Effects of Noonan Syndrome-Germline Mutations on Mitochondria and Energy Metabolism
Abstract
1. Introduction
2. The RAS/MAPK Pathway
3. Noonan Syndrome (NS) and Noonan Syndrome with Multiple Lentigines (NSML)
4. Energy Metabolism and the Oxidative Phosphorylation (OXPHOS) Machinery
5. Functional and Pathophysiological Effects of NS and NSML Mutations on Mitochondria and Energy Metabolism
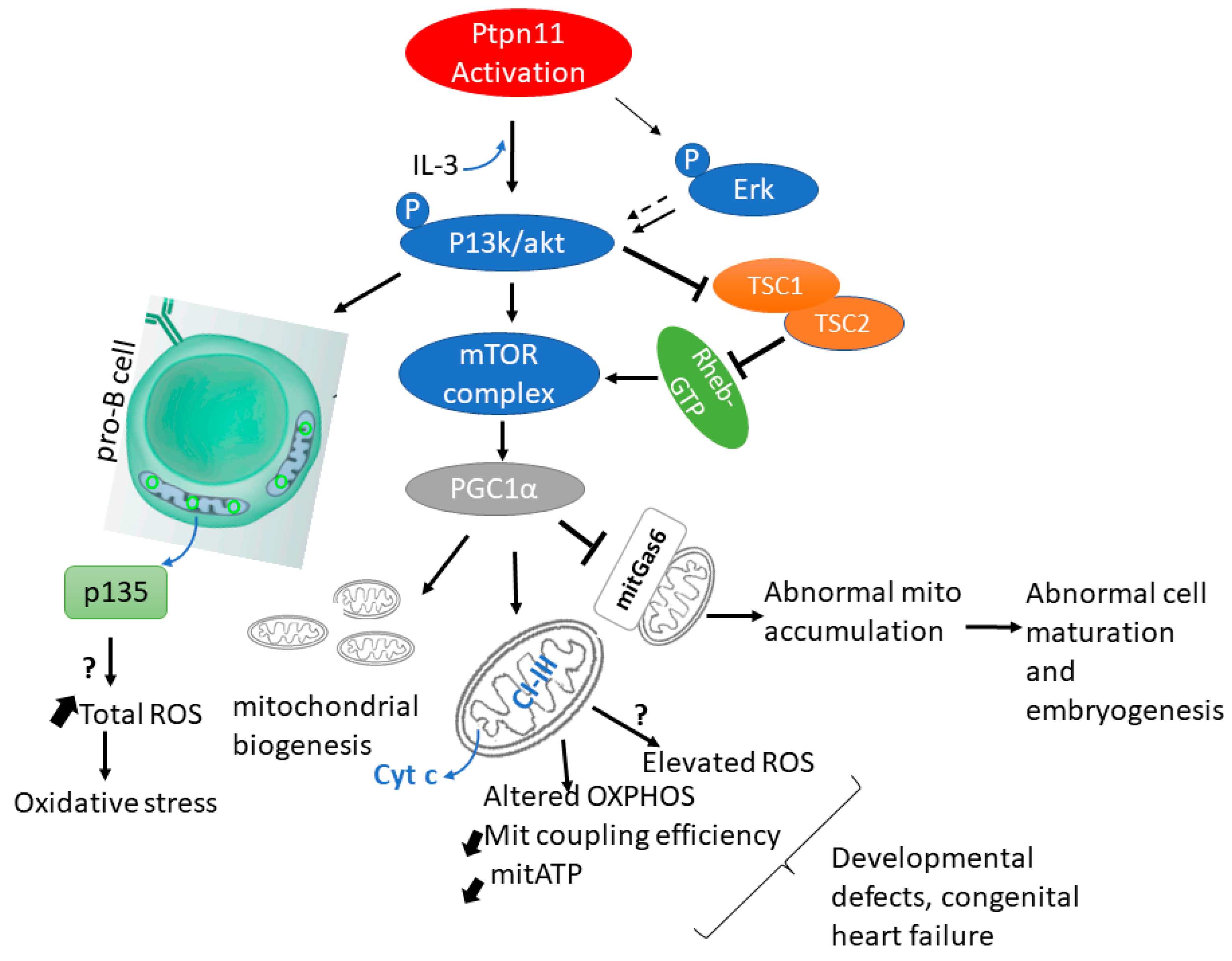
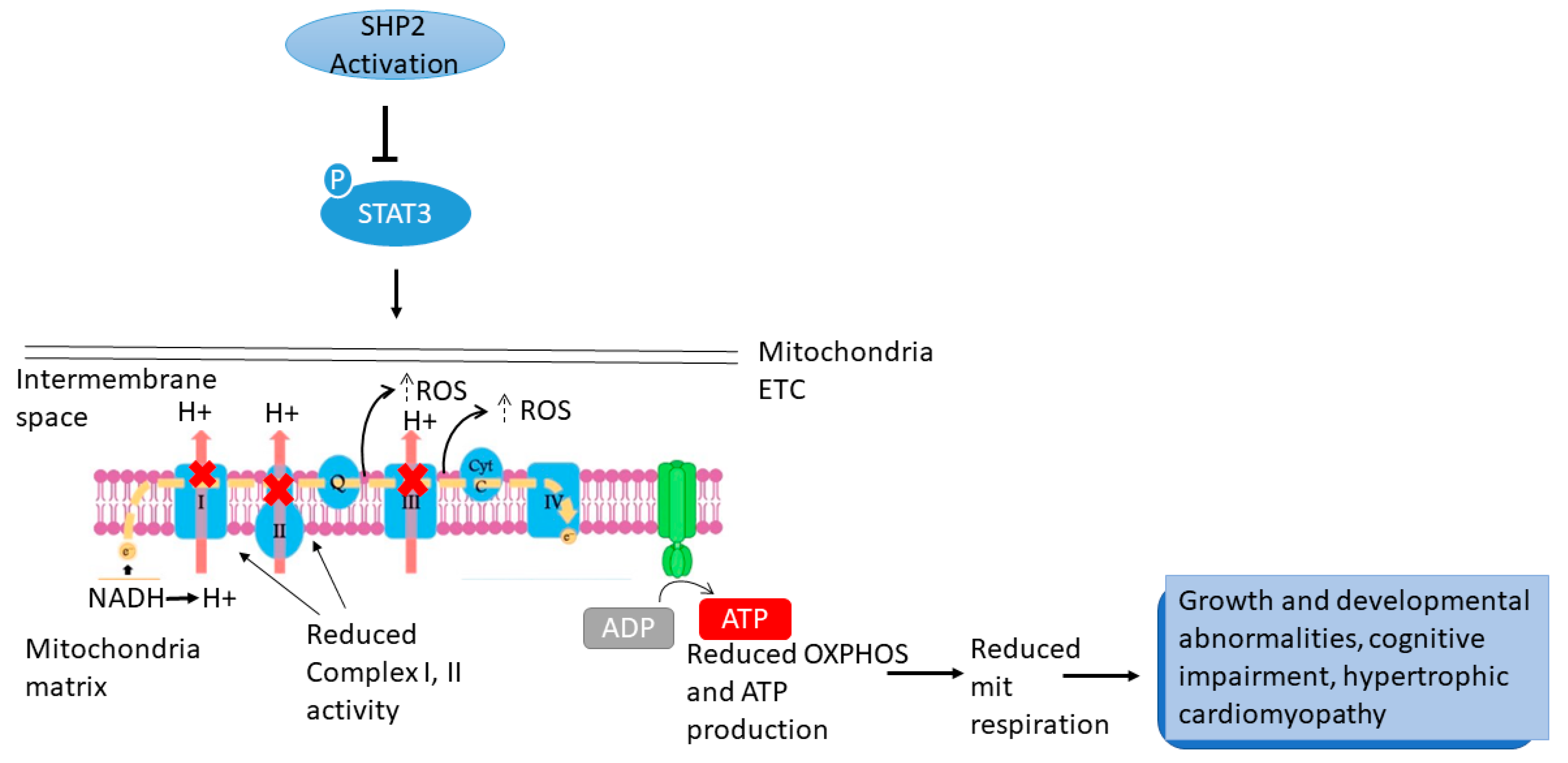
5.1. PTPN11/SHP2
5.2. KRAS
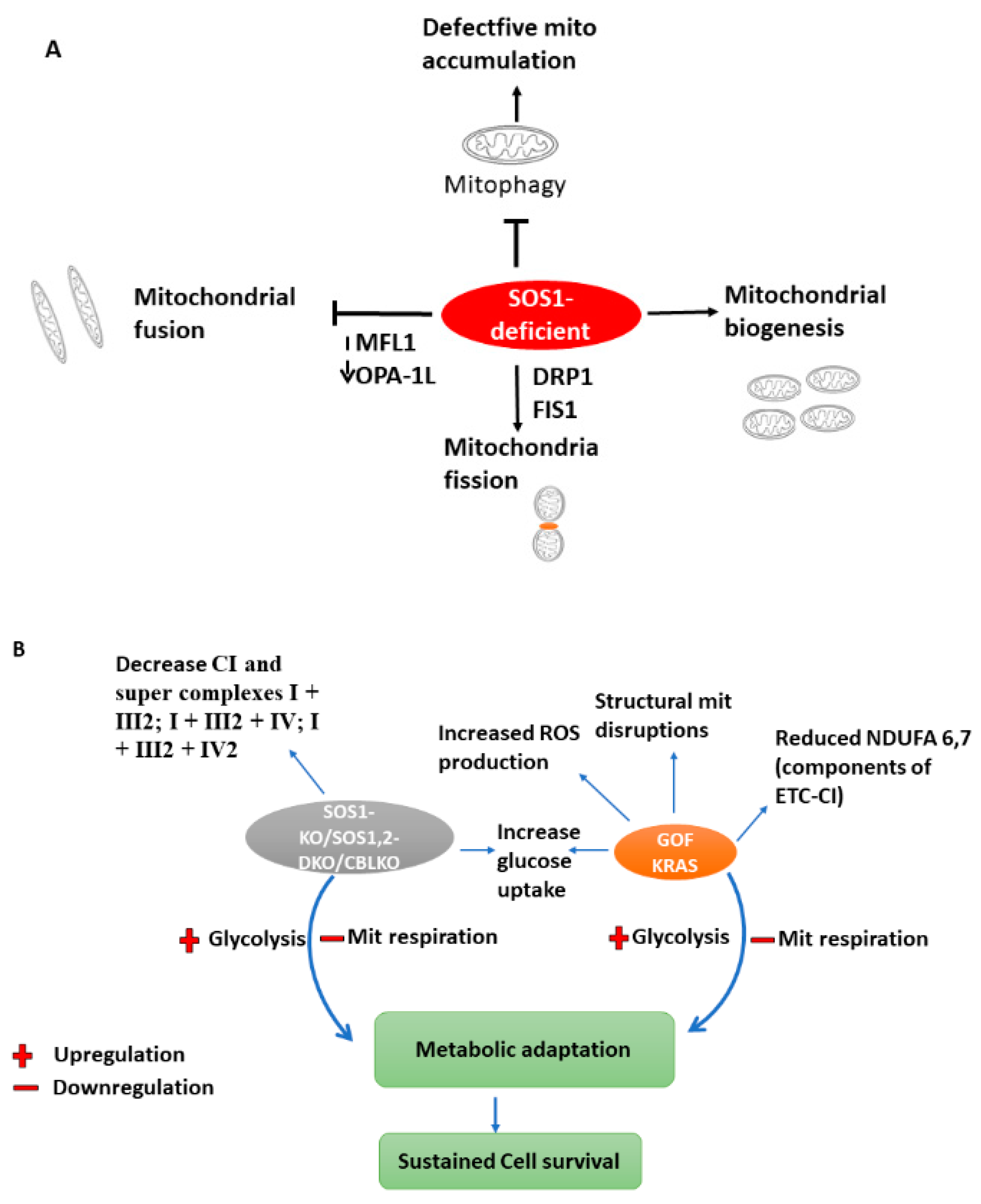
5.3. SOS1/SOS2 (Sons of the Sevenless)
5.4. CBL
5.5. RRAS
5.6. RAF
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2DG | 2-Deoxy-D-glucose |
| 2-NBDG | 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose |
| 3T3-L1 | 3-day transfer, inoculum 3 x 105 cells- fibroblast-like cell line |
| 8GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| ADP | Adenosine diphosphate |
| AMP | Adenosine monophosphate |
| AMPK | AMP-activated protein kinase |
| ATGL | Adipose triglyceride lipase |
| ATP | Adenosine triphosphate |
| BAD | BCL2 associated agonist of cell death |
| BAF3 | IL-3 dependent murine pro B cell |
| Bak | Bcl-2 homologous antagonist/killer |
| Bax | Bcl-2-like protein 4 |
| BCL-2 | B-cell lymphoma 2 |
| Beta-MHC | Beta-Myosin heavy chain |
| BIM | Bcl-2-like protein 11 |
| CAAX | C-Cysteine, AA-2 Aliphatic residues, X-any C-terminal amino acid |
| CBL-KO THP | Human CBL knockout HEK-293T cell line |
| CC3 | Cleaved caspase-3 |
| CcO | Cytochrome C oxidase |
| CI,II,III,IV | Complex I-IV |
| CMPs | common myeloid progenitors |
| COII | Cytochrome C oxidase subunit 2 |
| COS-1 | kidney fibroblast-like cell line /CV-1 (simian) in Origin, and carrying the SV40 genetic mate |
| Cyt c | Cytochrome c |
| DAPK | Death-Associated Protein Kinase |
| DRP1 | Dynamin-related protein 1 |
| ECAR | Extracellular acidification rate |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| EPO | Erythropoietin |
| ER | Endoplamic reticulum |
| ETC | Electron transport chain |
| FADH2 | flavin adenine dinucleotide |
| FCCP | Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone |
| FGF | Fibroblast growth factor |
| FIS1 | Mitochondrial fission 1 protein |
| FRS-2 | FGF receptor substrate 2 |
| Fru1,6bisP | Fructose-1-bisPhosphate |
| Gab1 | GRB2-associated-binding protein 1 |
| GDP | Guanosine diphosphate |
| GF | Growth factor |
| GLUT1 | Glucose transporter 1 |
| GLUT3 | Glucose transporter 3 |
| glycoATP | Glycolytic ATP |
| GM-CSF | Granulocyte macrophage-colony stimulating factor |
| GMPs | Granulocyte macrophage progenitors |
| GOF | Gain-of-function |
| GRB2 | Growth factor receptor-bound protein 2 |
| GSH | Glutathione |
| GSH/GSSG | Glutathione/Glutathione disulfide (reduced glutathione/Oxidised glutathione) ratio |
| GTP | Guanosine-5′-triphosphate |
| GTPase | GTP hydrolase |
| GW5074 | 3-(3,5-Dibromo-4-hydroxybenzyliden)-5-iodo-1,3-dihydroindol-2-one (a C-RAF inhibit) |
| HEK293 | Human embryonic kidney 293 cells |
| HFD | Hight fat diet |
| HGP | hepatic glucose production |
| HK1/HK2 | Hexokinase 1 and 2 |
| HSCs | Hematopoietic stem cells |
| HSL | Hormone-sensitive lipase |
| IL-3 | Interleukin 3 |
| IMS | intermembranous space |
| IRS-1 | Insulin receptor substrate 1 |
| Jak2 | Janus kinase 2 |
| JMML | Juvenile myelomonocytic leukemia |
| JNK | c-Jun N-terminal kinases |
| KO | Knock-out |
| LC3B | Light chain 3B (subunit of Microtubule-associated proteins 1A/1B) |
| LDHA | Lactate Dehydrogenase A |
| LIF | Leukemia inhibitory factor |
| LSHKO | Liver-specific Shp2 Knock-out |
| MAPK/ERK | Mitogen-activated protein kinase/Extracellular signal-regulated kinase 1/2 |
| MDC | Mitochondria disease criteria |
| MEDICA | MEthyl-substituted DICarboxylic Acids |
| MEF | Mouse embryonic fibroblast |
| MEK1 or MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1 |
| MEPs | Megakaryocyte erythroid progenitors |
| MFN1 | Mitofusin-1 protein |
| Mit | Mitochondria |
| mitoATP/mitATP | mitochondria ATP |
| MitoSOXTM | Red Mitochondrial Superoxide Indicator |
| MitoTrackerTM Red CMXRos | MitoTracker Red and Chloromethyl-X-rosamine |
| MMP | Mitochondrial membrane potential |
| mPTP | Mitochondrial permeability transition pores |
