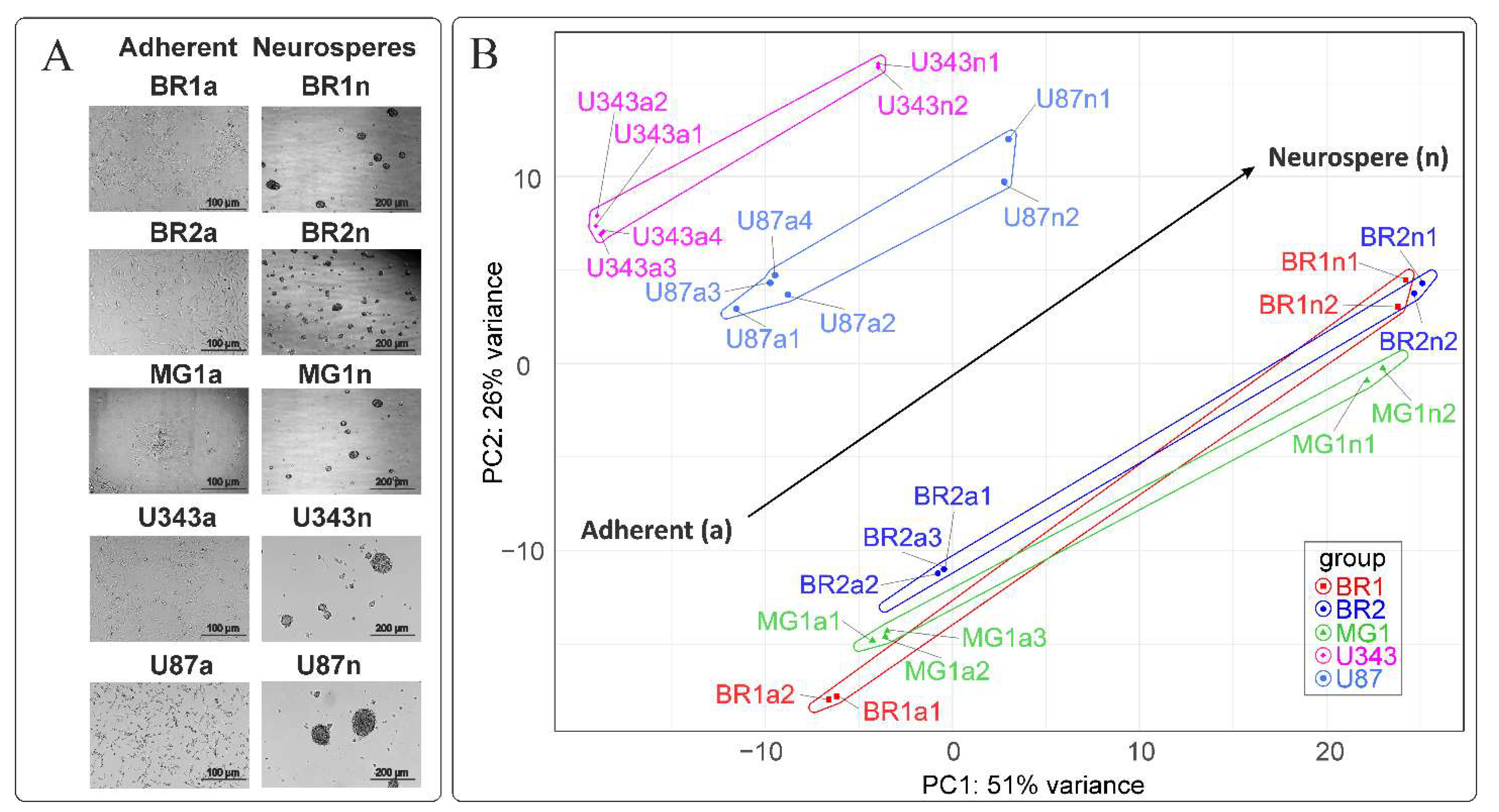
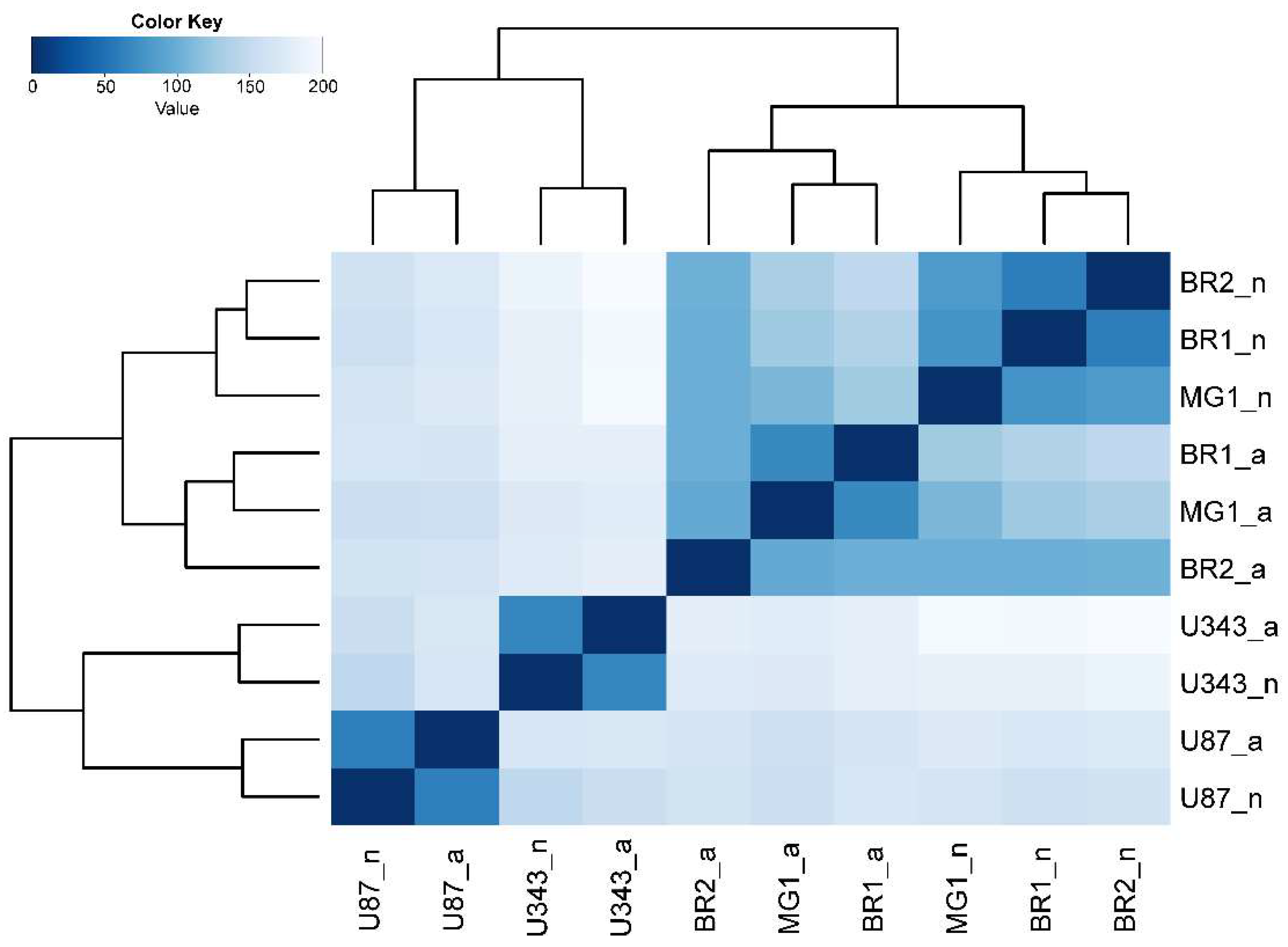
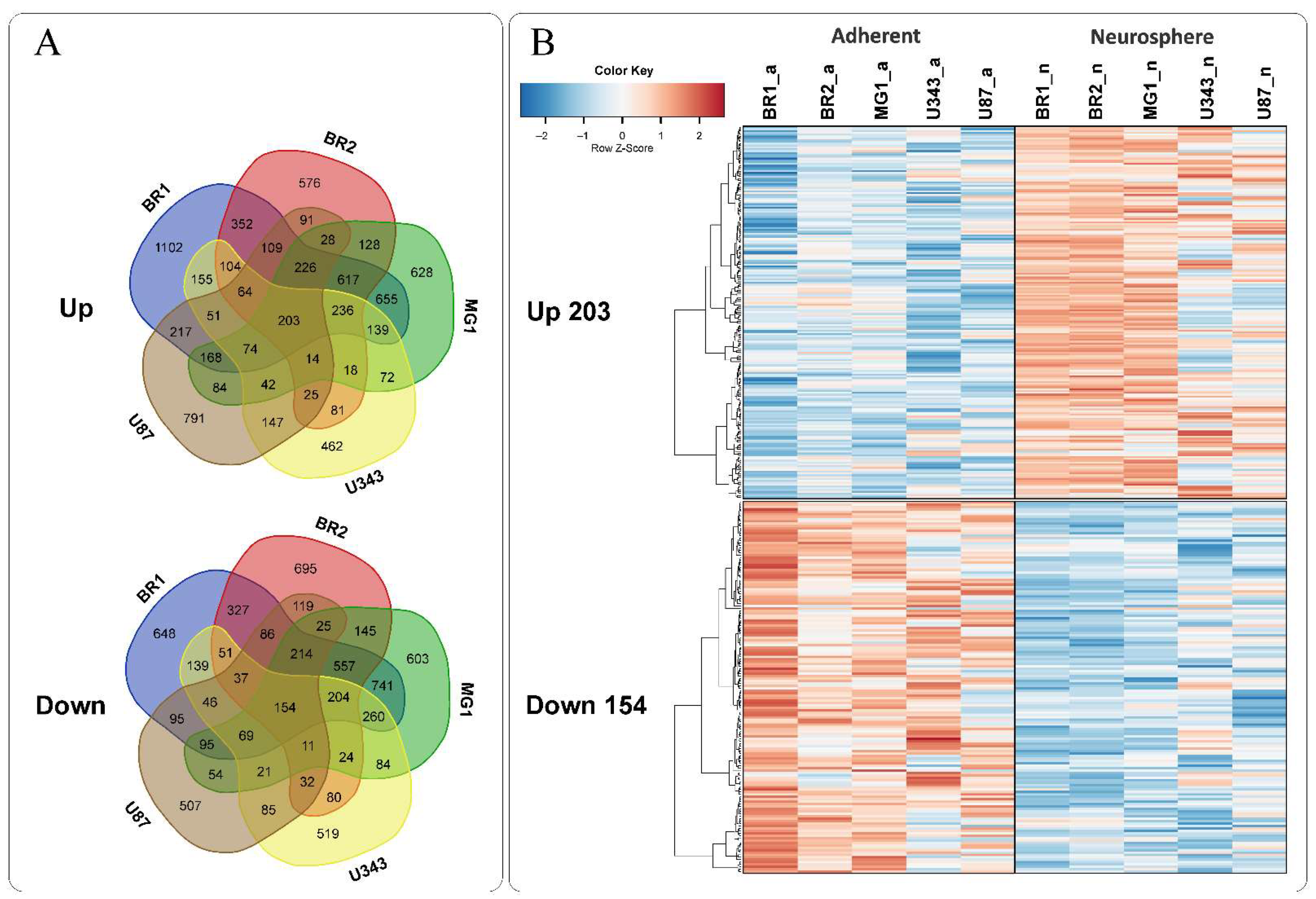
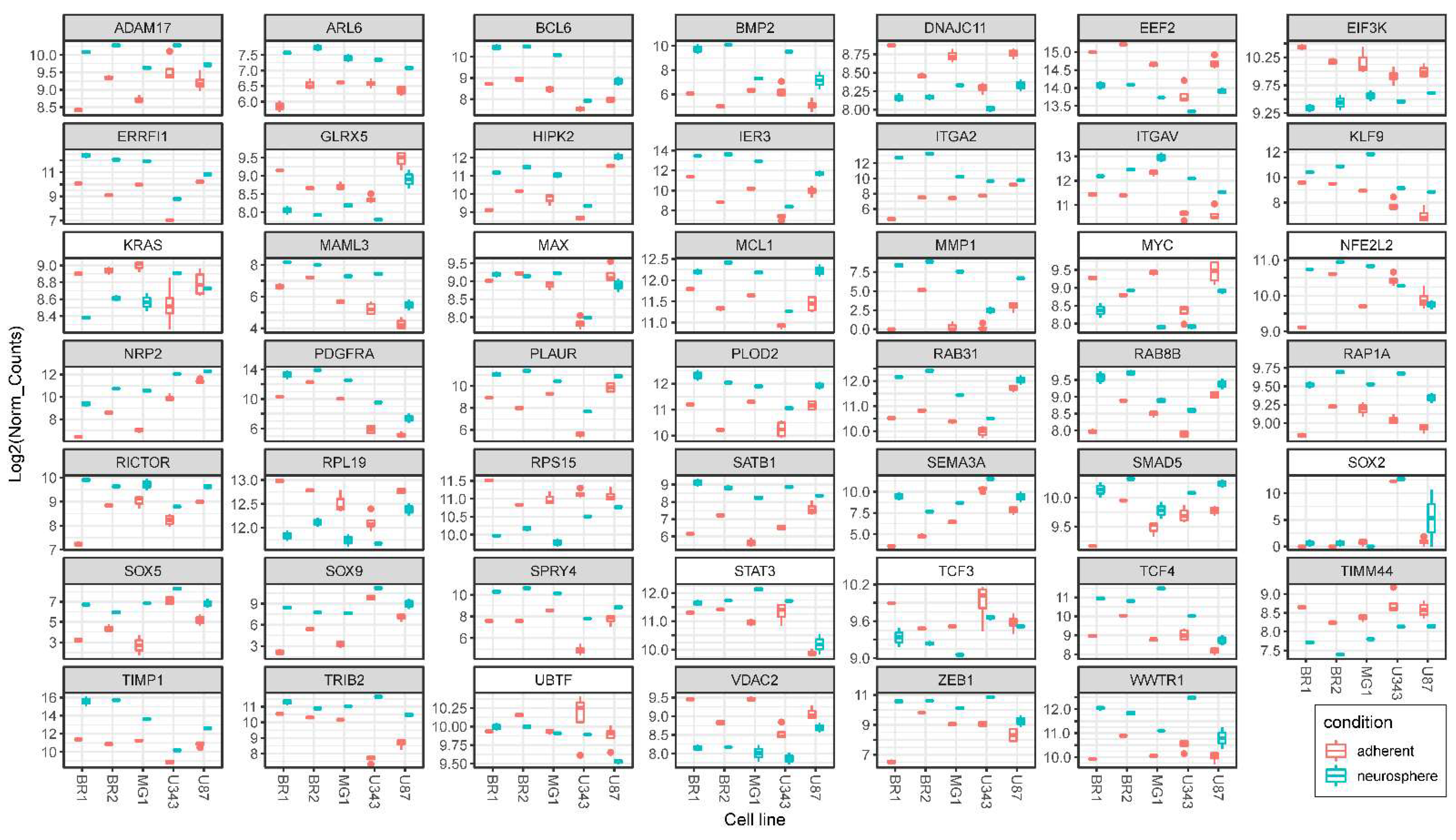


| Cell Culture | Histological Characteristic | Culture Conditions | NGS-Library * | Number of Replicates | Number of NGS-Sequencing Reads (106) *** |
|---|---|---|---|---|---|
| BR1 | GBM | MN | BR1a | 2 | 22.94 |
| NS | BR1n | 2 | 19.45 | ||
| BR2 | Diffuse astrocytoma | MN | BR2a | 3 | 35.47 |
| NS | BR2n | 2 | 19.68 | ||
| MG1 | GBM | MN | MG1a | 3 | 34.10 |
| NS | MG1n | 2 | 18.98 | ||
| U343 ** | GBM | MN | U343a | 4 | 46.60 |
| NS | U343n | 2 | 24.10 | ||
| U87 ** | GBM | MN | U87a | 4 | 46.35 |
| NS | U87n | 2 | 22.95 |
| Particular MN/NS Pairs | |||||
| BR1 | BR2 | MG1 | U343 | U87 | |
| Up * | 4472 | 2872 | 3332 | 1887 | 2334 |
| Down * | 3723 | 2761 | 3261 | 1816 | 1650 |
| Common (Overlapping) Genes | |||||
| All Up | 203 | ||||
| Up/Down ** | 121 | ||||
| All Down | 154 | ||||
| Rank | Overlapped | BR1 | BR2 | MG1 | U343 | U87 |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| 1 | SOX2 CHEA | UBTF ENCODE | UBTF ENCODE | UBTF ENCODE | SOX2 CHEA | UBTF ENCODE |
