Hedgehog Signaling as a Therapeutic Target for Airway Remodeling and Inflammation in Allergic Asthma
Abstract
1. Introduction
2. Material and Methods
2.1. Gain and Loss of Function Assays
2.2. Real-Time PCR
2.3. RNA Sequencing of Airway Epithelial Cells from Asthmatic and Non-Asthmatic Subjects
2.4. Immunofluorescence Microscopy on Differentiated Cell Culture
2.5. Mice
2.6. HDM Treatment Protocol
2.7. Cell Differential and Mouse Lung Tissue Assessments
2.8. Serum Cytokine Measurements
2.9. Statistical Analyses
3. Results
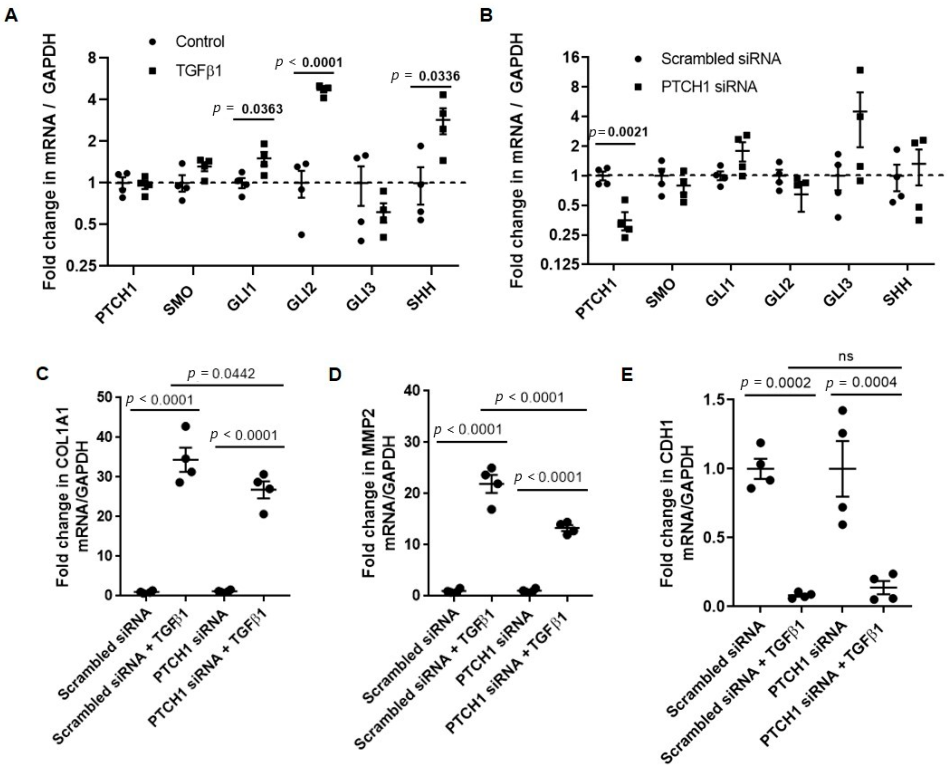
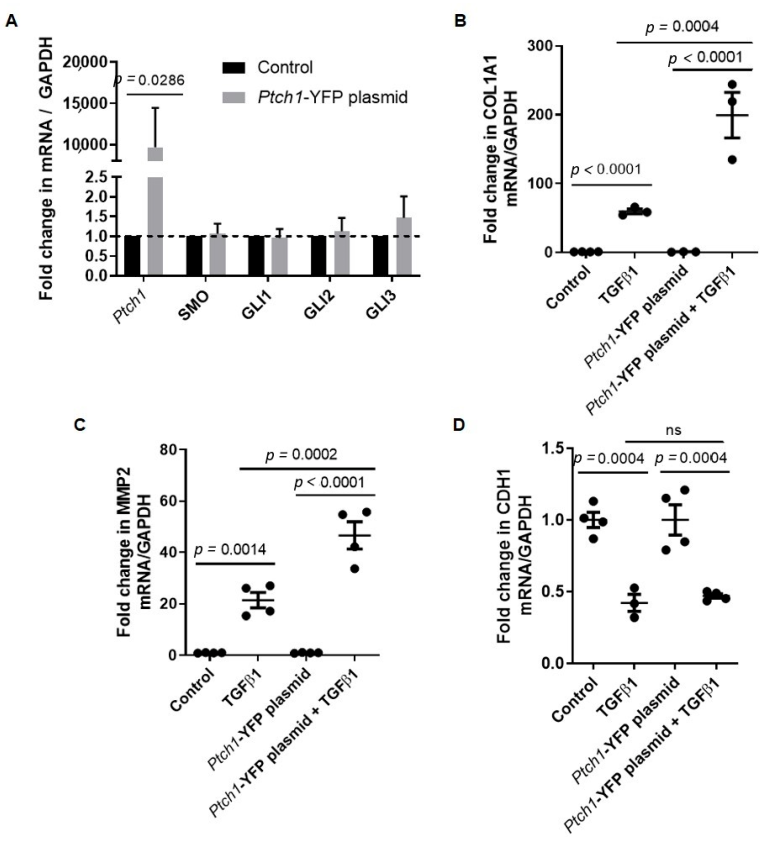
3.1. PTCH1 Knockdown Reduces TGFβ1-Induced COL1A1 and MMP2 Gene Expression in 1HAEo Cells
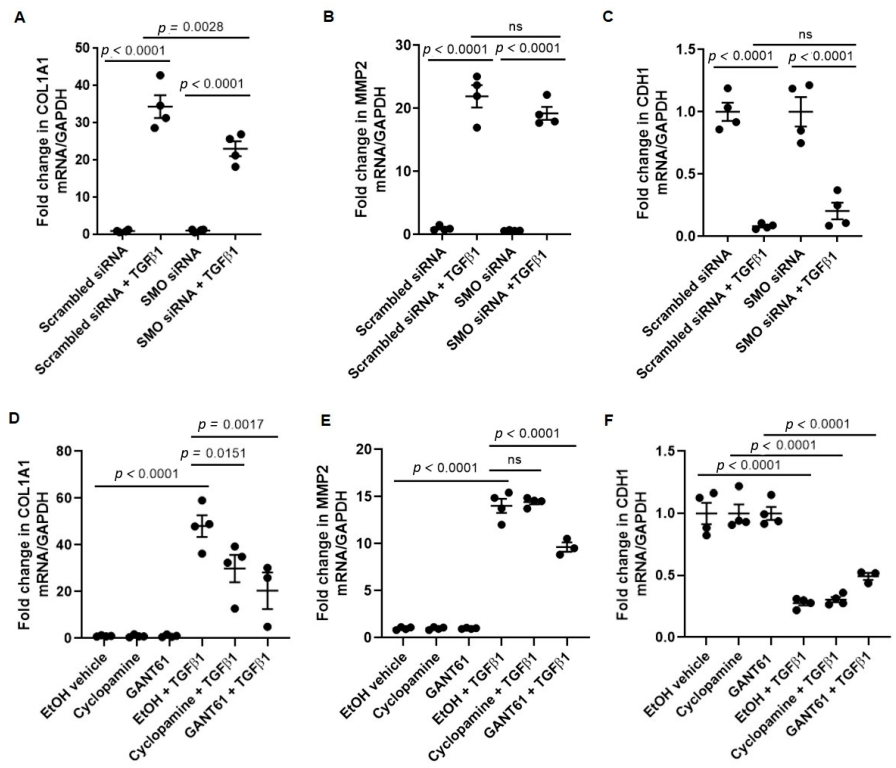
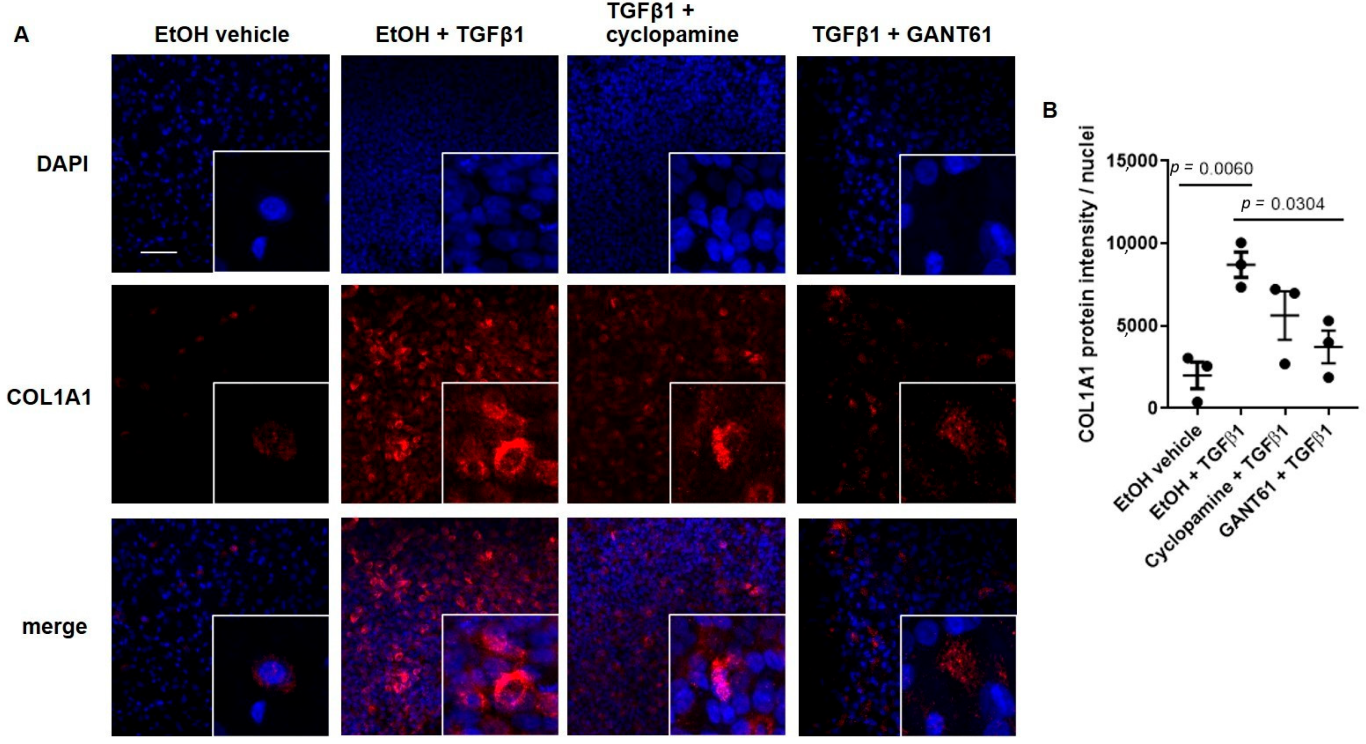
3.2. TGFβ1-Induced COL1A1 Gene Expression Signals through the SMO-GLI1/2 Axes
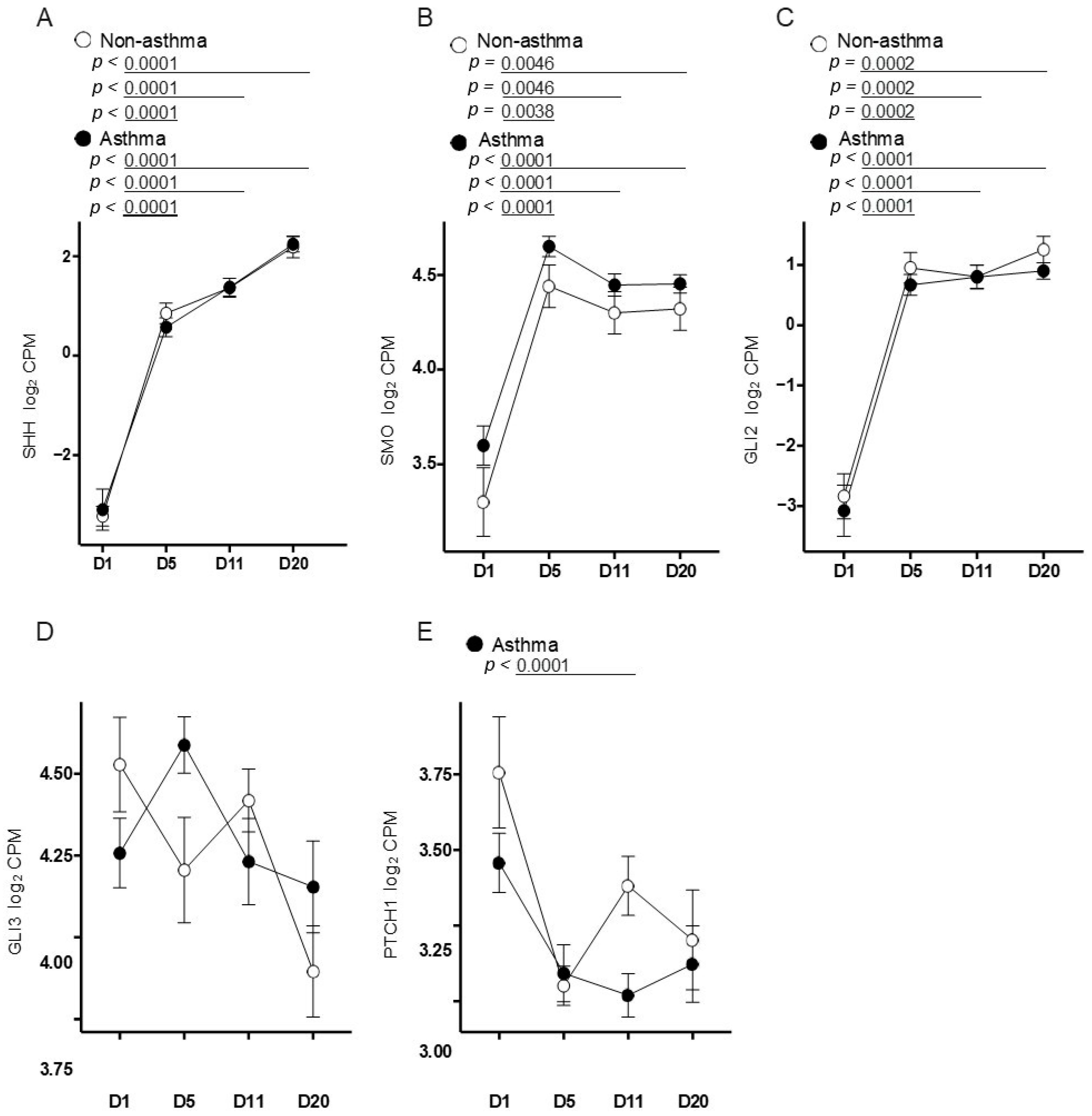
3.3. Primary Airway Epithelial Cells Exhibit Upregulation of Hedgehog Signaling
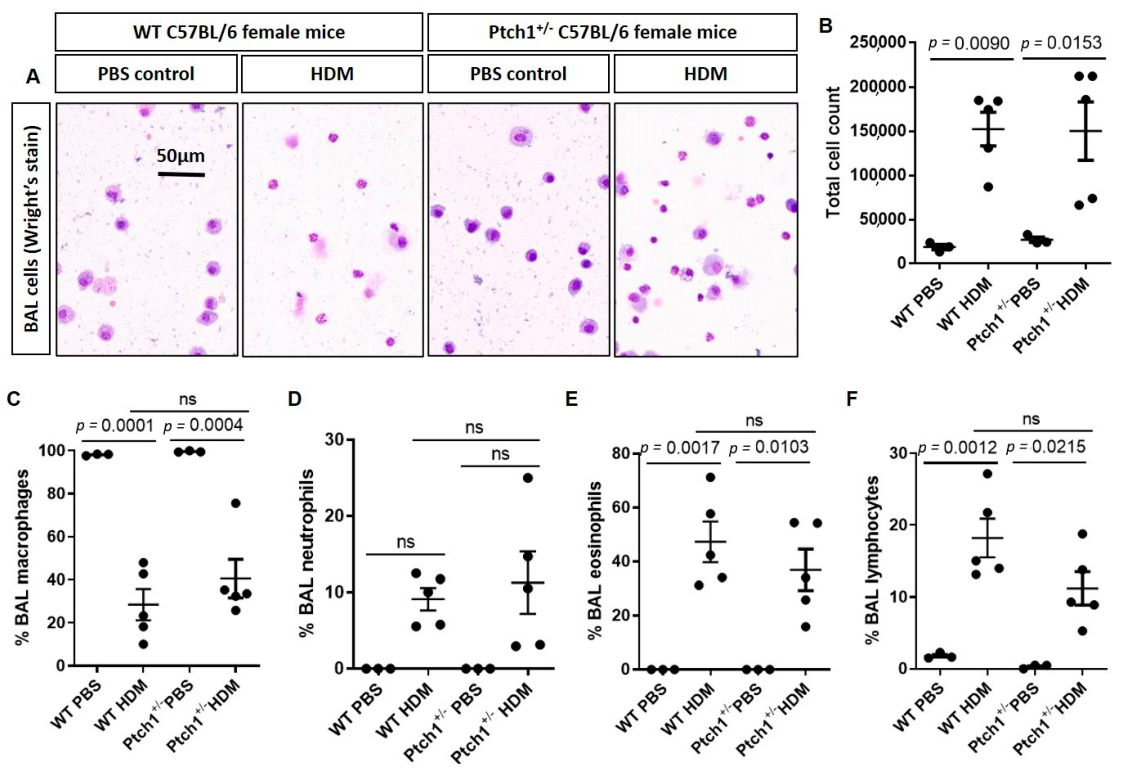
3.4. Ptch1+/− Mice Fail to Induce Total Airway Wall Collagen Protein Expression in Response to HDM Exposure
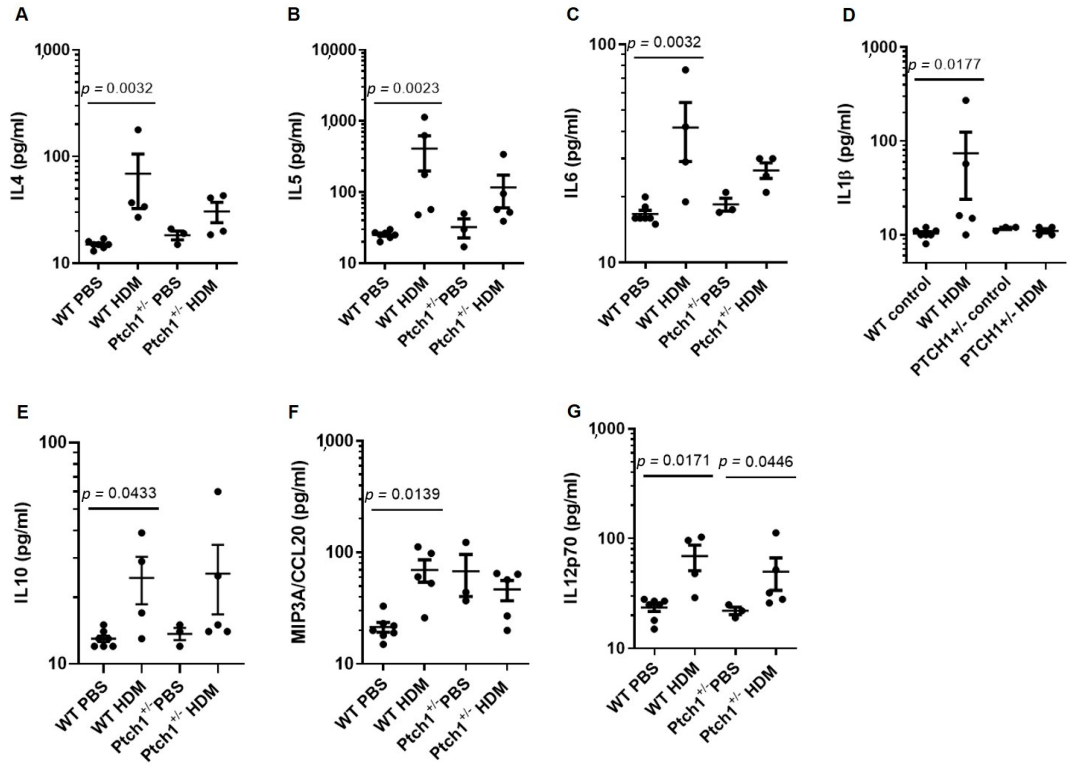
3.5. Ptch1+/− Mice Fail to Induce Il-4 and Il-5 Serum Protein Expression in Response to HDM Exposure
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Al-Alawi, M.; Hassan, T.; Chotirmall, S.H. Transforming growth factor beta and severe asthma: A perfect storm. Respir. Med. 2014, 108, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Sagara, H.; Okada, T.; Okumura, K.; Ogawa, H.; Ra, C.; Fukuda, T.; Nakao, A. Activation of TGF-beta/Smad2 signaling is associated with airway remodeling in asthma. J. Allergy Clin. Immunol. 