| MST2 | mammalian Sterile 20-like kinase 2 |
| mtDNA | Mitochondrial DNA |
| mTOR/S6K | mammalian target of rapamycin (mTOR)/ribosomal protein S6 kinase |
| mut | mutation |
| MYBPC3 | Cardiac myosin binding protein C3 |
| NADH | Nicotinamide adenine dinucleotide |
| NADP+ | Oxidised form of Nicotinamide adenine dinucleotide phosphate (NADPH) |
| ND | NADH Dehydrogenase |
| NDUSF3 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 |
| NLRP3 | NLR family pyrin domain containing 3 |
| NSC87877 | 8-Hydroxy-7-[(6-sulfo-2-naphthyl)azo]-5-quinolinesulfonic acid (Inhibitor of SHP2/SHP1) |
| OCR | Oxygen consumption rate |
| OGTT | Oral glucose tolerance test |
| OPA-1L | Dominant optic atrophy (long form of mitochondrial dynamin-like GTPase) |
| OXPHOS | Oxidative phosphorylation |
| pAMPK | phospho-AMP-activated protein kinase |
| Pdh | Pyruvate dehydrogenase |
| Pdhe1a | Pyruvate dehydrogenase E1 component subunit alpha |
| p-ERK | Phosphorylated ERK |
| PI3K-AKT-mTOR | The phosphatidylinositol-3-kinase (PI3K)/Akt and the mammalian target of rapamycin (mTOR) |
| PP1C | Protein phosphatase 1 catalytic subunit |
| PPP | Pentose phosphate pathway |
| RAS/MAPK | Rat sarcoma virus/Mitogen-activated protein kinase |
| RB2 | Retinoblastoma-like protein 2 |
| ROS | Reactive oxygen species |
| RTK | Receptor tyrosin kinase |
| SOD2 | Superoxide dismutase 2 |
| SOS1/2-DKO | Double knock-out SOS1/2 |
| SOS1-KO | Single Knock-out SOS1 |
| SOS2-KO | Single Knock-out SOS2 |
| SRC | Spare respiratory capacity |
| STAT3 | Signal transducer and activator of transcription 3 |
| TCA | Tricarboxylic acid cycle |
| THP-1 | Tamm-Horsfall Protein 1 (Spontaneously immortalized monocyte-like cell line) |
| TIM13 | Translocase of inner membrane 13 |
| TOM20 | Translocase of outer membrane 20 |
| TOMM40 | Translocase of outer mitochondria membrane 40 |
| UQCRC2 | Cytochrome b-c1 complex subunit 2 |
| VDAC | Voltage-dependent anion channels (mitochondrial porins) |
| WT | Wildtype |
References
- Dard, L.; Bellance, N.; Lacombe, D.; Rossignol, R. RAS signalling in energy metabolism and rare human diseases. Biochim. Biophys. Acta 2018, 1859, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Tajan, M.; Paccoud, R.; Branka, S.; Edouard, T.; Yart, A. The RASopathy Family: Consequences of Germline Activation of the RAS/MAPK Pathway. Endocr. Rev. 2018, 39, 676–700. [Google Scholar] [CrossRef]
- Rauen, K.A. The RASopathies. Annu. Rev. Genom. Hum. Genet. 2013, 14, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.P.; Dubois, S.M.; Agarwal, S.; Zon, L.I. Mitochondrial function in development and disease. Dis. Model. Mech. 2021, 14, dmm048912. [Google Scholar] [CrossRef]
- Salvi, M.; Stringaro, A.; Brunati, A.M.; Agostinelli, E.; Arancia, G.; Clari, G.; Toninello, A. Tyrosine phosphatase activity in mitochondria: Presence of Shp-2 phosphatase in mitochondria. Experientia 2004, 61, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Arachiche, A.; Augereau, O.; Decossas, M.; Pertuiset, C.; Gontier, E.; Letellier, T.; Dachary-Prigent, J. Localization of PTP-1B, SHP-2, and Src Exclusively in Rat Brain Mitochondria and Functional Consequences. J. Biol. Chem. 2008, 283, 24406–24411. [Google Scholar] [CrossRef]
- Lee, I.; Pecinova, A.; Pecina, P.; Neel, B.G.; Araki, T.; Kucherlapati, R.; Roberts, A.E.; Hüttemann, M. A suggested role for mitochondria in Noonan syndrome. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2010, 1802, 275–283. [Google Scholar] [CrossRef]
- Simanshu, D.K.; Nissley, D.V.; McCormick, F. RAS Proteins and Their Regulators in Human Disease. Cell 2017, 170, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Gelb, B.D.; Zenker, M. Noonan syndrome and clinically related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 161–179. [Google Scholar] [CrossRef]
- Tartaglia, M.; Zampino, G.; Gelb, B.D. Noonan Syndrome: Clinical Aspects and Molecular Pathogenesis. Mol. Syndromol. 2010, 1, 2–26. [Google Scholar] [CrossRef]
