Nucleolus and Nucleolar Stress: From Cell Fate Decision to Disease Development
Abstract
1. Introduction
2. Dynamic Regulation of Nucleolar Architecture
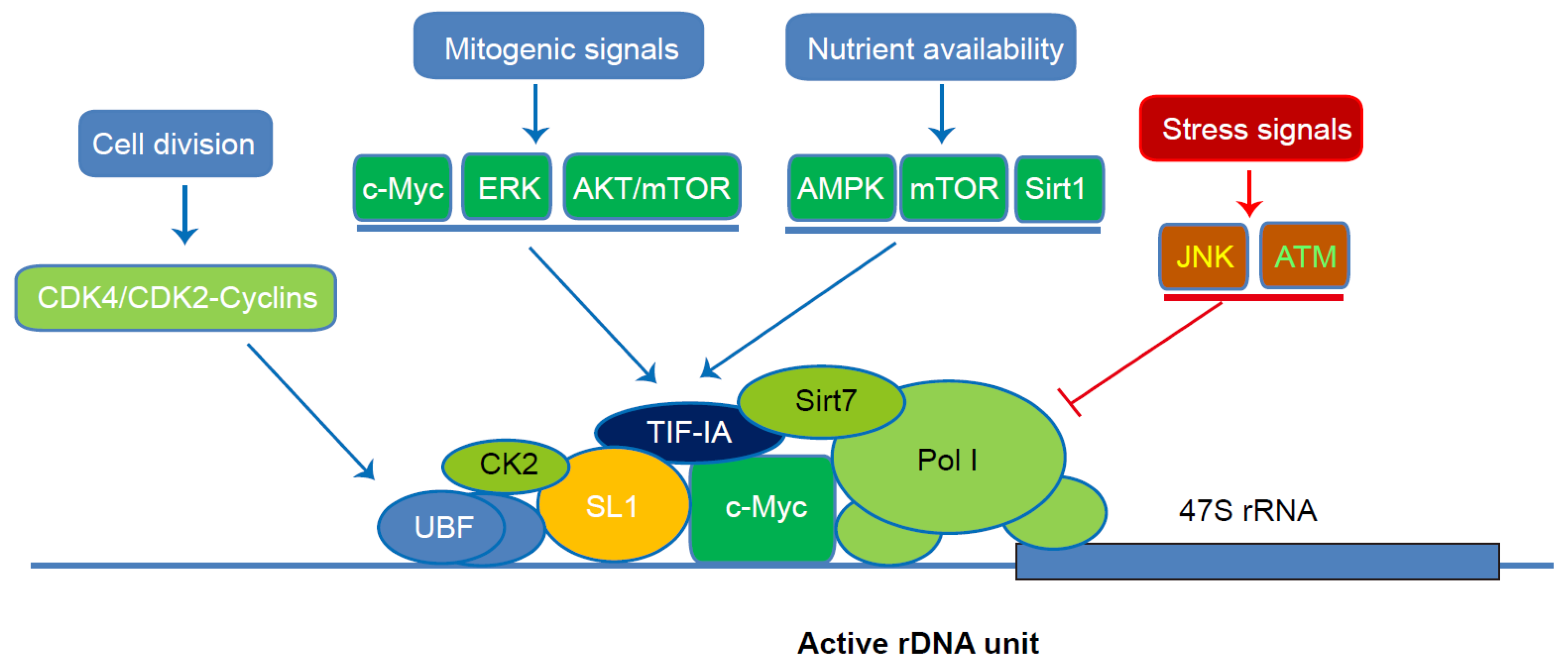
3. Coordinate Regulation of Pol I-Mediated rRNA Transcription
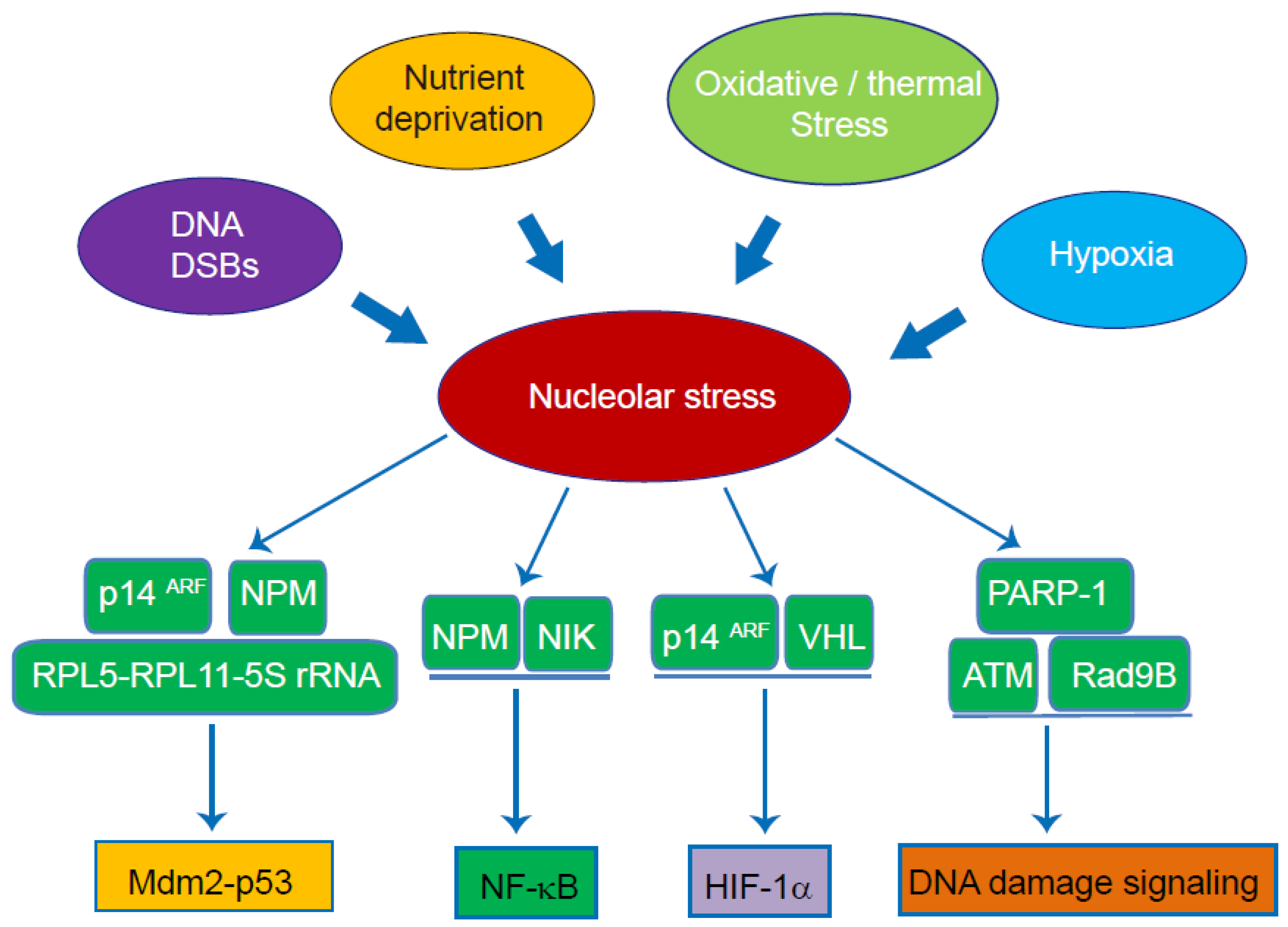
4. The Role of the Nucleolus as a Signal Hub and Stress Sensor
4.1. Mdm2-p53 Pathway
4.2. NF-κB Pathway
4.3. HIF-1α
4.4. DNA Damage-Induced Stress Signalling
4.5. Others
5. Nucleolus and Cell-Fate Decision
5.1. Cell Division
5.2. Stemness and Differentiation
5.3. Apoptosis
5.4. Senescence
5.5. Autophagy
6. Nucleolus, Nucleolar Stress and Disease Development
6.1. Cancer Development and Therapy
6.2. Neurodegenerative Diseases
6.3. Metabolic Diseases
6.4. Aging and Longevity
7. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Ohta, T.; Minoshima, S.; Kudoh, J.; Wang, Y.; de Jong, P.J.; Shimizu, N. Human ribosomal RNA gene cluster: Identification of the proximal end containing a novel tandem repeat sequence. Genomics 1995, 26, 521–526. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Puvion-Dutilleul, F.; Puvion, E.; Bachellerie, J.P. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5′ETS leader probe. Chromosoma 1997, 105, 496–505. [Google Scholar] [CrossRef]
- Schmidt, E.V. The role of c-myc in cellular growth control. Oncogene 1999, 18, 2988–2996. [Google Scholar] [CrossRef]
- Mahoney, S.J.; Dempsey, J.M.; Blenis, J. Cell signaling in protein synthesis ribosome biogenesis and translation initiation and elongation. Prog. Mol. Biol. Transl. Sci. 2009, 90, 53–107. [Google Scholar] [CrossRef]
- Németh, A.; Grummt, I. Dynamic regulation of nucleolar architecture. Curr. Opin. Cell Biol. 2018, 52, 105–111. [Google Scholar] [CrossRef]
- Derenzini, M.; Trere, D.; Pession, A.; Montanaro, L.; Sirri, V.; Ochs, R.L. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998, 152, 1291–1297. [Google Scholar]
- Pich, A.; Chiusa, L.; Margaria, E. Prognostic relevance of AgNORs in tumor pathology. Micron 2000, 31, 133–141. [Google Scholar] [CrossRef]
- Piffko, J.; Bankfalvi, A.; Ofner, D.; Bryne, M.; Rasch, D.; Joos, U.; Bocker, W.; Schmid, K.W. Prognostic value of histobiological factors (malignancy grading and AgNOR content) assessed at the invasive tumour front of oral squamous cell carcinomas. Br. J. Cancer 1997, 75, 1543–1546. [Google Scholar] [CrossRef]
- Bahadori, M.; Azizi, M.H.; Dabiri, S. Recent Advances on Nucleolar Functions in Health and Disease. Arch. Iran. Med. 2018, 21, 600–607. [Google Scholar]
- Weeks, S.E.; Metge, B.J.; Samant, R.S. The nucleolus: A central response hub for the stressors that drive cancer progression. Cell Mol. Life Sci. 2019, 76, 4511–4524. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Cellular stress and nucleolar function. Cell Cycle 2005, 4, 1036–1038. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Biggiogera, M.; Martin, T.E.; Gordon, J.; Amalric, F.; Fakan, S. Physiologically inactive nucleoli contain nucleoplasmic ribonucleoproteins: Immunoelectron microscopy of mouse spermatids and early embryos. Exp. Cell Res. 1994, 213, 55–63. [Google Scholar] [CrossRef]
- Engel, W.; Zenzes, M.T.; Schmid, M. Activation of mouse ribosomal RNA genes at the 2-cell stage. Hum. Genet. 1977, 38, 57–63. [Google Scholar] [CrossRef]
- Musinova, Y.R.; Kananykhina, E.Y.; Potashnikova, D.M.; Lisitsyna, O.M.; Sheval, E.V. A charge-dependent mechanism is responsible for the dynamic accumulation of proteins inside nucleoli. Biochim. Biophys. Acta 2015, 1853, 101–110. [Google Scholar] [CrossRef]
- Falahati, H.; Pelham-Webb, B.; Blythe, S.; Wieschaus, E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr. Biol. 2016, 26, 277–285. [Google Scholar] [CrossRef]
- Riback, J.A.; Zhu, L.; Ferrolino, M.C.; Tolbert, M.; Mitrea, D.M.; Sanders, D.W.; Wei, M.T.; Kriwacki, R.W.; Brangwynne, C.P. Composition-dependent thermodynamics of intracellular phase separation. Nature 2020, 581, 209–214. [Google Scholar] [CrossRef]
- Correction to Supporting Information for Falahati and Wieschaus, Independent active and thermodynamic processes govern the nucleolus assembly in vivo. Proc. Natl. Acad. Sci. USA 2017, 114, E3585. [CrossRef]
- Maggi, L.B., Jr.; Kuchenruether, M.; Dadey, D.Y.; Schwope, R.M.; Grisendi, S.; Townsend, R.R.; Pandolfi, P.P.; Weber, J.D. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol. Cell Biol. 2008, 28, 7050–7065. [Google Scholar] [CrossRef]
- Ni, C.; Buszczak, M. The homeostatic regulation of ribosome biogenesis. Semin. Cell Dev. Biol. 2022, in press. [Google Scholar] [CrossRef]
- Hempel, W.M.; Cavanaugh, A.H.; Hannan, R.D.; Taylor, L.; Rothblum, L.I. The species-specific RNA polymerase I transcription factor SL-1 binds to upstream binding factor. Mol. Cell. Biol. 1996, 16, 557–563. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanij, E.; Hannan, R.D. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 2009, 4, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, S.J.; Zomerdijk, J.C. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell. Biochem. 2013, 61, 211–236. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Zhao, J.; Yuan, X.; Grummt, I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004, 18, 423–434. [Google Scholar] [CrossRef]