2002, 110, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Halwani, R.; Al-Muhsen, S.; Al-Jahdali, H.; Hamid, Q. Role of transforming growth factor-beta in airway remodeling in asthma. Am. J. Respir. Cell Mol. Biol. 2011, 44, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Fahy, J.V. Type 2 inflammation in asthma—present in most, absent in many. Nat. Rev. Immunol. 2015, 15, 57–65. [Google Scholar] [CrossRef]
- Obeidat, M.; Hao, K.; Bosse, Y.; Nickle, D.C.; Nie, Y.; Postma, D.S.; Laviolette, M.; Sandford, A.J.; Daley, D.D.; Hogg, J.C.; et al. Molecular mechanisms underlying variations in lung function: A systems genetics analysis. Lancet Respir. Med. 2015, 3, 782–795. [Google Scholar] [CrossRef]
- Li, X.; Howard, T.D.; Moore, W.C.; Ampleford, E.J.; Li, H.; Busse, W.W.; Calhoun, W.J.; Castro, M.; Chung, K.F.; Erzurum, S.C.; et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J. Allergy Clin. Immunol. 2011, 127, 1457–1465. [Google Scholar] [CrossRef]
- Rubin, L.L.; de Sauvage, F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006, 5, 1026–1033. [Google Scholar] [CrossRef]
- Carballo, G.B.; Honorato, J.R.; de Lopes, G.P.F.; Spohr, T. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef]
- Brennan, D.; Chen, X.; Cheng, L.; Mahoney, M.; Riobo, N.A. Noncanonical Hedgehog signaling. Vitam. Horm. 2012, 88, 55–72. [Google Scholar] [CrossRef]