- Atay, O.; Skotheim, J.M. Spatial and temporal signal processing and decision making by MAPK pathways. J. Cell Biol. 2017, 216, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Dance, M.; Montagner, A.; Salles, J.-P.; Yart, A.; Raynal, P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell. Signal. 2008, 20, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Bunda, S.; Burrell, K.; Heir, P.; Zeng, L.; Alamsahebpour, A.; Kano, Y.; Raught, B.; Zhang, Z.Y.; Zadeh, G.; Ohh, M. Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nat. Commun. 2015, 6, 8859. [Google Scholar] [CrossRef] [PubMed]
- Avruch, J. Ras Activation of the Raf Kinase: Tyrosine Kinase Recruitment of the MAP Kinase Cascade. Recent Prog. Horm. Res. 2001, 56, 127–156. [Google Scholar] [CrossRef] [PubMed]
- Noonan, J.A. Hypertelorism With Turner Phenotype: A New Syndrome With Associated Congenital Heart Disease. Am. J. Dis. Child. 1968, 116, 373. [Google Scholar] [CrossRef] [PubMed]
- Allanson, J.E.; Bohring, A.; Dörr, H.-G.; Dufke, A.; Gillessen-Kaesbach, G.; Horn, D.; König, R.; Kratz, C.P.; Kutsche, K.; Pauli, S.; et al. The face of Noonan syndrome: Does phenotype predict genotype. Am. J. Med. Genet. Part A 2010, 152A, 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Digilio, M.C.; Conti, E.; Sarkozy, A.; Mingarelli, R.; Dottorini, T.; Marino, B.; Pizzuti, A.; Dallapiccola, B. Grouping of Multiple-Lentigines/LEOPARD and Noonan Syndromes on the PTPN11 Gene. Am. J. Hum. Genet. 2002, 71, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Digilio, M.C.; Sarkozy, A.; de Zorzi, A.; Pacileo, G.; Limongelli, G.; Mingarelli, R.; Calabrò, R.; Marino, B.; Dallapiccola, B. LEOPARD syndrome: Clinical diagnosis in the first year of life. Am. J. Med. Genet. Part A 2006, 140A, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Mehler, E.L.; Goldberg, R.; Zampino, G.; Brunner, H.G.; Kremer, H.; Van Der Burgt, I.; Crosby, A.H.; Ion, A.; Jeffery, S.; et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001, 29, 465–468. [Google Scholar] [CrossRef]
- Li, X.; Yao, R.; Tan, X.; Li, N.; Ding, Y.; Li, J.; Chang, G.; Chen, Y.; Ma, L.; Wang, J.; et al. Molecular and phenotypic spectrum of Noonan syndrome in Chinese patients. Clin. Genet. 2019, 96, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Sawada, M.; Kawakami, T.; Morita, M.; Aoki, Y. A Patient with Noonan Syndrome with a KRAS Mutation Who Presented Severe Nerve Root Hypertrophy. Case Rep. Neurol. 2021, 13, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, S.Y.; Yang, A.; Jang, J.-H.; Choi, Y.; Lee, J.-E.; Jin, D.-K. An atypical case of Noonan syndrome with KRAS mutation diagnosed by targeted exome sequencing. Ann. Pediatr. Endocrinol. Metab. 2017, 22, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Cirstea, I.C.; Kutsche, K.; Dvorsky, R.; Gremer, L.; Carta, C.; Horn, D.; Roberts, A.E.; Lepri, F.R.; Merbitz-Zahradnik, T.; König, R.; et al. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat. Genet. 2009, 42, 27–29. [Google Scholar] [CrossRef]
- Xu, S.; Fan, Y.; Sun, Y.; Wang, L.; Gu, X.; Yu, Y. Targeted/exome sequencing identified mutations in ten Chinese patients diagnosed with Noonan syndrome and related disorders. BMC Med. Genom. 2017, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, S.; Stellacci, E.; Pannone, L.; D’Agostino, D.; Consoli, F.; Lissewski, C.; Silvano, M.; Cencelli, G.; Lepri, F.; Maitz, S.; et al. Molecular Diversity and Associated Phenotypic Spectrum of Germline CBL Mutations. Hum. Mutat. 2015, 36, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Coe, R.R.; McKinnon, M.; Tarailo-Graovac, M.; Ross, C.J.; Wasserman, W.; Friedman, J.M.; Rogers, P.C.; van Karnebeek, C.D. A case of splenomegaly in CBL syndrome. Eur. J. Med. Genet. 2017, 60, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Vissers, L.; Bonetti, M.; Overman, J.P.; Nillesen, W.M.; Frints, S.G.; de Ligt, J.; Zampino, G.; Justino, A.; Machado, J.C.; Schepens, M.; et al. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur. J. Hum. Genet. 2014, 23, 317–324. [Google Scholar] [CrossRef]
- Yamamoto, G.L.; Aguena, M.; Gos, M.; Hung, C.; Pilch, J.; Fahiminiya, S.; Abramowicz, A.; Cristian, I.; Buscarilli, M.; Naslavsky, M.S.; et al. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J. Med. Genet. 2015, 52, 413–421. [Google Scholar] [CrossRef]
- Roberts, A.E. Noonan Syndrome. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. Available online: http://www.ncbi.nlm.nih.gov/books/NBK1124/ (accessed on 5 April 2022).