| 2 | UBTF ENCODE | NFE2L2 CHEA | NFE2L2 CHEA | NFE2L2 CHEA | SUZ12 CHEA | AR CHEA |
| 3 | FOXA2 ENCODE | AR CHEA | TAF1 ENCODE | SOX2 CHEA | NFE2L2 CHEA | NFE2L2 CHEA |
| 4 | NFE2L2 CHEA | SOX2 CHEA | CREB1 CHEA | SUZ12 CHEA | AR CHEA | SOX2 CHEA |
| 5 | TP53 CHEA | SMAD4 CHEA | SOX2 CHEA | ZBTB7A ENCODE | TCF3 CHEA | ZNF384 ENCODE |
| 6 | SALL4 CHEA | TCF7L2 ENCODE | BRCA1 ENCODE | GATA1 CHEA | TP63 CHEA | GATA1 CHEA |
| 7 | AR CHEA | FOXA2 ENCODE | PPARG CHEA | AR CHEA | UBTF ENCODE | SMAD4 CHEA |
| 8 | TCF3 ENCODE * | CHD1 ENCODE | CREB1 ENCODE | FOXA2 ENCODE | GATA2 CHEA | TCF3 ENCODE |
| 9 | TCF3 CHEA * | ZNF384 ENCODE | RUNX1 CHEA | TCF3 ENCODE | NANOG CHEA | CTCF ENCODE |
| 10 | VDR CHEA * | TCF3 ENCODE | GATA2 CHEA | ESR1 CHEA | STAT3 CHEA | TCF3 CHEA |
| Downregulated | ||||||
| 1 | MYC ENCODE | NFYB ENCODE | USF1 ENCODE | E2F4 ENCODE | USF1 ENCODE | E2F4 ENCODE |
| 2 | MAX ENCODE | MAX ENCODE | USF2 ENCODE | MAX ENCODE | MAX ENCODE | E2F6 ENCODE |
| 3 | MYC CHEA | MYC ENCODE | KLF4 CHEA | MYC ENCODE | USF2 ENCODE | MAX ENCODE |
| 4 | USF2 ENCODE | NFYA ENCODE | MAX ENCODE | TAF1 ENCODE | MYC ENCODE | MYC ENCODE |