- Ng, J.M.; Curran, T. The Hedgehog’s tale: Developing strategies for targeting cancer. Nat. Rev. Cancer 2011, 11, 493–501. [Google Scholar] [CrossRef]
- Cooper, M.K.; Porter, J.A.; Young, K.E.; Beachy, P.A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 1998, 280, 1603–1607. [Google Scholar] [CrossRef]
- Taipale, J.; Chen, J.K.; Cooper, M.K.; Wang, B.; Mann, R.K.; Milenkovic, L.; Scott, M.P.; Beachy, P.A. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 2000, 406, 1005–1009. [Google Scholar] [CrossRef]
- Meyers-Needham, M.; Lewis, J.A.; Gencer, S.; Sentelle, R.D.; Saddoughi, S.A.; Clarke, C.J.; Hannun, Y.A.; Norell, H.; da Palma, T.M.; Nishimura, M.; et al. Off-target function of the Sonic hedgehog inhibitor cyclopamine in mediating apoptosis via nitric oxide-dependent neutral sphingomyelinase 2/ceramide induction. Mol. Cancer Ther. 2012, 11, 1092–1102. [Google Scholar] [CrossRef]
- Lauth, M.; Bergstrom, A.; Shimokawa, T.; Toftgard, R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. USA 2007, 104, 8455–8460. [Google Scholar] [CrossRef]
- Xu, C.; Zou, C.; Hussain, M.; Shi, W.; Shao, Y.; Jiang, Z.; Wu, X.; Lu, M.; Wu, J.; Xie, Q.; et al. High expression of Sonic hedgehog in allergic airway epithelia contributes to goblet cell metaplasia. Mucosal. Immunol. 2018, 11, 1306–1315. [Google Scholar] [CrossRef]
- Johnson, J.R.; Roos, A.; Berg, T.; Nord, M.; Fuxe, J. Chronic respiratory aeroallergen exposure in mice induces epithelial-mesenchymal transition in the large airways. PLoS ONE 2011, 6, e16175. [Google Scholar] [CrossRef]