- Capri, Y.; Flex, E.; Krumbach, O.H.; Carpentieri, G.; Cecchetti, S.; Lißewski, C.; Adariani, S.R.; Schanze, D.; Brinkmann, J.; Piard, J.; et al. Activating Mutations of RRAS2 Are a Rare Cause of Noonan Syndrome. Am. J. Hum. Genet. 2019, 104, 1223–1232. [Google Scholar] [CrossRef]
- Flex, E.; Jaiswal, M.; Pantaleoni, F.; Martinelli, S.; Strullu, M.; Fansa, E.K.; Caye, A.; De Luca, A.; Lepri, F.; Dvorsky, R.; et al. Activating mutations in RRAS underlie a phenotype within the RASopathy spectrum and contribute to leukaemogenesis. Hum. Mol. Genet. 2014, 23, 4315–4327. [Google Scholar] [CrossRef]
- Aoki, Y.; Niihori, T.; Banjo, T.; Okamoto, N.; Mizuno, S.; Kurosawa, K.; Ogata, T.; Takada, F.; Yano, M.; Ando, T.; et al. Gain-of-Function Mutations in RIT1 Cause Noonan Syndrome, a RAS/MAPK Pathway Syndrome. Am. J. Hum. Genet. 2013, 93, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Lauriol, J.; Cabrera, J.R.; Roy, A.; Keith, K.; Hough, S.M.; Damilano, F.; Wang, B.; Segarra, G.C.; Flessa, M.E.; Miller, L.E.; et al. Developmental SHP2 dysfunction underlies cardiac hypertrophy in Noonan syndrome with multiple lentigines. J. Clin. Investig. 2016, 126, 2989–3005. [Google Scholar] [CrossRef] [PubMed]
- Conboy, E.; Dhamija, R.; Wang, M.; Xie, J.; Dyck, P.J.; Bridges, A.G.; Spinner, R.J.; Clayton, A.C.; Watson, R.E.; Messiaen, L.; et al. Paraspinal neurofibromas and hypertrophic neuropathy in Noonan syndrome with multiple lentigines. J. Med. Genet. 2015, 53, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, F.; Ponce, D.; Berward, F.J.; Lopetegui, B.; Cassorla, F.; Aracena, M. RAF1 variant in a patient with Noonan syndrome with multiple lentigines and craniosynostosis. Am. J. Med. Genet. Part A 2019, 179, 1598–1602. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Thompson, C.B. Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us? Cell 2012, 148, 1132–1144. [Google Scholar] [CrossRef]
- Benard, G.; Bellance, N.; Jose, C.; Melser, S.; Nouette-Gaulain, K.; Rossignol, R. Multi-site control and regulation of mitochondrial energy production. Biochim. Biophys. Acta 2010, 1797, 698–709. [Google Scholar] [CrossRef]
- Walsh, C.T.; Tu, B.P.; Tang, Y. Eight Kinetically Stable but Thermodynamically Activated Molecules that Power Cell Metabolism. Chem. Rev. 2017, 118, 1460–1494. [Google Scholar] [CrossRef]
- Sazanov, L. A giant molecular proton pump: Structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 2015, 16, 375–388. [Google Scholar] [CrossRef]
- Chen, Q.; Vazquez, E.J.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Production of Reactive Oxygen Species by Mitochondria. J. Biol. Chem. 2003, 278, 36027–36031. [Google Scholar] [CrossRef]
- Feramisco, J.R.; Gross, M.; Kamata, T.; Rosenberg, M.; Sweet, R.W. Microinjection of the oncogene form of the human H-ras (t-24) protein results in rapid proliferation of quiescent cells. Cell 1984, 38, 109–117. [Google Scholar] [CrossRef]
- Posada, I.M.D.; Lectez, B.; Siddiqui, F.A.; Oetken-Lindholm, C.; Sharma, M.; Abankwa, D. Opposite feedback from mTORC1 to H-ras and K-ras4B downstream of SREBP1. Sci. Rep. 2017, 7, 8944. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, V.; Alliouachene, S.; Sotiropoulos, A.; Sobering, A.; Athea, Y.; Djouadi, F.; Miraux, S.; Thiaudière, E.; Foretz, M.; Viollet, B.; et al. S6 Kinase Deletion Suppresses Muscle Growth Adaptations to Nutrient Availability by Activating AMP Kinase. Cell Metab. 2007, 5, 476–487. [Google Scholar] [CrossRef] [PubMed]
- DiMauro, S.; Schon, E.A. Mitochondrial Respiratory-Chain Diseases. New Engl. J. Med. 2003, 348, 2656–2668. [Google Scholar] [CrossRef] [PubMed]
- Bugiardini, E.; Bottani, E.; Marchet, S.; Poole, O.V.; Beninca, C.; Horga, A.; Woodward, C.; Lam, A.; Hargreaves, I.; Chalasani, A.; et al. Expanding the Molecular and Phenotypic Spectrum of Truncating MT-ATP6 Mutations. Neurol. Genet. 2020, 6, e381. [Google Scholar] [CrossRef] [PubMed]
- Inak, G.; Rybak-Wolf, A.; Lisowski, P.; Pentimalli, T.M.; Jüttner, R.; Glažar, P.; Uppal, K.; Bottani, E.; Brunetti, D.; Secker, C.; et al. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat. Commun. 2021, 12, 1929. [Google Scholar] [CrossRef]
- Burbulla, L.F.; Song, P.; Mazzulli, J.R.; Zampese, E.; Wong, Y.C.; Jeon, S.; Santos, D.P.; Blanz, J.; Obermaier, C.D.; Strojny, C.; et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 2017, 357, 1255–1261. [Google Scholar] [CrossRef]
- Dolci, S.; Mannino, L.; Bottani, E.; Campanelli, A.; Di Chio, M.; Zorzin, S.; D’Arrigo, G.; Amenta, A.; Segala, A.; Paglia, G.; et al. Therapeutic induction of energy metabolism reduces neural tissue damage and increases microglia activation in severe spinal cord injury. Pharmacol. Res. 2022, 178, 106149. [Google Scholar] [CrossRef]
- Brunetti, D.; Bottani, E.; Segala, A.; Marchet, S.; Rossi, F.; Orlando, F.; Malavolta, M.; Carruba, M.O.; Lamperti, C.; Provinciali, M.; et al. Targeting Multiple Mitochondrial Processes by a Metabolic Modulator Prevents Sarcopenia and Cognitive Decline in SAMP8 Mice. Front. Pharmacol. 2020, 11, 1171. [Google Scholar] [CrossRef]
- Bottani, E.; Cerutti, R.; Harbour, M.E.; Ravaglia, S.; Dogan, S.A.; Giordano, C.; Fearnley, I.M.; D’Amati, G.; Viscomi, C.; Fernandez-Vizarra, E.; et al. TTC19 Plays a Husbandry Role on UQCRFS1 Turnover in the Biogenesis of Mitochondrial Respiratory Complex III. Mol. Cell 2017, 67, 96–105.e4. [Google Scholar] [CrossRef]
- Luo, Y.; Ma, J.; Lu, W. The Significance of Mitochondrial Dysfunction in Cancer. Int. J. Mol. Sci. 2020, 21, 5598. [Google Scholar] [CrossRef]
- Carmona-Carmona, C.A.; Pozza, E.D.; Ambrosini, G.; Cisterna, B.; Palmieri, M.; Decimo, I.; Cuezva, J.M.; Bottani, E.; Dando, I. Mitochondrial Elongation and OPA1 Play Crucial Roles during the Stemness Acquisition Process in Pancreatic Ductal Adenocarcinoma. Cancers 2022, 14, 3432. [Google Scholar] [CrossRef] [PubMed]
- Viscomi, C.; Bottani, E.; Civiletto, G.; Cerutti, R.; Moggio, M.; Fagiolari, G.; Schon, E.A.; Lamperti, C.; Zeviani, M. In Vivo Correction of COX Deficiency by Activation of the AMPK/PGC-1α Axis. Cell Metab. 2011, 14, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.K. The SHP-2 tyrosine phosphatase: Signaling mechanisms and biological functions. Cell Res. 2000, 10, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.; Park, K.-S. SHP2 is a downstream target of ZAP70 to regulate JAK1/STAT3 and ERK signaling pathways in mouse embryonic stem cells. FEBS Lett. 2010, 584, 4241–4246. [Google Scholar] [CrossRef] [PubMed]
- Annerén, C.; Cowan, C.A.; Melton, D.A. The Src Family of Tyrosine Kinases Is Important for Embryonic Stem Cell Self-renewal. J. Biol. Chem. 2004, 279, 31590–31598. [Google Scholar] [CrossRef] [PubMed]
- Xu, D. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008, 13, 4925–4932. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Nawa, H.; Neel, B.G. Tyrosyl Phosphorylation of Shp2 Is Required for Normal ERK Activation in Response to Some, but Not All, Growth Factors. J. Biol. Chem. 2003, 278, 41677–41684. [Google Scholar] [CrossRef]