| 5 | TAF1 ENCODE | TAF1 ENCODE | BHLHE40 ENCODE | NFYB ENCODE | GABPA ENCODE | BRCA1 ENCODE |
| 6 | USF1 ENCODE | BRCA1 ENCODE | E2F6 ENCODE | MYC CHEA | ZBTB7A ENCODE | TAF1 ENCODE |
| 7 | ATF2 ENCODE | GABPA ENCODE | CTCF ENCODE | E2F6 ENCODE | YY1 ENCODE | NFYB ENCODE |
| 8 | PML ENCODE | MYC CHEA | ZBTB7A ENCODE | NFYA ENCODE | ELF1 ENCODE | ATF2 ENCODE |
| 9 | NFYA ENCODE | E2F4 ENCODE | SMC3 ENCODE | SIN3A ENCODE | TAF1 ENCODE | CREB1 ENCODE |
| 10 | ZBTB7A ENCODE | YY1 ENCODE | NFYB ENCODE | E2F1 CHEA | CREB1 CHEA | YY1 ENCODE |
| # * | Overlapped | BR1 | BR2 | MG1 | U343 | U87 |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| 1 | KRAS Signaling Up | Interferon Gamma Response | TNF-alpha Signaling via NF-kB | TNF-alpha Signaling via NF-kB | TNF-alpha Signaling via NF-kB | EMT ** |
| 2 | TNF-alpha Signaling via NF-kB | Interferon Alpha Response | Interferon Gamma Response | Interferon Gamma Response | Cholesterol Homeostasis | UV Response Dn |
| 3 | TGF-beta Signaling | KRAS Signaling Up | EMT ** | Interferon Alpha Response | Apoptosis | Cholesterol Homeostasis |