- Gregory, L.G.; Causton, B.; Murdoch, J.R.; Mathie, S.A.; O’Donnell, V.; Thomas, C.P.; Priest, F.M.; Quint, D.J.; Lloyd, C.M. Inhaled house dust mite induces pulmonary T helper 2 cytokine production. Clin. Exp. Allergy 2009, 39, 1597–1610. [Google Scholar] [CrossRef]
- Standing, A.S.I.; Yanez, D.C.; Ross, R.; Crompton, T.; Furmanski, A.L. Frontline Science: Shh production and Gli signaling is activated in vivo in lung, enhancing the Th2 response during a murine model of allergic asthma. J. Leukoc. Biol. 2017, 102, 965–976. [Google Scholar] [CrossRef]
- Wang, X.; Xu, C.; Ji, J.; Cai, Y.; Shu, Y.; Chao, Y.; Wu, X.; Zou, C.; Wu, X.; Tang, L. IL-4/IL-13 upregulates Sonic hedgehog expression to induce allergic airway epithelial remodeling. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2020, 318, L888–L899. [Google Scholar] [CrossRef]
- Hackett, T.L.; Warner, S.M.; Stefanowicz, D.; Shaheen, F.; Pechkovsky, D.V.; Murray, L.A.; Argentieri, R.; Kicic, A.; Stick, S.M.; Bai, T.R.; et al. Induction of epithelial-mesenchymal transition in primary airway epithelial cells from patients with asthma by transforming growth factor-beta1. Am. J. Respir. Crit. Care Med. 2009, 180, 122–133. [Google Scholar] [CrossRef]
- Guo, X.; Wang, X.F. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009, 19, 71–88. [Google Scholar] [CrossRef]
- Javelaud, D.; Pierrat, M.J.; Mauviel, A. Crosstalk between TGF-beta and hedgehog signaling in cancer. FEBS Lett. 2012, 586, 2016–2025. [Google Scholar] [CrossRef]