- Chong, Z.Z.; Maiese, K. The Src homology 2 domain tyrosine phosphatases SHP-1 and SHP-2: Diversified control of cell growth, inflammation, and injury. Histol. Histopathol. 2007, 22, 1251–1267. [Google Scholar] [CrossRef]
- Saxton, T.M.; Henkemeyer, M.; Gasca, S.; Shen, R.; Rossi, D.J.; Shalaby, F.; Feng, G.-S.; Pawson, T. Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J. 1997, 16, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Klaman, L.D.; Chen, B.; Araki, T.; Harada, H.; Thomas, S.M.; George, E.L.; Neel, B.G. An Shp2/SFK/Ras/Erk Signaling Pathway Controls Trophoblast Stem Cell Survival. Dev. Cell 2006, 10, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Mohi, M.G.; Ismat, F.A.; Bronson, R.T.; Williams, I.R.; Kutok, J.L.; Yang, W.; Pao, L.I.; Gilliland, D.G.; Epstein, J.A.; et al. Mouse model of Noonan syndrome reveals cell type—And gene dosage—Dependent effects of Ptpn11 mutation. Nat. Med. 2004, 10, 849–857. [Google Scholar] [CrossRef]
- Mohi, M.G.; Williams, I.R.; Dearolf, C.R.; Chan, G.; Kutok, J.L.; Cohen, S.; Morgan, K.; Boulton, C.; Shigematsu, H.; Keilhack, H.; et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell 2005, 7, 179–191. [Google Scholar] [CrossRef]
- Chan, R.J.; Leedy, M.B.; Munugalavadla, V.; Voorhorst, C.S.; Li, Y.; Yu, M.; Kapur, R. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood 2005, 105, 3737–3742. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-M.; Daino, H.; Chen, J.; Bunting, K.D.; Qu, C.-K. Effects of a Leukemia-associated Gain-of-Function Mutation of SHP-2 Phosphatase on Interleukin-3 Signaling. J. Biol. Chem. 2006, 281, 5426–5434. [Google Scholar] [CrossRef] [PubMed]
- Keilhack, H.; David, F.S.; McGregor, M.; Cantley, L.; Neel, B.G. Diverse Biochemical Properties of Shp2 Mutants. J. Biol. Chem. 2005, 280, 30984–30993. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Brunati, A.M.; Bordin, L.; La Rocca, N.; Clari, G.; Toninello, A. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim. Biophys. Acta 2002, 1589, 181–195. [Google Scholar] [CrossRef]
- Augereau, O.; Claverol, S.; Boudes, N.; Basurko, M.-J.; Bonneu, M.; Rossignol, R.; Mazat, J.-P.; Letellier, T.; Dachary-Prigent, J. Identification of tyrosine-phosphorylated proteins of the mitochondrial oxidative phosphorylation machinery. Experientia 2005, 62, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Tajan, M.; Batut, A.; Cadoudal, T.; Deleruyelle, S.; Le Gonidec, S.; Laurent, C.S.; Vomscheid, M.; Wanecq, E.; Tréguer, K.; Serra-Nédélec, A.D.R.; et al. LEOPARD syndrome-associated SHP2 mutation confers leanness and protection from diet-induced obesity. Proc. Natl. Acad. Sci. USA 2014, 111, E4494–E4503. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, S.S.; Meng, Q.; Li, S.; Zhu, H.H.; Raquil, M.-A.; Alderson, N.; Zhang, H.; Wu, J.; Rui, L.; et al. Shp2 Controls Female Body Weight and Energy Balance by Integrating Leptin and Estrogen Signals. Mol. Cell. Biol. 2012, 32, 1867–1878. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krajewska, M.; Banares, S.; Zhang, E.E.; Huang, X.; Scadeng, M.; Jhala, U.S.; Feng, G.-S.; Krajewski, S. Development of Diabesity in Mice with Neuronal Deletion of Shp2 Tyrosine Phosphatase. Am. J. Pathol. 2008, 172, 1312–1324. [Google Scholar] [CrossRef]
- Zhang, E.E.; Chapeau, E.; Hagihara, K.; Feng, G.-S. Neuronal Shp2 tyrosine phosphatase controls energy balance and metabolism. Proc. Natl. Acad. Sci. USA 2004, 101, 16064–16069. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Han, D.; Meng, X.; Xu, M.; Zheng, C.; Xia, Q. Activating Mutation of SHP2 Establishes a Tumorigenic Phonotype Through Cell-Autonomous and Non-Cell-Autonomous Mechanisms. Front. Cell Dev. Biol. 2021, 9, 630712. [Google Scholar] [CrossRef] [PubMed]
- Wortmann, S.B.; Kluijtmans, L.A.; Engelke, U.F.H.; Wevers, R.A.; Morava, E. The 3-methylglutaconic acidurias: What’s new? J. Inherit. Metab. Dis. 2010, 35, 13–22. [Google Scholar] [CrossRef]
- Kleefstra, T.; Wortmann, S.B.; Rodenburg, R.J.T.; Bongers, E.M.H.F.; Hadzsiev, K.; Noordam, C.; Heuvel, L.P.V.D.; Nillesen, W.M.; Hollódy, K.; Gillessen-Kaesbach, G.; et al. Mitochondrial dysfunction and organic aciduria in five patients carrying mutations in the Ras-MAPK pathway. Eur. J. Hum. Genet. 2010, 19, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Aeby, A.; Sznajer, Y.; Cavé, H.; Rebuffat, E.; Van Coster, R.; Rigal, O.; Van Bogaert, P. Cardiofaciocutaneous (CFC) syndrome associated with muscular coenzyme Q10 deficiency. J. Inherit. Metab. Dis. 2007, 30, 827. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zheng, H.; Yu, W.-M.; Qu, C.-K. Activating Mutations in Protein Tyrosine Phosphatase Ptpn11 (Shp2) Enhance Reactive Oxygen Species Production That Contributes to Myeloproliferative Disorder. PLoS ONE 2013, 8, e63152. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, R.A.; O’Marcaigh, A.; Wardak, Z.; Zhang, Y.-Y.; Dranoff, G.; Jacks, T.; Clapp, D.; Shannon, K.M. Nf1 and Gmcsf Interact in Myeloid Leukemogenesis. Mol. Cell 2000, 5, 189–195. [Google Scholar] [CrossRef]
- Kadenbach, B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim. Biophys. Acta 2003, 1604, 77–94. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kim, E.-Y.; Ko, J.-J.; Lee, K.-A. Gas6 is a reciprocal regulator of mitophagy during mammalian oocyte maturation. Sci. Rep. 2019, 9, 10343. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kim, E.-Y.; Lee, S.-Y.; Ko, J.-J.; Lee, K.-A. Oocyte Cytoplasmic Gas6 and Heparan Sulfate (HS) are Required to Establish the Open Chromatin State in Nuclei During Remodeling and Reprogramming. Cell. Physiol. Biochem. 2017, 45, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Li, S.; Hsu, P.; Qu, C.-K. Induction of a Tumor-associated Activating Mutation in Protein Tyrosine Phosphatase Ptpn11 (Shp2) Enhances Mitochondrial Metabolism, Leading to Oxidative Stress and Senescence. J. Biol. Chem. 2013, 288, 25727–25738. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Nicholls, D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011, 435, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jin, C.; Hou, Y.; Zhang, L.; Li, S.; Zhang, L.; Wu, B.; Li, Q.; Xu, C.; Tian, Y.; et al. Overexpression of Shp-2 attenuates apoptosis in neonatal rat cardiac myocytes through the ERK pathway. Exp. Mol. Pathol. 2012, 93, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Matsuo, K.; Bettaieb, A.; Bakke, J.; Matsuo, I.; Graham, J.; Xi, Y.; Liu, S.; Tomilov, A.; Tomilova, N.; et al. Hepatic Src Homology Phosphatase 2 Regulates Energy Balance in Mice. Endocrinology 2012, 153, 3158–3169. [Google Scholar] [CrossRef][Green Version]
- Zhang, W.; Chan, R.J.; Chen, H.; Yang, Z.; He, Y.; Zhang, X.; Luo, Y.; Yin, F.; Moh, A.; Miller, L.C.; et al. Negative Regulation of Stat3 by Activating PTPN11 Mutants Contributes to the Pathogenesis of Noonan Syndrome and Juvenile Myelomonocytic Leukemia. J. Biol. Chem. 2009, 284, 22353–22363. [Google Scholar] [CrossRef]