- Fath, S.; Milkereit, P.; Peyroche, G.; Riva, M.; Carles, C.; Tschochner, H. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. USA 2001, 98, 14334–14339. [Google Scholar] [CrossRef]
- Filipowicz, W.; Pelczar, P.; Pogacic, V.; Dragon, F. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 1999, 46, 377–389. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Komatsu, W.; Hayano, T.; Miura, Y.; Homma, K.; Izumikawa, K.; Ishikawa, H.; Miyazawa, N.; Tachikawa, H.; Yamauchi, Y.; et al. Splicing factor 2-associated protein p32 participates in ribosome biogenesis by regulating the binding of Nop52 and fibrillarin to preribosome particles. Mol. Cell. Proteom. 2011, 10, M110.006148. [Google Scholar] [CrossRef]
- Rodgers, M.L.; Woodson, S.A. A roadmap for rRNA folding and assembly during transcription. Trends Biochem. Sci. 2021, 46, 889–901. [Google Scholar] [CrossRef]
- Zhang, Q.; Shalaby, N.A.; Buszczak, M. Changes in rRNA Transcription Influence Proliferation and Cell Fate Within a Stem Cell Lineage. Science 2014, 343, 298–301. [Google Scholar] [CrossRef]
- Larson, D.E.; Xie, W.; Glibetic, M.; O’Mahony, D.; Sells, B.H.; Rothblum, L.I. Coordinated decreases in rRNA gene transcription factors and rRNA synthesis during muscle cell differentiation. Proc. Natl. Acad. Sci. USA 1993, 90, 7933–7936. [Google Scholar] [CrossRef]
- Lempiainen, H.; Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009, 21, 855–863. [Google Scholar] [CrossRef]
- Brown, I.N.; Lafita-Navarro, M.C.; Conacci-Sorrell, M. Regulation of Nucleolar Activity by MYC. Cells 2022, 11, 574. [Google Scholar] [CrossRef]
- Arabi, A.; Wu, S.; Ridderstrale, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlen, S.; Hydbring, P.; Soderberg, O.; Grummt, I.; et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef]
- Stefanovsky, V.; Langlois, F.; Gagnon-Kugler, T.; Rothblum, L.I.; Moss, T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 2006, 21, 629–639. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, X.; Frodin, M.; Grummt, I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 2003, 11, 405–413. [Google Scholar] [CrossRef]
- Stefanovsky, V.Y.; Langlois, F.; Bazett-Jones, D.; Pelletier, G.; Moss, T. ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry 2006, 45, 3626–3634. [Google Scholar] [CrossRef]
- Nguyen, L.X.T.; Mitchell, B.S. Akt activation enhances ribosomal RNA synthesis through casein kinase II and TIF-IA. Proc. Natl. Acad. Sci. USA 2013, 110, 20681–20686. [Google Scholar] [CrossRef]
- Wu, C.; You, J.; Fu, J.; Wang, X.; Zhang, Y. Phosphatidylinositol 3-Kinase/Akt Mediates Integrin Signaling To Control RNA Polymerase I Transcriptional Activity. Mol. Cell Biol. 2016, 36, 1555–1568. [Google Scholar] [CrossRef]
- Voit, R.; Hoffmann, M.; Grummt, I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999, 18, 1891–1899. [Google Scholar] [CrossRef]
- Hoppe, S.; Bierhoff, H.; Cado, I.; Weber, A.; Tiebe, M.; Grummt, I.; Voit, R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. USA 2009, 106, 17781–17786. [Google Scholar] [CrossRef] [PubMed]
- James, M.J.; Zomerdijk, J.C. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J. Biol. Chem. 2004, 279, 8911–8918. [Google Scholar] [CrossRef] [PubMed]
- Muth, V.; Nadaud, S.; Grummt, I.; Voit, R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001, 20, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, G.; Stefanovsky, V.Y.; Faubladier, M.; Hirschler-Laszkiewicz, I.; Savard, J.; Rothblum, L.I.; Cote, J.; Moss, T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell 2000, 6, 1059–1066. [Google Scholar] [CrossRef]
- Houston, R.; Sekine, S.; Calderon, M.J.; Seifuddin, F.; Wang, G.; Kawagishi, H.; Malide, D.A.; Li, Y.; Gucek, M.; Pirooznia, M.; et al. Acetylation-mediated remodeling of the nucleolus regulates cellular acetyl-CoA responses. PLoS Biol. 2020, 18, e3000981. [Google Scholar] [CrossRef]
- Murayama, A.; Ohmori, K.; Fujimura, A.; Minami, H.; Yasuzawa-Tanaka, K.; Kuroda, T.; Oie, S.; Daitoku, H.; Okuwaki, M.; Nagata, K.; et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 2008, 133, 627–639. [Google Scholar] [CrossRef]
- Kruhlak, M.; Crouch, E.E.; Orlov, M.; Montano, C.; Gorski, S.A.; Nussenzweig, A.; Misteli, T.; Phair, R.D.; Casellas, R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 2007, 447, 730–734. [Google Scholar] [CrossRef]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef]
- Brombin, A.; Joly, J.S.; Jamen, F. New tricks for an old dog: Ribosome biogenesis contributes to stem cell homeostasis. Curr. Opin. Genet. Dev. 2015, 34, 61–70. [Google Scholar] [CrossRef]
- Meng, L.; Zhu, Q.; Tsai, R.Y. Nucleolar trafficking of nucleostemin family proteins: Common versus protein-specific mechanisms. Mol. Cell Biol. 2007, 27, 8670–8682. [Google Scholar] [CrossRef]
- Colombo, E.; Marine, J.C.; Danovi, D.; Falini, B.; Pelicci, P.G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002, 4, 529–533. [Google Scholar] [CrossRef]