- Gruenert, D.C.; Basbaum, C.B.; Welsh, M.J.; Li, M.; Finkbeiner, W.E.; Nadel, J.A. Characterization of human tracheal epithelial cells transformed by an origin-defective simian virus 40. Proc. Natl. Acad. Sci. USA 1988, 85, 5951–5955. [Google Scholar] [CrossRef]
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376. [Google Scholar] [CrossRef]
- Tam, A.; Wadsworth, S.; Dorscheid, D.; Man, S.F.; Sin, D.D. Estradiol increases mucus synthesis in bronchial epithelial cells. PLoS ONE 2014, 9, e100633. [Google Scholar] [CrossRef]
- Tam, A.; Hughes, M.; McNagny, K.M.; Obeidat, M.; Hackett, T.L.; Leung, J.M.; Shaipanich, T.; Dorscheid, D.R.; Singhera, G.K.; Yang, C.W.T.; et al. Hedgehog signaling in the airway epithelium of patients with chronic obstructive pulmonary disease. Sci. Rep. 2019, 9, 3353. [Google Scholar] [CrossRef]
- Osei, E.T.; Mostaço-Guidolin, L.B.; Hsieh, A.; Warner, S.M.; Al-Fouadi, M.; Wang, M.; Cole, D.J.; Maksym, G.N.; Hallstrand, T.S.; Timens, W.; et al. Epithelial-interleukin-1 inhibits collagen formation by airway fibroblasts: Implications for asthma. Sci. Rep. 2020, 10, 8721. [Google Scholar] [CrossRef]
- Hackett, T.L.; Singhera, G.K.; Shaheen, F.; Hayden, P.; Jackson, G.R.; Hegele, R.G.; Van Eeden, S.; Bai, T.R.; Dorscheid, D.R.; Knight, D.A. Intrinsic phenotypic differences of asthmatic epithelium and its inflammatory responses to respiratory syncytial virus and air pollution. Am. J. Respir. Cell Mol. Biol. 2011, 45, 1090–1100. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Goodrich, L.V.; Milenkovic, L.; Higgins, K.M.; Scott, M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997, 277, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Gold, M.J.; Antignano, F.; Halim, T.Y.; Hirota, J.A.; Blanchet, M.R.; Zaph, C.; Takei, F.; McNagny, K.M. Group 2 innate lymphoid cells facilitate sensitization to local, but not systemic, TH2-inducing allergen exposures. J. Allergy Clin. Immunol. 2014, 133, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, H.A.; Cool, C.; Szefler, S.J.; Covar, R.; Brugman, S.; Gelfand, E.W.; Spahn, J.D. Histopathology of severe childhood asthma: A case series. Chest 2003, 124, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Minafra, L.; Bravata, V.; Forte, G.I.; Cammarata, F.P.; Gilardi, M.C.; Messa, C. Gene expression profiling of epithelial-mesenchymal transition in primary breast cancer cell culture. Anticancer. Res. 2014, 34, 2173–2183. [Google Scholar]
- Le Cras, T.D.; Acciani, T.H.; Mushaben, E.M.; Kramer, E.L.; Pastura, P.A.; Hardie, W.D.; Korfhagen, T.R.; Sivaprasad, U.; Ericksen, M.; Gibson, A.M.; et al. Epithelial EGF receptor signaling mediates airway hyperreactivity and remodeling in a mouse model of chronic asthma. Am. J. Physiol. Lung. Cell Mol. Physiol. 2011, 300, L414–L421. [Google Scholar] [CrossRef]
- Sharma, P.; Yi, R.; Nayak, A.P.; Wang, N.; Tang, F.; Knight, M.J.; Pan, S.; Oliver, B.; Deshpande, D.A. Bitter Taste Receptor Agonists Mitigate Features of Allergic Asthma in Mice. Sci. Rep. 2017, 7, 46166. [Google Scholar] [CrossRef]
- Zou, Y.; Song, W.; Zhou, L.; Mao, Y.; Hong, W. House dust mite induces Sonic hedgehog signaling that mediates epithelialmesenchymal transition in human bronchial epithelial cells. Mol. Med. Rep. 2019, 20, 4674–4682. [Google Scholar] [CrossRef]