| 4 | EMT ** | TNF-alpha Signaling via NF-kB | Inflammatory Response | KRAS Signaling Up | EMT ** | KRAS Signaling Up |
| 5 | Apoptosis | EMT ** | KRAS Signaling Up | Inflammatory Response | Hypoxia | TGF-beta Signaling |
| 6 | Coagulation | TGF-beta Signaling | Interferon Alpha Response | EMT ** | p53 Pathway | TNF-alpha Signaling via NF-kB |
| 7 | Complement | Inflammatory Response | TGF-beta Signaling | TGF-beta Signaling | Inflammatory Response | Complement |
| 8 | Androgen Response | IL-6/JAK/STAT3 Signaling | IL-6/JAK/STAT3 Signaling | IL-6/JAK/STAT3 Signaling | KRAS Signaling Up | Hypoxia |
| 9 | Estrogen Response Early | IL-2/STAT5 Signaling | Apoptosis | Estrogen Response Early | Angiogenesis | Coagulation |
| 10 | Angiogenesis | UV Response Dn | IL-2/STAT5 Signaling | Angiogenesis | Estrogen Response Early | Androgen Response |
| Downregulated | ||||||
| 1 | Apical Junction | Myc Targets V1 | Cholesterol Homeostasis | E2F Targets | Myc Targets V2 | E2F Targets |
| 2 | Oxidative Phosphorylation | Oxidative Phosphorylation | Fatty Acid Metabolism | Myc Targets V1 | Apical Junction | Oxidative Phosphorylation |