- Korsholm, L.M.; Gal, Z.; Nieto, B.; Quevedo, O.; Boukoura, S.; Lund, C.C.; Larsen, D.H. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2020, 48, 9449–9461. [Google Scholar] [CrossRef]
- Gorner, W.; Durchschlag, E.; Wolf, J.; Brown, E.L.; Ammerer, G.; Ruis, H.; Schuller, C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002, 21, 135–144. [Google Scholar] [CrossRef]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef]
- Lindstrom, M.S.; Bartek, J.; Maya-Mendoza, A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022, 29, 972–982. [Google Scholar] [CrossRef]
- Shadfan, M.; Lopez-Pajares, V.; Yuan, Z.M. MDM2 and MDMX: Alone and together in regulation of p53. Transl. Cancer Res. 2012, 1, 88–89. [Google Scholar]
- Rubbi, C.P.; Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef]
- Yuan, X.; Zhou, Y.; Casanova, E.; Chai, M.; Kiss, E.; Grone, H.J.; Schutz, G.; Grummt, I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell 2005, 19, 77–87. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Chen, L.; Chen, J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006, 25, 5614–5625. [Google Scholar] [CrossRef]
- Daftuar, L.; Zhu, Y.; Jacq, X.; Prives, C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE 2013, 8, e68667. [Google Scholar] [CrossRef]
- Sloan, K.E.; Bohnsack, M.T.; Watkins, N.J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013, 5, 237–247. [Google Scholar] [CrossRef]
- Donati, G.; Peddigari, S.; Mercer, C.A.; Thomas, G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013, 4, 87–98. [Google Scholar] [CrossRef]
- Llanos, S.; Clark, P.A.; Rowe, J.; Peters, G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 2001, 3, 445–452. [Google Scholar] [CrossRef]
- Lee, C.; Smith, B.A.; Bandyopadhyay, K.; Gjerset, R.A. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 2005, 65, 9834–9842. [Google Scholar] [CrossRef] [PubMed]
- Kurki, S.; Peltonen, K.; Latonen, L.; Kiviharju, T.M.; Ojala, P.M.; Meek, D.; Laiho, M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 2004, 5, 465–475. [Google Scholar] [CrossRef]
- Chen, J.; Stark, L.A. Crosstalk between NF-kappaB and Nucleoli in the Regulation of Cellular Homeostasis. Cells 2018, 7, 157. [Google Scholar] [CrossRef] [PubMed]
- Verma, I.M. Nuclear factor (NF)-kappaB proteins: Therapeutic targets. Ann. Rheum. Dis. 2004, 63 (Suppl. 2), ii57–ii61. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.K.; Lynn, B.C.; Daosukho, C.; St Clair, D.K. Identification of nucleophosmin as an NF-kappaB co-activator for the induction of the human SOD2 gene. J. Biol. Chem. 2004, 279, 28209–28219. [Google Scholar] [CrossRef]
- Birbach, A.; Bailey, S.T.; Ghosh, S.; Schmid, J.A. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-kappaB inducing kinase NIK. J. Cell Sci. 2004, 117, 3615–3624. [Google Scholar] [CrossRef]
- Stark, L.A.; Dunlop, M.G. Nucleolar sequestration of RelA (p65) regulates NF-kappaB-driven transcription and apoptosis. Mol. Cell Biol. 2005, 25, 5985–6004. [Google Scholar] [CrossRef]
- Thoms, H.C.; Loveridge, C.J.; Simpson, J.; Clipson, A.; Reinhardt, K.; Dunlop, M.G.; Stark, L.A. Nucleolar targeting of RelA(p65) is regulated by COMMD1-dependent ubiquitination. Cancer Res. 2010, 70, 139–149. [Google Scholar] [CrossRef]
- Khandelwal, N.; Simpson, J.; Taylor, G.; Rafique, S.; Whitehouse, A.; Hiscox, J.; Stark, L.A. Nucleolar NF-kappaB/RelA mediates apoptosis by causing cytoplasmic relocalization of nucleophosmin. Cell Death Differ. 2011, 18, 1889–1903. [Google Scholar] [CrossRef]
- Michel, J.; Nolin, F.; Wortham, L.; Lalun, N.; Tchelidze, P.; Banchet, V.; Terryn, C.; Ploton, D. Various Nucleolar Stress Inducers Result in Highly Distinct Changes in Water, Dry Mass and Elemental Content in Cancerous Cell Compartments: Investigation Using a Nano-Analytical Approach. Nanotheranostics 2019, 3, 179–195. [Google Scholar] [CrossRef]
- Liu, X.F.; Xiang, L.; Zhou, Q.; Carralot, J.P.; Prunotto, M.; Niederfellner, G.; Pastan, I. Actinomycin D enhances killing of cancer cells by immunotoxin RG7787 through activation of the extrinsic pathway of apoptosis. Proc. Natl. Acad. Sci. USA 2016, 113, 10666–10671. [Google Scholar] [CrossRef]
- Chen, J.; Lobb, I.T.; Morin, P.; Novo, S.M.; Simpson, J.; Kennerknecht, K.; von Kriegsheim, A.; Batchelor, E.E.; Oakley, F.; Stark, L.A. Identification of a novel TIF-IA-NF-kappaB nucleolar stress response pathway. Nucleic Acids Res. 2018, 46, 6188–6205. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, J.; Han, Y.; Chen, Y.; Guo, X.; Cui, X.; Cheng, M.; Gao, H.; Wang, J.; Jiang, F. CX-5461 is a potent immunosuppressant which inhibits T cell-mediated alloimmunity via p53-DUSP5. Pharmacol. Res. 2022, 177, 106120. [Google Scholar] [CrossRef]
- Fatyol, K.; Szalay, A.A. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha) and inhibits HIF-1-mediated transcription. J. Biol. Chem. 2001, 276, 28421–28429. [Google Scholar] [CrossRef]