- Cigna, N.; Farrokhi Moshai, E.; Brayer, S.; Marchal-Somme, J.; Wemeau-Stervinou, L.; Fabre, A.; Mal, H.; Leseche, G.; Dehoux, M.; Soler, P.; et al. The hedgehog system machinery controls transforming growth factor-beta-dependent myofibroblastic differentiation in humans: Involvement in idiopathic pulmonary fibrosis. Am. J. Pathol. 2012, 181, 2126–2137. [Google Scholar] [CrossRef]
- Hu, B.; Liu, J.; Wu, Z.; Liu, T.; Ullenbruch, M.R.; Ding, L.; Henke, C.A.; Bitterman, P.B.; Phan, S.H. Reemergence of hedgehog mediates epithelial-mesenchymal crosstalk in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2015, 52, 418–428. [Google Scholar] [CrossRef]
- Cao, H.; Chen, X.; Hou, J.; Wang, C.; Xiang, Z.; Shen, Y.; Han, X. The Shh/Gli signaling cascade regulates myofibroblastic activation of lung-resident mesenchymal stem cells via the modulation of Wnt10a expression during pulmonary fibrogenesis. Lab. Investig. 2020, 100, 363–377. [Google Scholar] [CrossRef]
- Miller, L.A.; Wert, S.E.; Clark, J.C.; Xu, Y.; Perl, A.K.; Whitsett, J.A. Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Dev. Dyn. 2004, 231, 57–71. [Google Scholar] [CrossRef]
- Moshai, E.F.; Wemeau-Stervinou, L.; Cigna, N.; Brayer, S.; Somme, J.M.; Crestani, B.; Mailleux, A.A. Targeting the hedgehog-glioma-associated oncogene homolog pathway inhibits bleomycin-induced lung fibrosis in mice. Am. J. Respir. Cell Mol. Biol. 2014, 51, 11–25. [Google Scholar] [CrossRef]