| 3 | Hypoxia | E2F Targets | p53 Pathway | G2-M Checkpoint | Mitotic Spindle | Myc Targets V1 |
| 4 | Myc Targets V1 | G2-M Checkpoint | Myogenesis | Oxidative Phosphorylation | EMT ** | G2-M Checkpoint |
| 5 | Androgen Response | mTORC1 Signaling | mTORC1 Signaling | mTORC1 Signaling | Unfolded Protein Response | DNA Repair |
| 6 | PI3K/AKT/mTOR Signaling | DNA Repair | Adipogenesis | Mitotic Spindle | UV Response Dn | Mitotic Spindle |
| 7 | *** | Adipogenesis | Mitotic Spindle | Glycolysis | Adipogenesis | Estrogen Response Late |
| 8 | *** | Fatty Acid Metabolism | UV Response Dn | Myc Targets V2 | PI3K/AKT/mTOR Signaling | p53 Pathway |
| 9 | *** | Myc Targets V2 | Hypoxia | Hypoxia | Estrogen Response Early | Adipogenesis |
| 10 | *** | Cholesterol Homeostasis | Oxidative Phosphorylation | DNA Repair | Myc Targets V1 | Myc Targets V2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasileva, N.S.; Kuligina, E.V.; Dymova, M.A.; Savinovskaya, Y.I.; Zinchenko, N.D.; Ageenko, A.B.; Mishinov, S.V.; Dome, A.S.; Stepanov, G.A.; Richter, V.A.; et al. Transcriptome Changes in Glioma Cells Cultivated under Conditions of Neurosphere Formation. Cells 2022, 11, 3106. https://doi.org/10.3390/cells11193106