- Mekhail, K.; Gunaratnam, L.; Bonicalzi, M.E.; Lee, S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004, 6, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Han, Y.; Wang, Y.; Sun, X.; Yan, S.; Yeh, E.T.; Chen, Y.; Cang, H.; Li, H.; Shi, G.; et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009, 28, 2748–2762. [Google Scholar] [CrossRef]
- Mekhail, K.; Rivero-Lopez, L.; Khacho, M.; Lee, S. Restriction of rRNA synthesis by VHL maintains energy equilibrium under hypoxia. Cell Cycle 2006, 5, 2401–2413. [Google Scholar] [CrossRef]
- Yang, K.; Wang, M.; Zhao, Y.; Sun, X.; Yang, Y.; Li, X.; Zhou, A.; Chu, H.; Zhou, H.; Xu, J.; et al. A redox mechanism underlying nucleolar stress sensing by nucleophosmin. Nat. Commun. 2016, 7, 13599. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, E.; Chmurciakova, N.; Cmarko, D. Human rDNA and Cancer. Cells 2021, 10, 3452. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol. Life Sci. 2011, 68, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, L.M.; Baserga, S.J. Crosstalk between the nucleolus and the DNA damage response. Mol. Biosyst. 2017, 13, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Rancourt, A.; Satoh, M.S. Delocalization of nucleolar poly(ADP-ribose) polymerase-1 to the nucleoplasm and its novel link to cellular sensitivity to DNA damage. DNA Repair 2009, 8, 286–297. [Google Scholar] [CrossRef]
- Harding, S.M.; Boiarsky, J.A.; Greenberg, R.A. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep. 2015, 13, 251–259. [Google Scholar] [CrossRef]
- van Sluis, M.; McStay, B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015, 29, 1151–1163. [Google Scholar] [CrossRef]
- Korsholm, L.M.; Gal, Z.; Lin, L.; Quevedo, O.; Ahmad, D.A.; Dulina, E.; Luo, Y.; Bartek, J.; Larsen, D.H. Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair. Nucleic Acids Res. 2019, 47, 8019–8035. [Google Scholar] [CrossRef]
- Perez-Castro, A.J.; Freire, R. Rad9B responds to nucleolar stress through ATR and JNK signalling, and delays the G1-S transition. J. Cell Sci. 2012, 125, 1152–1164. [Google Scholar] [CrossRef]
- Lindstrom, M.S.; Jurada, D.; Bursac, S.; Orsolic, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef]
- Xuan, J.; Gitareja, K.; Brajanovski, N.; Sanij, E. Harnessing the Nucleolar DNA Damage Response in Cancer Therapy. Genes 2021, 12, 1156. [Google Scholar] [CrossRef]
- Jayaraman, S.; Chittiboyina, S.; Bai, Y.; Abad, P.C.; Vidi, P.A.; Stauffacher, C.V.; Lelievre, S.A. The nuclear mitotic apparatus protein NuMA controls rDNA transcription and mediates the nucleolar stress response in a p53-independent manner. Nucleic Acids Res. 2017, 45, 11725–11742. [Google Scholar] [CrossRef]
- Russo, A.; Russo, G. Ribosomal Proteins Control or Bypass p53 during Nucleolar Stress. Int. J. Mol. Sci. 2017, 18, 140. [Google Scholar] [CrossRef]
- Juan, G.; Cordon-Cardo, C. Intranuclear compartmentalization of cyclin E during the cell cycle: Disruption of the nucleoplasm-nucleolar shuttling of cyclin E in bladder cancer. Cancer Res. 2001, 61, 1220–1226. [Google Scholar]
- Liu, J.; Hebert, M.D.; Ye, Y.; Templeton, D.J.; Kung, H.; Matera, A.G. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J. Cell Sci. 2000, 113 Pt 9, 1543–1552. [Google Scholar] [CrossRef]
- Hu, J.; Cai, X.F.; Yan, G. Alphavirus M1 induces apoptosis of malignant glioma cells via downregulation and nucleolar translocation of p21WAF1/CIP1 protein. Cell Cycle 2009, 8, 3328–3339. [Google Scholar] [CrossRef]
- Hu, B.; Hua, L.; Ni, W.; Wu, M.; Yan, D.; Chen, Y.; Lu, C.; Chen, B.; Wan, C. Nucleostemin/GNL3 promotes nucleolar polyubiquitylation of p27(kip1) to drive hepatocellular carcinoma progression. Cancer Lett. 2017, 388, 220–229. [Google Scholar] [CrossRef]
- Bhaskaran, N.; van Drogen, F.; Ng, H.F.; Kumar, R.; Ekholm-Reed, S.; Peter, M.; Sangfelt, O.; Reed, S.I. Fbw7alpha and Fbw7gamma collaborate to shuttle cyclin E1 into the nucleolus for multiubiquitylation. Mol. Cell Biol. 2013, 33, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Welcker, M.; Orian, A.; Grim, J.E.; Eisenman, R.N.; Clurman, B.E. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 2004, 14, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.A.; Gruppuso, P.A. Nucleolar localization of hepatic c-Myc: A potential mechanism for c-Myc regulation. Biochim. Biophys. Acta 2005, 1743, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar] [CrossRef]
- Tasselli, L.; Zheng, W.; Chua, K.F. SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol. Metab. 2017, 28, 168–185. [Google Scholar] [CrossRef]
- Tang, B.L. SIRT7 and hepatic lipid metabolism. Front. Cell Dev. Biol. 2015, 3, 1. [Google Scholar] [CrossRef]
- Kreiner, G.; Sonmez, A.; Liss, B.; Parlato, R. Integration of the Deacetylase SIRT1 in the Response to Nucleolar Stress: Metabolic Implications for Neurodegenerative Diseases. Front. Mol. Neurosci. 2019, 12, 106. [Google Scholar] [CrossRef]
- Rieker, C.; Engblom, D.; Kreiner, G.; Domanskyi, A.; Schober, A.; Stotz, S.; Neumann, M.; Yuan, X.; Grummt, I.; Schutz, G.; et al. Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J. Neurosci. 2011, 31, 453–460. [Google Scholar] [CrossRef]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Korner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef]