- Arensdorf, A.M.; Marada, S.; Ogden, S.K. Smoothened Regulation: A Tale of Two Signals. Trends Pharmacol. Sci. 2016, 37, 62–72. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tam, A.; Osei, E.T.; Cheung, C.Y.; Hughes, M.; Yang, C.X.; McNagny, K.M.; Dorscheid, D.R.; Singhera, G.K.; Hallstrand, T.S.; Warner, S.; et al. Hedgehog Signaling as a Therapeutic Target for Airway Remodeling and Inflammation in Allergic Asthma. Cells 2022, 11, 3016. https://doi.org/10.3390/cells11193016
Tam A, Osei ET, Cheung CY, Hughes M, Yang CX, McNagny KM, Dorscheid DR, Singhera GK, Hallstrand TS, Warner S, et al. Hedgehog Signaling as a Therapeutic Target for Airway Remodeling and Inflammation in Allergic Asthma. Cells. 2022; 11(19):3016. https://doi.org/10.3390/cells11193016
Chicago/Turabian StyleTam, Anthony, Emmanuel Twumasi Osei, Chung Y. Cheung, Michael Hughes, Chen X. Yang, Kelly M. McNagny, Delbert R. Dorscheid, Gurpreet K. Singhera, Teal S. Hallstrand, Stephanie Warner, and et al. 2022. "Hedgehog Signaling as a Therapeutic Target for Airway Remodeling and Inflammation in Allergic Asthma" Cells 11, no. 19: 3016. https://doi.org/10.3390/cells11193016
APA StyleTam, A., Osei, E. T., Cheung, C. Y., Hughes, M., Yang, C. X., McNagny, K. M., Dorscheid, D. R., Singhera, G. K., Hallstrand, T. S., Warner, S., Hogg, J. C., Hackett, T. L., Lim, C. J., & Sin, D. D. (2022). Hedgehog Signaling as a Therapeutic Target for Airway Remodeling and Inflammation in Allergic Asthma. Cells, 11(19), 3016. https://doi.org/10.3390/cells11193016