- Bard-Chapeau, E.A.; Li, S.; Ding, J.; Zhang, S.S.; Zhu, H.H.; Princen, F.; Fang, D.D.; Han, T.; Bailly-Maitre, B.; Poli, V.; et al. Ptpn11/Shp2 Acts as a Tumor Suppressor in Hepatocellular Carcinogenesis. Cancer Cell 2011, 19, 629–639. [Google Scholar] [CrossRef]
- Wegrzyn, J.; Potla, R.; Chwae, Y.-J.; Sepuri, N.B.V.; Zhang, Q.; Koeck, T.; Derecka, M.; Szczepanek, K.; Szelag, M.; Gornicka, A.; et al. Function of Mitochondrial Stat3 in Cellular Respiration. Science 2009, 323, 793–797. [Google Scholar] [CrossRef]
- Mantel, C.; Messina-Graham, S.; Moh, A.; Cooper, S.; Hangoc, G.; Fu, X.-Y.; Broxmeyer, H.E. Mouse hematopoietic cell–targeted STAT3 deletion: Stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging–like phenotype. Blood 2012, 120, 2589–2599. [Google Scholar] [CrossRef]
- Princen, F.; Bard, E.; Sheikh, F.; Zhang, S.S.; Wang, J.; Zago, W.M.; Wu, D.; Trelles, R.D.; Bailly-Maitre, B.; Kahn, C.R.; et al. Deletion of Shp2 Tyrosine Phosphatase in Muscle Leads to Dilated Cardiomyopathy, Insulin Resistance, and Premature Death. Mol. Cell. Biol. 2009, 29, 378–388. [Google Scholar] [CrossRef]
- Bard-Chapeau, E.A.; Yuan, J.; Droin, N.; Long, S.; Zhang, E.; Nguyen, T.V.; Feng, G.-S. Concerted Functions of Gab1 and Shp2 in Liver Regeneration and Hepatoprotection. Mol. Cell. Biol. 2006, 26, 4664–4674. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT Signaling: From Interferons to Cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G. Moonlighting in Mitochondria. Science 2009, 323, 723–724. [Google Scholar] [CrossRef] [PubMed]
- Cirstea, I.C.; Gremer, L.; Dvorsky, R.; Zhang, S.-C.; Piekorz, R.P.; Zenker, M.; Ahmadian, M.R. Diverging gain-of-function mechanisms of two novel KRAS mutations associated with Noonan and cardio-facio-cutaneous syndromes. Hum. Mol. Genet. 2012, 22, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Razzaque, A.; Komoike, Y.; Nishizawa, T.; Inai, K.; Furutani, M.; Higashinakagawa, T.; Matsuoka, R. Characterization of a novel KRAS mutation identified in Noonan syndrome. Am. J. Med. Genet. Part A 2012, 158A, 524–532. [Google Scholar] [CrossRef]
- Hu, Y.; Lu, W.; Chen, G.; Wang, P.; Chen, Z.; Zhou, Y.; Ogasawara, M.; Trachootham, D.; Feng, L.; Pelicano, H.; et al. K-rasG12V transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2011, 22, 399–412. [Google Scholar] [CrossRef]
- Baracca, A.; Chiaradonna, F.; Sgarbi, G.; Solaini, G.; Alberghina, L.; Lenaz, G. Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim. Biophys. Acta BBA Bioenerg. 2010, 1797, 314–323. [Google Scholar] [CrossRef]
- Mazat, J.-P.; Devin, A.; Ransac, S. Modelling mitochondrial ROS production by the respiratory chain. Cell. Mol. Life Sci. 2019, 77, 455–465. [Google Scholar] [CrossRef]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef]
- Ying, H.; Kimmelman, A.C.; Lyssiotis, C.A.; Hua, S.; Chu, G.C.; Fletcher-Sananikone, E.; Locasale, J.W.; Son, J.; Zhang, H.; Coloff, J.L.; et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell 2012, 149, 656–670. [Google Scholar] [CrossRef]
- Kerr, E.M.; Gaude, E.; Turrell, F.K.; Frezza, C.; Martins, C.P. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nat. Cell Biol. 2016, 531, 110–113. [Google Scholar] [CrossRef]
- Gaglio, D.; Metallo, C.M.; Gameiro, P.A.; Hiller, K.; Danna, L.S.; Balestrieri, C.; Alberghina, L.; Stephanopoulos, G.; Chiaradonna, F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011, 7, 523. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Lyssiotis, C.A.; Ying, H.; Wang, X.; Hua, S.; Ligorio, M.; Perera, R.M.; Ferrone, C.R.; Mullarky, E.; Shyh-Chang, N.; et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013, 496, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, J.; Zheng, L.; Meissl, K.; Chaneton, B.; Selivanov, V.; Mackay, G.; Van Der Burg, S.H.; Verdegaal, E.M.E.; Cascante, M.; Shlomi, T.; et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 2013, 498, 109–112. [Google Scholar] [CrossRef] [PubMed]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef]
- Bryant, K.L.; Mancias, J.D.; Kimmelman, A.C.; Der, C.J. KRAS: Feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014, 39, 91–100. [Google Scholar] [CrossRef]
- Pierre, S.; Bats, A.-S.; Coumoul, X. Understanding SOS (Son of Sevenless). Biochem. Pharmacol. 2011, 82, 1049–1056. [Google Scholar] [CrossRef]
- Bandaru, P.; Kondo, Y.; Kuriyan, J. The Interdependent Activation of Son-of-Sevenless and Ras. Cold Spring Harb. Perspect. Med. 2018, 9, a031534. [Google Scholar] [CrossRef]
- Toma-Fukai, S.; Shimizu, T. Structural Insights into the Regulation Mechanism of Small GTPases by GEFs. Molecules 2019, 24, 3308. [Google Scholar] [CrossRef]
- Rojas, J.M.; Oliva, J.L.; Santos, E. Mammalian Son of Sevenless Guanine Nucleotide Exchange Factors: Old Concepts and New Perspectives. Genes Cancer 2011, 2, 298–305. [Google Scholar] [CrossRef]
- Cordeddu, V.; Yin, J.C.; Gunnarsson, C.; Virtanen, C.; Drunat, S.; Lepri, F.; De Luca, A.; Rossi, C.; Ciolfi, A.; Pugh, T.J.; et al. Activating Mutations Affecting the Dbl Homology Domain of SOS2 Cause Noonan Syndrome. Hum. Mutat. 2015, 36, 1080–1087. [Google Scholar] [CrossRef]
- Baltanás, F.C.; Pérez-Andrés, M.; Ginel-Picardo, A.; Diaz, D.; Jimeno, D.; Liceras-Boillos, P.; Kortum, R.L.; Samelson, L.E.; Orfao, A.; Santos, E. Functional Redundancy of Sos1 and Sos2 for Lymphopoiesis and Organismal Homeostasis and Survival. Mol. Cell. Biol. 2013, 33, 4562–4578. [Google Scholar] [CrossRef] [PubMed]
- Liceras-Boillos, P.; García-Navas, R.; Ginel-Picardo, A.; Anta, B.; Pérez-Andrés, M.; Lillo, C.; Gómez, C.; Jimeno, D.; Fernández-Medarde, A.; Baltanás, F.C.; et al. Sos1 disruption impairs cellular proliferation and viability through an increase in mitochondrial oxidative stress in primary MEFs. Oncogene 2016, 35, 6389–6402. [Google Scholar] [CrossRef]
- Peng, J.-Y.; Lin, C.-C.; Chen, Y.-J.; Kao, L.-S.; Liu, Y.-C.; Chou, C.-C.; Huang, Y.-H.; Chang, F.-R.; Wu, Y.-C.; Tsai, Y.-S.; et al. Automatic Morphological Subtyping Reveals New Roles of Caspases in Mitochondrial Dynamics. PLOS Comput. Biol. 2011, 7, e1002212. [Google Scholar] [CrossRef] [PubMed]
- Iershov, A.; Nemazanyy, I.; Alkhoury, C.; Girard, M.; Barth, E.; Cagnard, N.; Montagner, A.; Chretien, D.; Rugarli, E.I.; Guillou, H.; et al. The class 3 PI3K coordinates autophagy and mitochondrial lipid catabolism by controlling nuclear receptor PPARα. Nat. Commun. 2019, 10, 1566. [Google Scholar] [CrossRef] [PubMed]
- García-Navas, R.; Liceras-Boillos, P.; Gómez, C.; Baltanás, F.C.; Calzada, N.; Nuevo-Tapioles, C.; Cuezva, J.M.; Santos, E. Critical requirement of SOS1 RAS-GEF function for mitochondrial dynamics, metabolism, and redox homeostasis. Oncogene 2021, 40, 4538–4551. [Google Scholar] [CrossRef]
- Ong, S.-B.; Kalkhoran, S.B.; Cabrera-Fuentes, H.A.; Hausenloy, D.J. Mitochondrial fusion and fission proteins as novel therapeutic targets for treating cardiovascular disease. Eur. J. Pharmacol. 2015, 763, 104–114. [Google Scholar] [CrossRef]
- Schrepfer, E.; Scorrano, L. Mitofusins, from Mitochondria to Metabolism. Mol. Cell 2016, 61, 683–694. [Google Scholar] [CrossRef]