- Azkanaz, M.; Lopez, A.R.; de Boer, B.; Huiting, W.; Angrand, P.O.; Vellenga, E.; Kampinga, H.H.; Bergink, S.; Martens, J.H.; Schuringa, J.J.; et al. Protein quality control in the nucleolus safeguards recovery of epigenetic regulators after heat shock. Elife 2019, 8, e45205. [Google Scholar] [CrossRef]
- Amer-Sarsour, F.; Ashkenazi, A. The Nucleolus as a Proteostasis Regulator. Trends Cell Biol. 2019, 29, 849–851. [Google Scholar] [CrossRef]
- Golstein, P. Conserved nucleolar stress at the onset of cell death. FEBS J. 2017, 284, 3791–3800. [Google Scholar] [CrossRef]
- Zhang, W.; Cheng, W.; Parlato, R.; Guo, X.; Cui, X.; Dai, C.; Xu, L.; Zhu, J.; Zhu, M.; Luo, K.; et al. Nucleolar stress induces a senescence-like phenotype in smooth muscle cells and promotes development of vascular degeneration. Aging 2020, 12, 22174–22198. [Google Scholar] [CrossRef]
- Kalita, K.; Makonchuk, D.; Gomes, C.; Zheng, J.J.; Hetman, M. Inhibition of nucleolar transcription as a trigger for neuronal apoptosis. J. Neurochem. 2008, 105, 2286–2299. [Google Scholar] [CrossRef]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef]
- Donati, G.; Brighenti, E.; Vici, M.; Mazzini, G.; Trere, D.; Montanaro, L.; Derenzini, M. Selective inhibition of rRNA transcription downregulates E2F-1: A new p53-independent mechanism linking cell growth to cell proliferation. J. Cell Sci. 2011, 124, 3017–3028. [Google Scholar] [CrossRef]
- Iadevaia, V.; Caldarola, S.; Biondini, L.; Gismondi, A.; Karlsson, S.; Dianzani, I.; Loreni, F. PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene 2010, 29, 5490–5499. [Google Scholar] [CrossRef]
- Dai, M.S.; Arnold, H.; Sun, X.X.; Sears, R.; Lu, H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 2007, 26, 3332–3345. [Google Scholar] [CrossRef]
- Lee, H.C.; Wang, H.; Baladandayuthapani, V.; Lin, H.; He, J.; Jones, R.J.; Kuiatse, I.; Gu, D.; Wang, Z.; Ma, W.; et al. RNA Polymerase I Inhibition with CX-5461 as a Novel Therapeutic Strategy to Target MYC in Multiple Myeloma. Br. J. Haematol. 2017, 177, 80–94. [Google Scholar] [CrossRef]
- Gupta, S.; Santoro, R. Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization. Stem Cell Rep. 2020, 15, 1206–1219. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef]
- Meshorer, E.; Misteli, T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006, 7, 540–546. [Google Scholar] [CrossRef]
- Savic, N.; Bar, D.; Leone, S.; Frommel, S.C.; Weber, F.A.; Vollenweider, E.; Ferrari, E.; Ziegler, U.; Kaech, A.; Shakhova, O.; et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 2014, 15, 720–734. [Google Scholar] [CrossRef]
- Zentner, G.E.; Balow, S.A.; Scacheri, P.C. Genomic characterization of the mouse ribosomal DNA locus. Genes Genomes Genet. 2014, 4, 243–254. [Google Scholar] [CrossRef]
- Zaidi, S.K.; Boyd, J.R.; Grandy, R.A.; Medina, R.; Lian, J.B.; Stein, G.S.; Stein, J.L. Expression of Ribosomal RNA and Protein Genes in Human Embryonic Stem Cells Is Associated With the Activating H3K4me3 Histone Mark. J. Cell Physiol. 2016, 231, 2007–2013. [Google Scholar] [CrossRef]
- Tsai, R.Y.; McKay, R.D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002, 16, 2991–3003. [Google Scholar] [CrossRef]
- Yang, A.; Shi, G.; Zhou, C.; Lu, R.; Li, H.; Sun, L.; Jin, Y. Nucleolin maintains embryonic stem cell self-renewal by suppression of p53 protein-dependent pathway. J. Biol. Chem. 2011, 286, 43370–43382. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Susaki, K.; Takada, H.; Enomoto, K.; Miwata, K.; Ishimine, H.; Intoh, A.; Ohtaka, M.; Nakanishi, M.; Sugino, H.; Asashima, M.; et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 2014, 32, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, Z.; Lu, J.Y.; Huang, B.; Zhou, H.; Xie, W.; Wang, J.; Shen, X. DEAD-Box Helicase 18 Counteracts PRC2 to Safeguard Ribosomal DNA in Pluripotency Regulation. Cell Rep. 2020, 30, 81–97.e7. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Kuroda, T.; Kishimoto, H.; Wang, C.; Iwama, A.; Kimura, K. Downregulation of rRNA transcription triggers cell differentiation. PLoS ONE 2014, 9, e98586. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, H.; Lau, T.C.; Tsang, S.Y.; Lau, K.F.; Chan, H.Y. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl. Acad. Sci. USA 2012, 109, 13428–13433. [Google Scholar] [CrossRef]
- Makhale, A.; Nanayakkara, D.; Raninga, P.; Khanna, K.K.; Kalimutho, M. CX-5461 Enhances the Efficacy of APR-246 via Induction of DNA Damage and Replication Stress in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2021, 22, 5782. [Google Scholar] [CrossRef]
- Pan, M.; Wright, W.C.; Chapple, R.H.; Zubair, A.; Sandhu, M.; Batchelder, J.E.; Huddle, B.C.; Low, J.; Blankenship, K.B.; Wang, Y.; et al. The chemotherapeutic CX-5461 primarily targets TOP2B and exhibits selective activity in high-risk neuroblastoma. Nat. Commun. 2021, 12, 6468. [Google Scholar] [CrossRef]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef]
- d’Adda di Fagagna, F. Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef]