Vasileva NS, Kuligina EV, Dymova MA, Savinovskaya YI, Zinchenko ND, Ageenko AB, Mishinov SV, Dome AS, Stepanov GA, Richter VA, et al. Transcriptome Changes in Glioma Cells Cultivated under Conditions of Neurosphere Formation. Cells. 2022; 11(19):3106. https://doi.org/10.3390/cells11193106
Chicago/Turabian StyleVasileva, Natalia S., Elena V. Kuligina, Maya A. Dymova, Yulya I. Savinovskaya, Nikita D. Zinchenko, Alisa B. Ageenko, Sergey V. Mishinov, Anton S. Dome, Grigory A. Stepanov, Vladimir A. Richter, and et al. 2022. "Transcriptome Changes in Glioma Cells Cultivated under Conditions of Neurosphere Formation" Cells 11, no. 19: 3106. https://doi.org/10.3390/cells11193106
APA StyleVasileva, N. S., Kuligina, E. V., Dymova, M. A., Savinovskaya, Y. I., Zinchenko, N. D., Ageenko, A. B., Mishinov, S. V., Dome, A. S., Stepanov, G. A., Richter, V. A., & Semenov, D. V. (2022). Transcriptome Changes in Glioma Cells Cultivated under Conditions of Neurosphere Formation. Cells, 11(19), 3106. https://doi.org/10.3390/cells11193106