- Serasinghe, M.N.; Chipuk, J.E. Mitochondrial Fission in Human Diseases. Antibiotics 2016, 240, 159–188. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, T.-E.; Chen, M.; Xu, D.; Zhu, Y.; Hu, B.-Y.; Lin, Z.-F.; Pan, J.-J.; Wang, X.; Wu, C.; et al. MFN1-dependent alteration of mitochondrial dynamics drives hepatocellular carcinoma metastasis by glucose metabolic reprogramming. Br. J. Cancer 2019, 122, 209–220. [Google Scholar] [CrossRef]
- Buday, L.; Downward, J. Many faces of Ras activation. Biochim. Biophys. Acta 2008, 1786, 178–187. [Google Scholar] [CrossRef]
- Seo, B.J.; Choi, J.; La, H.; Habib, O.; Choi, Y.; Hong, K.; Do, J.T. Role of mitochondrial fission-related genes in mitochondrial morphology and energy metabolism in mouse embryonic stem cells. Redox Biol. 2020, 36, 101599. [Google Scholar] [CrossRef]
- Babu, D.; Leclercq, G.; Motterlini, R.; Lefebvre, R.A. Differential Effects of CORM-2 and CORM-401 in Murine Intestinal Epithelial MODE-K Cells under Oxidative Stress. Front. Pharmacol. 2017, 8, 31. [Google Scholar] [CrossRef]
- Maus, M.; Cuk, M.; Patel, B.; Lian, J.; Ouimet, M.; Kaufmann, U.; Yang, J.; Horvath, R.; Hornig-Do, H.-T.; Chrzanowska-Lightowlers, Z.; et al. Store-Operated Ca2+ Entry Controls Induction of Lipolysis and the Transcriptional Reprogramming to Lipid Metabolism. Cell Metab. 2017, 25, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Gu, J.; Zong, S.; Wu, M.; Yang, M. Structure and mechanism of mitochondrial electron transport chain. Biomed. J. 2018, 41, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.O.; Dieteren, C.E.J.; Heuvel, L.P.W.J.V.D.; Willems, P.H.; Smeitink, J.A.M.; Koopman, W.J.H.; Nijtmans, L.G.J. Identification of Mitochondrial Complex I Assembly Intermediates by Tracing Tagged NDUFS3 Demonstrates the Entry Point of Mitochondrial Subunits. J. Biol. Chem. 2007, 282, 7582–7590. [Google Scholar] [CrossRef]
- Wang, D.-W.; Su, F.; Zhang, T.; Yang, T.-C.; Wang, H.-Q.; Yang, L.-J.; Zhou, F.-F.; Feng, M.-H. The miR-370/UQCRC2 axis facilitates tumorigenesis by regulating epithelial-mesenchymal transition in Gastric Cancer. J. Cancer 2020, 11, 5042–5055. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Wang, X.; Auwerx, J. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE). Curr. Protoc. Mouse Biol. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Baker, N.; Patel, J.; Khacho, M. Linking mitochondrial dynamics, cristae remodeling and supercomplex formation: How mitochondrial structure can regulate bioenergetics. Mitochondrion 2019, 49, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Arismendi-Morillo, G.; Mukherjee, P.; Chinopoulos, C. On the Origin of ATP Synthesis in Cancer. iScience 2020, 23, 101761. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Nagdas, S.; Kashatus, J.A.; Nascimento, A.; Hussain, S.S.; Trainor, R.E.; Pollock, S.; Adair, S.J.; Michaels, A.D.; Sesaki, H.; Stelow, E.B.; et al. Drp1 Promotes KRas-Driven Metabolic Changes to Drive Pancreatic Tumor Growth. Cell Rep. 2019, 28, 1845–1859.e5. [Google Scholar] [CrossRef]
- Ren, Y.; Shen, H.-M. Critical role of AMPK in redox regulation under glucose starvation. Redox Biol. 2019, 25, 101154. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Baskfield, A.; Lin, Y.; Beers, J.; Zou, J.; Liu, C.; Jaffré, F.; Roberts, A.E.; Ottinger, E.A.; Kontaridis, M.I.; et al. Generation of an induced pluripotent stem cell line (TRNDi003-A) from a Noonan syndrome with multiple lentigines (NSML) patient carrying a p.Q510P mutation in the PTPN11 gene. Stem Cell Res. 2018, 34, 101374. [Google Scholar] [CrossRef] [PubMed]
- Tekendo-Ngongang, C.; Agenbag, G.; Bope, C.D.; Esterhuizen, A.I.; Wonkam, A. Noonan Syndrome in South Africa: Clinical and Molecular Profiles. Front. Genet. 2019, 10, 333. [Google Scholar] [CrossRef]
- Mohapatra, B.; Ahmad, G.; Nadeau, S.; Zutshi, N.; An, W.; Scheffe, S.; Dong, L.; Feng, D.; Goetz, B.; Arya, P.; et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of Cbl-family ubiquitin ligases. Biochim. Biophys. Acta 2012, 1833, 122–139. [Google Scholar] [CrossRef]
- Chung, I.-C.; Yuan, S.-N.; Ouyang, C.-N.; Lin, H.-C.; Huang, K.-Y.; Chen, Y.-J.; Chung, A.-K.; Chu, C.-L.; Ojcius, D.M.; Chang, Y.-S.; et al. Src-family kinase-Cbl axis negatively regulates NLRP3 inflammasome activation. Cell Death Dis. 2018, 9, 1109. [Google Scholar] [CrossRef]
- Molero, J.C.; Jensen, T.E.; Withers, P.C.; Couzens, M.; Herzog, H.; Thien, C.B.; Langdon, W.Y.; Walder, K.; Murphy, M.A.; Bowtell, D.D.; et al. c-Cbl–deficient mice have reduced adiposity, higher energy expenditure, and improved peripheral insulin action. J. Clin. Investig. 2004, 114, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-C.; Chen, Y.-J.; Wei, Y.-H.; Chuang, Y.-T.; Hsieh, S.-H.; Hsieh, J.-Y.; Hsieh, Y.-L.; Ojcius, D.M.; Huang, K.-Y.; Chung, I.-C.; et al. Cbl Negatively Regulates NLRP3 Inflammasome Activation through GLUT1-Dependent Glycolysis Inhibition. Int. J. Mol. Sci. 2020, 21, 5104. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef]
- Muñoz-Planillo, R.; Kuffa, P.; Martínez-Colón, G.; Smith, B.L.; Rajendiran, T.M.; Núñez, G. K+ Efflux Is the Common Trigger of NLRP3 Inflammasome Activation by Bacterial Toxins and Particulate Matter. Immunity 2013, 38, 1142–1153. [Google Scholar] [CrossRef]
- Anand, P.K.; Malireddi, R.K.; Kanneganti, T.-D. Role of the Nlrp3 Inflammasome in Microbial Infection. Front. Microbiol. 2011, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Freemerman, A.J.; Johnson, A.R.; Sacks, G.N.; Milner, J.J.; Kirk, E.L.; Troester, M.A.; Macintyre, A.N.; Goraksha-Hicks, P.; Rathmell, J.C.; Makowski, L. Metabolic reprogramming of macrophages: Glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 2014, 289, 7884–7896. [Google Scholar] [CrossRef]
- Alcover-Sanchez, B.; Garcia-Martin, G.; Escudero-Ramirez, J.; Gonzalez-Riano, C.; Lorenzo, P.; Gimenez-Cassina, A.; Formentini, L.; de la Villa-Polo, P.; Pereira, M.P.; Wandosell, F.; et al. Absence of R-Ras1 and R-Ras2 causes mitochondrial alterations that trigger axonal degeneration in a hypomyelinating disease model. Glia 2020, 69, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Galmiche, A.; Fueller, J.; Santel, A.; Krohne, G.; Wittig, I.; Doye, A.; Rolando, M.; Flatau, G.; Lemichez, E.; Rapp, U.R. Isoform-specific Interaction of C-RAF with Mitochondria. J. Biol. Chem. 2008, 283, 14857–14866. [Google Scholar] [CrossRef] [PubMed]
- Szabadkai, G.; Simoni, A.; Bianchi, K.; De Stefani, D.; Leo, S.; Wieckowski, M.; Rizzuto, R. Mitochondrial dynamics and Ca2+ signaling. Biochim. Biophys. Acta 2006, 1763, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Campello, S.; Lacalle, R.A.; Bettella, M.; Manñes, S.; Scorrano, L.; Viola, A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 2006, 203, 2879–2886. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-G.; Rapp, U.R.; Reed, J.C. Bcl-2 Targets the Protein Kinase Raf-1 to Mitochondria. Cell 1996, 87, 629–638. [Google Scholar] [CrossRef]