- Pathak, R.U.; Soujanya, M.; Mishra, R.K. Deterioration of nuclear morphology and architecture: A hallmark of senescence and aging. Ageing Res. Rev. 2021, 67, 101264. [Google Scholar] [CrossRef]
- Pinho, M.; Macedo, J.C.; Logarinho, E.; Pereira, P.S. NOL12 Repression Induces Nucleolar Stress-Driven Cellular Senescence and Is Associated with Normative Aging. Mol. Cell Biol. 2019, 39, e00099-19. [Google Scholar] [CrossRef]
- Zanotti, S.; Vanhauwaert, S.; Van Neste, C.; Olexiouk, V.; Van Laere, J.; Verschuuren, M.; Van der Meulen, J.; Mus, L.M.; Durinck, K.; Tilleman, L.; et al. MYCN-induced nucleolar stress drives an early senescence-like transcriptional program in hTERT-immortalized RPE cells. Sci. Rep. 2021, 11, 14454. [Google Scholar] [CrossRef]
- Nishimura, K.; Kumazawa, T.; Kuroda, T.; Katagiri, N.; Tsuchiya, M.; Goto, N.; Furumai, R.; Murayama, A.; Yanagisawa, J.; Kimura, K. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 2015, 10, 1310–1323. [Google Scholar] [CrossRef]
- Wu, H.C.; Rerolle, D.; Berthier, C.; Hleihel, R.; Sakamoto, T.; Quentin, S.; Benhenda, S.; Morganti, C.; Wu, C.; Conte, L.; et al. Actinomycin D Targets NPM1c-Primed Mitochondria to Restore PML-Driven Senescence in AML Therapy. Cancer Discov. 2021, 11, 3198–3213. [Google Scholar] [CrossRef]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef]
- Pfister, A.S. Emerging Role of the Nucleolar Stress Response in Autophagy. Front. Cell Neurosci. 2019, 13, 156. [Google Scholar] [CrossRef]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Cloonan, S.M.; Choi, A.M. Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol. Cells 2013, 36, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, N.; Kuroda, T.; Kishimoto, H.; Hayashi, Y.; Kumazawa, T.; Kimura, K. The nucleolar protein nucleophosmin is essential for autophagy induced by inhibiting Pol I transcription. Sci. Rep. 2015, 5, 8903. [Google Scholar] [CrossRef] [PubMed]
- Dannheisig, D.P.; Schimansky, A.; Donow, C.; Pfister, A.S. Nucleolar Stress Functions Upstream to Stimulate Expression of Autophagy Regulators. Cancers 2021, 13, 6220. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Zhao, J.; Fan, S.; Wang, L.; Li, X. CX-5461 induces autophagy and inhibits tumor growth via mammalian target of rapamycin-related signaling pathways in osteosarcoma. Onco Targets Ther. 2016, 9, 5985–5997. [Google Scholar] [CrossRef]
- Okamoto, S.; Miyano, K.; Kajikawa, M.; Yamauchi, A.; Kuribayashi, F. The rRNA synthesis inhibitor CX-5461 may induce autophagy that inhibits anticancer drug-induced cell damage to leukemia cells. Biosci. Biotechnol. Biochem. 2020, 84, 2319–2326. [Google Scholar] [CrossRef]
- Lafita-Navarro, M.C.; Conacci-Sorrell, M. Nucleolar stress: From development to cancer. Semin. Cell Dev. Biol. 2022; in press. [Google Scholar] [CrossRef]
- Parlato, R.; Kreiner, G. Nucleolar activity in neurodegenerative diseases: A missing piece of the puzzle? J. Mol. Med. 2013, 91, 541–547. [Google Scholar] [CrossRef]
- Hein, N.; Hannan, K.M.; George, A.J.; Sanij, E.; Hannan, R.D. The nucleolus: An emerging target for cancer therapy. Trends Mol. Med. 2013, 19, 643–654. [Google Scholar] [CrossRef]
- Chen, S.; He, H.; Wang, Y.; Liu, L.; Liu, Y.; You, H.; Dong, Y.; Lyu, J. Poor prognosis of nucleophosmin overexpression in solid tumors: A meta-analysis. BMC Cancer 2018, 18, 838. [Google Scholar] [CrossRef]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487, 114–118. [Google Scholar] [CrossRef]
- Okamoto, N.; Yasukawa, M.; Nguyen, C.; Kasim, V.; Maida, Y.; Possemato, R.; Shibata, T.; Ligon, K.L.; Fukami, K.; Hahn, W.C.; et al. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc. Natl. Acad. Sci. USA 2011, 108, 20388–20393. [Google Scholar] [CrossRef]
- Blank, M.F.; Grummt, I. The seven faces of SIRT7. Transcription 2017, 8, 67–74. [Google Scholar] [CrossRef]
- Bywater, M.J.; Poortinga, G.; Sanij, E.; Hein, N.; Peck, A.; Cullinane, C.; Wall, M.; Cluse, L.; Drygin, D.; Anderes, K.; et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 2012, 22, 51–65. [Google Scholar] [CrossRef]
- Pecoraro, A.; Carotenuto, P.; Russo, G.; Russo, A. Ribosomal protein uL3 targets E2F1 and Cyclin D1 in cancer cell response to nucleolar stress. Sci. Rep. 2019, 9, 15431. [Google Scholar] [CrossRef]
- Haddach, M.; Schwaebe, M.K.; Michaux, J.; Nagasawa, J.; O’Brien, S.E.; Whitten, J.P.; Pierre, F.; Kerdoncuff, P.; Darjania, L.; Stansfield, R.; et al. Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. ACS Med. Chem. Lett. 2012, 3, 602–606. [Google Scholar] [CrossRef]
- Cornelison, R.; Biswas, K.; Llaneza, D.C.; Harris, A.R.; Sosale, N.G.; Lazzara, M.J.; Landen, C.N. CX-5461 Treatment Leads to Cytosolic DNA-Mediated STING Activation in Ovarian Cancer. Cancers 2021, 13, 5056. [Google Scholar] [CrossRef]
- Hilton, J.; Gelmon, K.; Bedard, P.L.; Tu, D.; Xu, H.; Tinker, A.V.; Goodwin, R.; Laurie, S.A.; Jonker, D.; Hansen, A.R.; et al. Results of the phase I CCTG IND.231 trial of CX-5461 in patients with advanced solid tumors enriched for DNA-repair deficiencies. Nat. Commun. 2022, 13, 3607. [Google Scholar] [CrossRef]
- Khot, A.; Brajanovski, N.; Cameron, D.P.; Hein, N.; Maclachlan, K.H.; Sanij, E.; Lim, J.; Soong, J.; Link, E.; Blombery, P.; et al. First-in-Human RNA Polymerase I Transcription Inhibitor CX-5461 in Patients with Advanced Hematologic Cancers: Results of a Phase I Dose-Escalation Study. Cancer Discov. 2019, 9, 1036–1049. [Google Scholar] [CrossRef]
- Lee, J.; Hwang, Y.J.; Ryu, H.; Kowall, N.W.; Ryu, H. Nucleolar dysfunction in Huntington’s disease. Biochim. Biophys. Acta 2014, 1842, 785–790. [Google Scholar] [CrossRef][Green Version]
- Arogundade, O.A.; Nguyen, S.; Leung, R.; Wainio, D.; Rodriguez, M.; Ravits, J. Nucleolar stress in C9orf72 and sporadic ALS spinal motor neurons precedes TDP-43 mislocalization. Acta Neuropathol. Commun. 2021, 9, 26. [Google Scholar] [CrossRef]
- Pietrzak, M.; Rempala, G.; Nelson, P.T.; Zheng, J.J.; Hetman, M. Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS ONE 2011, 6, e22585. [Google Scholar] [CrossRef]
- Caudle, W.M.; Kitsou, E.; Li, J.; Bradner, J.; Zhang, J. A role for a novel protein, nucleolin, in Parkinson’s disease. Neurosci. Lett. 2009, 459, 11–15. [Google Scholar] [CrossRef]
- Vilotti, S.; Codrich, M.; Dal Ferro, M.; Pinto, M.; Ferrer, I.; Collavin, L.; Gustincich, S.; Zucchelli, S. Parkinson’s disease DJ-1 L166P alters rRNA biogenesis by exclusion of TTRAP from the nucleolus and sequestration into cytoplasmic aggregates via TRAF6. PLoS ONE 2012, 7, e35051. [Google Scholar] [CrossRef]
- Lee, J.; Hwang, Y.J.; Boo, J.H.; Han, D.; Kwon, O.K.; Todorova, K.; Kowall, N.W.; Kim, Y.; Ryu, H. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington’s disease. Cell Death Differ. 2011, 18, 1726–1735. [Google Scholar] [CrossRef]
- Sonmez, A.; Mustafa, R.; Ryll, S.T.; Tuorto, F.; Wacheul, L.; Ponti, D.; Litke, C.; Hering, T.; Kojer, K.; Koch, J.; et al. Nucleolar stress controls mutant Huntington toxicity and monitors Huntington’s disease progression. Cell Death Dis. 2021, 12, 1139. [Google Scholar] [CrossRef] [PubMed]
- Latonen, L. Nucleolar aggresomes as counterparts of cytoplasmic aggresomes in proteotoxic stress. Proteasome inhibitors induce nuclear ribonucleoprotein inclusions that accumulate several key factors of neurodegenerative diseases and cancer. Bioessays 2011, 33, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Wang, H.; Xia, Q.; Li, K.; Jiang, X.; Xu, G.; Wang, G.; Ying, Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 2015, 24, 2426–2441. [Google Scholar] [CrossRef] [PubMed]
- White, M.R.; Mitrea, D.M.; Zhang, P.; Stanley, C.B.; Cassidy, D.E.; Nourse, A.; Phillips, A.H.; Tolbert, M.; Taylor, J.P.; Kriwacki, R.W. C9orf72 Poly(PR) Dipeptide Repeats Disturb Biomolecular Phase Separation and Disrupt Nucleolar Function. Mol. Cell 2019, 74, 713–728.e716. [Google Scholar] [CrossRef]
- Maina, M.B.; Bailey, L.J.; Wagih, S.; Biasetti, L.; Pollack, S.J.; Quinn, J.P.; Thorpe, J.R.; Doherty, A.J.; Serpell, L.C. The involvement of tau in nucleolar transcription and the stress response. Acta Neuropathol. Commun. 2018, 6, 70. [Google Scholar] [CrossRef]
- Marquez-Lona, E.M.; Tan, Z.; Schreiber, S.S. Nucleolar stress characterized by downregulation of nucleophosmin: A novel cause of neuronal degeneration. Biochem. Biophys. Res. Commun. 2012, 417, 514–520. [Google Scholar] [CrossRef]
- Herrmann, D.; Parlato, R. C9orf72-associated neurodegeneration in ALS-FTD: Breaking new ground in ribosomal RNA and nucleolar dysfunction. Cell Tissue Res. 2018, 373, 351–360. [Google Scholar] [CrossRef]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011, 14, 33–44. [Google Scholar] [CrossRef]
- Schaffer, J.E. Death by lipids: The role of small nucleolar RNAs in metabolic stress. J. Biol. Chem. 2020, 295, 8628–8635. [Google Scholar] [CrossRef]
- Dupuis-Sandoval, F.; Poirier, M.; Scott, M.S. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA 2015, 6, 381–397. [Google Scholar] [CrossRef]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef]
- Bratkovic, T.; Rogelj, B. The many faces of small nucleolar RNAs. Biochim. Biophys. Acta 2014, 1839, 438–443. [Google Scholar] [CrossRef]
- Holley, C.L.; Li, M.W.; Scruggs, B.S.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 2015, 290, 11741–11748. [Google Scholar] [CrossRef]
- Lee, J.; Harris, A.N.; Holley, C.L.; Mahadevan, J.; Pyles, K.D.; Lavagnino, Z.; Scherrer, D.E.; Fujiwara, H.; Sidhu, R.; Zhang, J.; et al. Rpl13a small nucleolar RNAs regulate systemic glucose metabolism. J. Clin. Investig. 2016, 126, 4616–4625. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, X.; Li, Y.; Zhu, T.; Zhang, J.; Zhang, Z.; Zhang, L.; Zhang, Y.; Wang, Y.; Zou, X.; et al. PHA-4/FoxA senses nucleolar stress to regulate lipid accumulation in Caenorhabditis elegans. Nat. Commun. 2018, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Karim, M.F.; Sato, Y.; Senokuchi, T.; Miyata, K.; Fukuda, T.; Go, C.; Tasaki, M.; Uchimura, K.; Kadomatsu, T.; et al. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014, 19, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; He, M.; Liu, Y.; Paredes, S.; Villanova, L.; Brown, K.; Qiu, X.; Nabavi, N.; Mohrin, M.; Wojnoonski, K.; et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013, 5, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.; Jo, Y.S.; Lo Sasso, G.; Stein, S.; Zhang, H.; Perino, A.; Lee, J.U.; Zeviani, M.; Romand, R.; Hottiger, M.O.; et al. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 2014, 20, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Mohrin, M.; Shin, J.; Liu, Y.; Brown, K.; Luo, H.; Xi, Y.; Haynes, C.M.; Chen, D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015, 347, 1374–1377. [Google Scholar] [CrossRef]