- Chinton, J.; Huckstadt, V.; Moresco, A.; Gravina, L.P.; Obregon, M.G. Clinical and molecular characterization of children with Noonan syndrome and other RASopathies in Argentina. Arch. Argent. de Pediatr. 2019, 117, 330–337. [Google Scholar] [CrossRef]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Carr, H.S.; Dan, I.; Ruvolo, P.P.; Frost, J.A. p21 activated kinase 5 activates Raf-1 and targets it to mitochondria. J. Cell. Biochem. 2008, 105, 167–175. [Google Scholar] [CrossRef]
- Kim, S.Y. RKIP Downregulation Induces the HBx-Mediated Raf-1 Mitochondrial Translocation. J. Microbiol. Biotechnol. 2011, 21, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Alavi, A.; Hood, J.D.; Frausto, R.; Stupack, D.G.; Cheresh, D.A. Role of Raf in Vascular Protection from Distinct Apoptotic Stimuli. Science 2003, 301, 94–96. [Google Scholar] [CrossRef]
- O’Neill, E.; Rushworth, L.; Baccarini, M.; Kolch, W. Role of the Kinase MST2 in Suppression of Apoptosis by the Proto-Oncogene Product Raf-1. Science 2004, 306, 2267–2270. [Google Scholar] [CrossRef] [PubMed]
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; Frederick, D.T.; Hurley, A.D.; Nellore, A.; Kung, A.L.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 2013, 23, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Zebisch, A.; Troppmair, J. Back to the roots: The remarkable RAF oncogene story. Experientia 2006, 63, 1314–1330. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-T.; Chuang, M.-J.; Tang, S.-H.; Wu, S.-T.; Chen, Y.-C.; Sun, G.-H.; Hsiao, P.-W.; Huang, S.-M.; Lee, H.-J.; Yu, C.-P.; et al. Novel Cancer Therapeutics with Allosteric Modulation of the Mitochondrial C-Raf–DAPK Complex by Raf Inhibitor Combination Therapy. Cancer Res. 2015, 75, 3568–3582. [Google Scholar] [CrossRef] [PubMed]
- Samovski, D.; Kalderon, B.; Yehuda-Shnaidman, E.; Bar-Tana, J. Gating of the Mitochondrial Permeability Transition Pore by Long Chain Fatty Acyl Analogs in Vivo. J. Biol. Chem. 2010, 285, 6879–6890. [Google Scholar] [CrossRef] [PubMed]
- Wellbrock, C.; Karasarides, M.; Marais, R. The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 2004, 5, 875–885. [Google Scholar] [CrossRef]
- Koshkin, V.; Dai, F.F.; Robson-Doucette, C.A.; Chan, C.; Wheeler, M. Limited Mitochondrial Permeabilization Is an Early Manifestation of Palmitate-induced Lipotoxicity in Pancreatic β-Cells. J. Biol. Chem. 2008, 283, 7936–7948. [Google Scholar] [CrossRef]
- Grefte, S.; Wagenaars, J.A.; Jansen, R.; Willems, P.H.; Koopman, W.J. Rotenone inhibits primary murine myotube formation via Raf-1 and ROCK2. Biochim. Biophys. Acta 2015, 1853, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Chávez, M.D.; Tse, H.M. Targeting Mitochondrial-Derived Reactive Oxygen Species in T Cell-Mediated Autoimmune Diseases. Front. Immunol. 2021, 12, 703972. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Li, S.; Jiang, N.; Shao, X.; Zhang, M.; Jin, H.; Zhang, Z.; Shen, J.; Zhou, Y.; Zhou, W.; et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019, 26, 101254. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kim, H.Y.; Lee, Y.-K.; Yoon, Y.-S.; Xu, W.G.; Yoon, J.-K.; Choi, S.-E.; Ko, Y.-G.; Kim, M.-J.; Lee, S.-J.; et al. Involvement of mitophagy in oncogenic K-Ras-induced transformation. Autophagy 2011, 7, 1187–1198. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Brooks, A.; Burns, A.; Burkitt-Wright, E.; Kerr, B.; Huson, S.; Emsley, R.; Green, J. Autism spectrum disorder and other neurobehavioural comorbidities in rare disorders of the Ras/MAPK pathway. Dev. Med. Child Neurol. 2017, 59, 544–549. [Google Scholar] [CrossRef]
- Levy, A.; Xiao, X.; Shaw, J.; Devi, S.P.S.; Katrancha, S.M.; Bennett, A.M.; Greer, C.A.; Howe, J.R.; Machida, K.; Koleske, A.J. Noonan Syndrome-Associated SHP2 Dephosphorylates GluN2B to Regulate NMDA Receptor Function. Cell Rep. 2018, 24, 1523–1535. [Google Scholar] [CrossRef]
- Schofield, J.; Schafer, Z.T. Mitochondrial Reactive Oxygen Species and Mitophagy: A Complex and Nuanced Relationship. Antioxid. Redox Signal. 2021, 34, 517–530. [Google Scholar] [CrossRef]
- Kontaridis, M.I.; Chennappan, S. Mitochondria and the future of RASopathies: The emergence of bioenergetics. J. Clin. Investig. 2022, 132, 1–5. [Google Scholar] [CrossRef]


| Disease | Gene | Location | Amino Acid Change | Mutation Rate (%) | Protein Name | Protein Class | REF |
|---|---|---|---|---|---|---|---|
| NS | PTPN11 | 12q24 | p.D61G, p.G60A | 50–60 | SHP2: Protein tyrosine phosphatase non-receptor type 11; Src Homology 2 | Phosphatase | [7,19] |
| SOS1 | 2p22-p21 | p.A1654G | 10–13 | Son of sevenless (SOS) homolog 1 | RasGEF | [20] | |
| RAF1 | 3p25.2 | p.R89L | 5–10 | v-Raf-1 murine leukemia viral oncogene homolog 1 | Kinase | [20] | |
| KRAS | 12p12.1 | p.G12X (X = any amino acid) c.A458T p.D153V p.Y71D | 10 | V-Ki-Ras2 Kirsten rat sarcoma viral oncogene homolog | GTPase | [20,21,22] | |
| NRAS | 1p13.2 | p. G12X p.Q61X | <1 | Neuroblastoma RAS viral (v-RAS) oncogene homolog | GTPase | [20,23,24] | |
| SHOC2 | 10q25.2 | p.S2G | 12 | soc-2 suppressor of clear homolog | Scaffolding | [20] | |
| CBL | 11q23.3 | p.Y371X p.C404R p.W408R p.G415V p.L380P p.C840W | <1 | Casitas B-lineage lymphoma | Ubiquitin Ligase | [20,25,26] | |
| BRAF | 7q34 | p.V600E | <1 | Serine/Threonine-Protein Kinase B-Raf | Kinase | [24] | |
| A2ML1 | 12p13.31 | p.R802H, p.R592L, p.R802L | <1 | α-2-macroglobulin-like 1 | Protease inhibitor | [27] | |
| SOS 2 | 2p22.1 | p.T376SL | <1 | Son of sevenless homolog 2 | RasGEF | [28] | |
| LZTR1 | 22q11.21 | p.R284C, p.H287Y, p.Y119C, p.G248R, p.S247 N | <1 | Leucine Zipper-Like Transcription Regulator 1 | Adaptor protein | [20,28] | |
| RASA2 | 3q23 | p.Y326C, p.Y326N, p.R511C | <1 | Ras P21 Protein Activator 2 | RasGAP | [29] | |
| RRAS | 19q13.33 | p.G39dup, p.V55M | <1 | Related RAS Viral (R-Ras) Oncogene Homolog | GTPase | [30,31] | |
| RIT1 | 1q22 | p.S35 T, p.A57G, p.E81G, p.F82V, p.F82L, p.T83P, p.Y89H, p.M90I, p.G95A | 7 | 10 Ras-Like Without CAAX 1 | GTPase | [20,32] | |
| NSML | PTPN11 | 12q24 | p.Q510P | 90 | SHP2: Protein tyrosine phosphatase non-receptor type 11; Src Homology 2 | Phosphatase | [20,33,34] |
| RAF1 | 3p25.2 | p.V263G p.S257 L | 5 | v-Raf-1 murine leukemia viral oncogene homolog 1 | Kinase | [24,35] | |
| BRAF | 7q34 | <1 | Serine/Threonine-Protein Kinase B-Raf | Kinase | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajia, D.; Bottani, E.; Derwich, K. Effects of Noonan Syndrome-Germline Mutations on Mitochondria and Energy Metabolism. Cells 2022, 11, 3099. https://doi.org/10.3390/cells11193099
Bajia D, Bottani E, Derwich K. Effects of Noonan Syndrome-Germline Mutations on Mitochondria and Energy Metabolism. Cells. 2022; 11(19):3099. https://doi.org/10.3390/cells11193099
Chicago/Turabian StyleBajia, Donald, Emanuela Bottani, and Katarzyna Derwich. 2022. "Effects of Noonan Syndrome-Germline Mutations on Mitochondria and Energy Metabolism" Cells 11, no. 19: 3099. https://doi.org/10.3390/cells11193099
APA StyleBajia, D., Bottani, E., & Derwich, K. (2022). Effects of Noonan Syndrome-Germline Mutations on Mitochondria and Energy Metabolism. Cells, 11(19), 3099. https://doi.org/10.3390/cells11193099