- Liu, Y.; He, Y.; Jin, A.; Tikunov, A.P.; Zhou, L.; Tollini, L.A.; Leslie, P.; Kim, T.H.; Li, L.O.; Coleman, R.A.; et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. USA 2014, 111, E2414–E2422. [Google Scholar] [CrossRef]
- Guarente, L. Link between aging and the nucleolus. Genes Dev. 1997, 11, 2449–2455. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Gotta, M.; Sinclair, D.A.; Mills, K.; McNabb, D.S.; Murthy, M.; Pak, S.M.; Laroche, T.; Gasser, S.M.; Guarente, L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 1997, 89, 381–391. [Google Scholar] [CrossRef]
- Tiku, V.; Jain, C.; Raz, Y.; Nakamura, S.; Heestand, B.; Liu, W.; Spath, M.; Suchiman, H.E.D.; Muller, R.U.; Slagboom, P.E.; et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2017, 8, 16083. [Google Scholar] [CrossRef]
- Buchwalter, A.; Hetzer, M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017, 8, 328. [Google Scholar] [CrossRef]
- Gensous, N.; Ravaioli, F.; Pirazzini, C.; Gramignoli, R.; Ellis, E.; Storci, G.; Capri, M.; Strom, S.; Laconi, E.; Franceschi, C.; et al. Aging and Caloric Restriction Modulate the DNA Methylation Profile of the Ribosomal RNA Locus in Human and Rat Liver. Nutrients 2020, 12, 277. [Google Scholar] [CrossRef]
- Smith, D.L., Jr.; Li, C.; Matecic, M.; Maqani, N.; Bryk, M.; Smith, J.S. Calorie restriction effects on silencing and recombination at the yeast rDNA. Aging Cell 2009, 8, 633–642. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef]
- Morlot, S.; Song, J.; Leger-Silvestre, I.; Matifas, A.; Gadal, O.; Charvin, G. Excessive rDNA Transcription Drives the Disruption in Nuclear Homeostasis during Entry into Senescence in Budding Yeast. Cell Rep. 2019, 28, 408–422.e404. [Google Scholar] [CrossRef]
- Perry, R.P. Nucleolus: Structure and function. Science 1966, 153, 214–219. [Google Scholar] [CrossRef]
- Derenzini, M.; Trere, D.; Pession, A.; Govoni, M.; Sirri, V.; Chieco, P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 2000, 191, 181–186. [Google Scholar] [CrossRef]
- Hariharan, N.; Sussman, M.A. Stressing on the nucleolus in cardiovascular disease. Biochim. Biophys. Acta 2014, 1842, 798–801. [Google Scholar] [CrossRef]
- Calo, E.; Gu, B.; Bowen, M.E.; Aryan, F.; Zalc, A.; Liang, J.; Flynn, R.A.; Swigut, T.; Chang, H.Y.; Attardi, L.D.; et al. Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 2018, 554, 112–117. [Google Scholar] [CrossRef]
- Bianco, C.; Mohr, I. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. Elife 2019, 8, e49551. [Google Scholar] [CrossRef]
- Matos-Perdomo, E.; Machin, F. Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer. Cells 2019, 8, 779. [Google Scholar] [CrossRef] [PubMed]
- Parlato, R.; Liss, B. How Parkinson’s disease meets nucleolar stress. Biochim. Biophys. Acta 2014, 1842, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Latonen, L. Phase-to-Phase With Nucleoli—Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell Neurosci. 2019, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Avitabile, D.; Bailey, B.; Cottage, C.T.; Sundararaman, B.; Joyo, A.; McGregor, M.; Gude, N.; Truffa, S.; Zarrabi, A.; Konstandin, M.; et al. Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc. Natl. Acad. Sci. USA 2011, 108, 6145–6150. [Google Scholar] [CrossRef]
- Pfister, J.A.; D’Mello, S.R. Insights into the regulation of neuronal viability by nucleophosmin/B23. Exp. Biol. Med. 2015, 240, 774–786. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, H.; Jiang, B.; Liang, P.; Liu, M.; Deng, G.; Xiao, X. Nucleolin/C23 is a negative regulator of hydrogen peroxide-induced apoptosis in HUVECs. Cell Stress Chaperones 2010, 15, 249–257. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, L.; Yan, D.; Wan, C.; Hu, B. Nucleolus and Nucleolar Stress: From Cell Fate Decision to Disease Development. Cells 2022, 11, 3017. https://doi.org/10.3390/cells11193017
Hua L, Yan D, Wan C, Hu B. Nucleolus and Nucleolar Stress: From Cell Fate Decision to Disease Development. Cells. 2022; 11(19):3017. https://doi.org/10.3390/cells11193017
Chicago/Turabian StyleHua, Lu, Daliang Yan, Chunhua Wan, and Baoying Hu. 2022. "Nucleolus and Nucleolar Stress: From Cell Fate Decision to Disease Development" Cells 11, no. 19: 3017. https://doi.org/10.3390/cells11193017
APA StyleHua, L., Yan, D., Wan, C., & Hu, B. (2022). Nucleolus and Nucleolar Stress: From Cell Fate Decision to Disease Development. Cells, 11(19), 3017. https://doi.org/10.3390/cells11193017




