Retinoids Promote Mouse Bone Marrow-Derived Macrophage Differentiation and Efferocytosis via Upregulating Bone Morphogenetic Protein-2 and Smad3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals
2.3. Bone Marrow-Derived Macrophage Cell Culture and Treatment
2.4. Generation of Apoptotic Thymocytes
2.5. mRNA Sequencing
2.6. Functional Analysis of Differentially Expressed Genes (DEGs)
2.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis of mRNA Expressions
2.8. Efferocytosis Assays
2.9. Fluorescent Microscopy
2.10. Statistical Analysis
3. Results
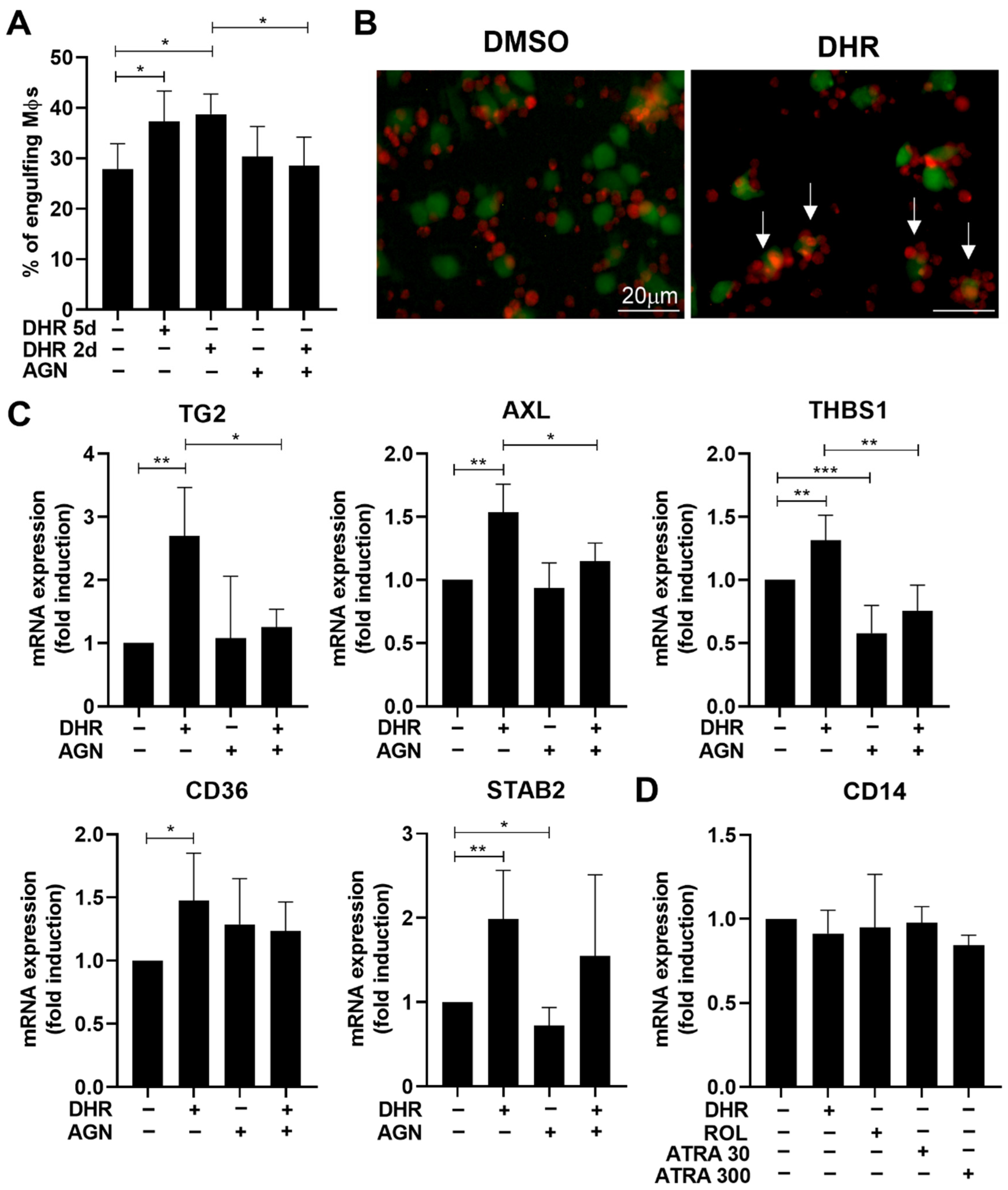
3.1. Dihydroretinol Administered during Monocyte Differentiation Enhances Efferocytosis of Macrophages by Upregulating the Expression of Several Efferocytosis-Related Molecules
3.2. DHR Administered during Monocyte Differentiation Promotes Efferocytosis via Directly Affecting RARs
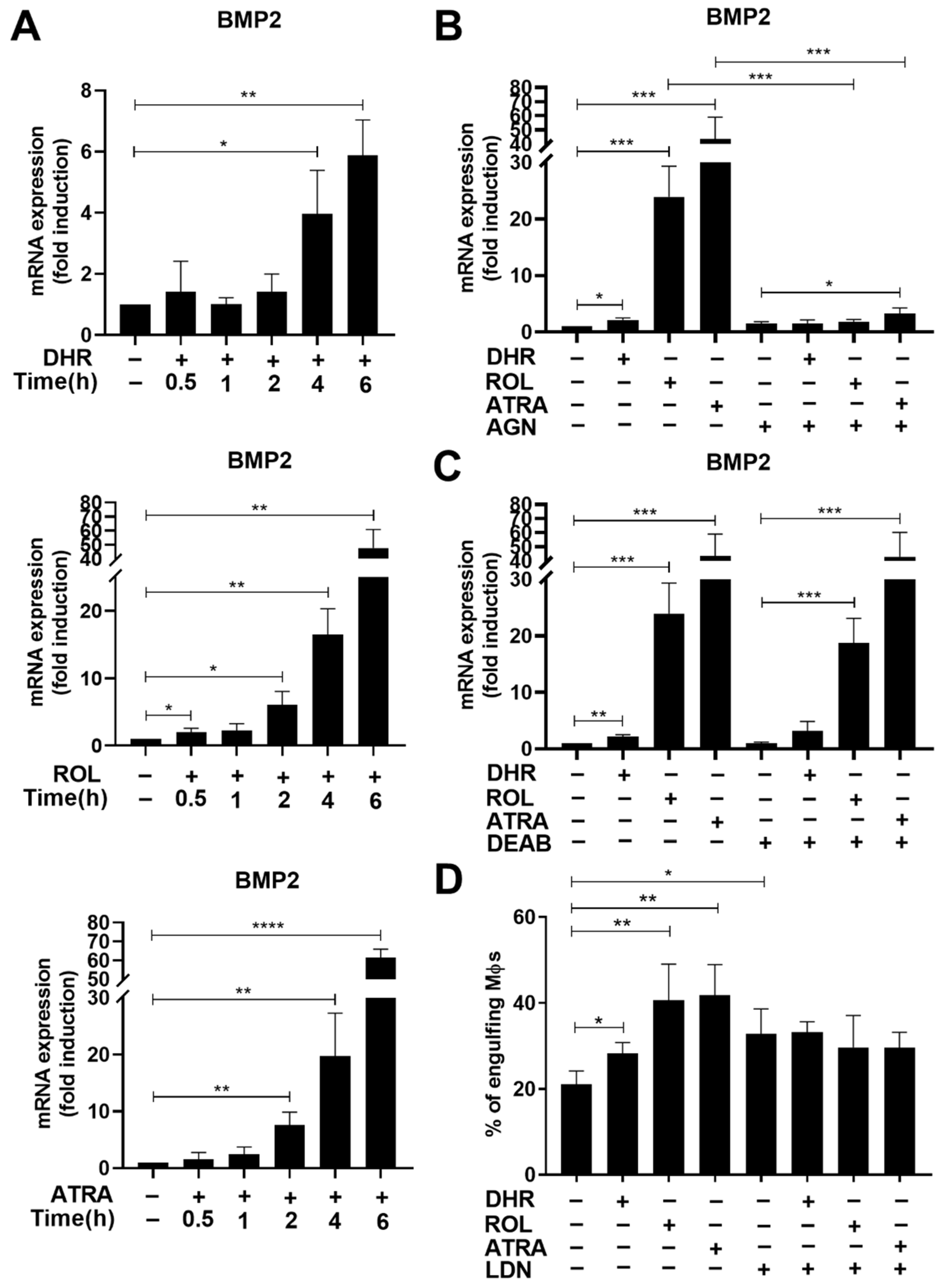
3.3. Retinoids Added from Day 4 of Monocyte Differentiation Induce the Expression of Bone Morphogenetic Protein 2 (BMP-2)
3.4. BMP-2 Contributes to the Retinoid-Induced Efferocytic Capacity in Differentiating Macrophages
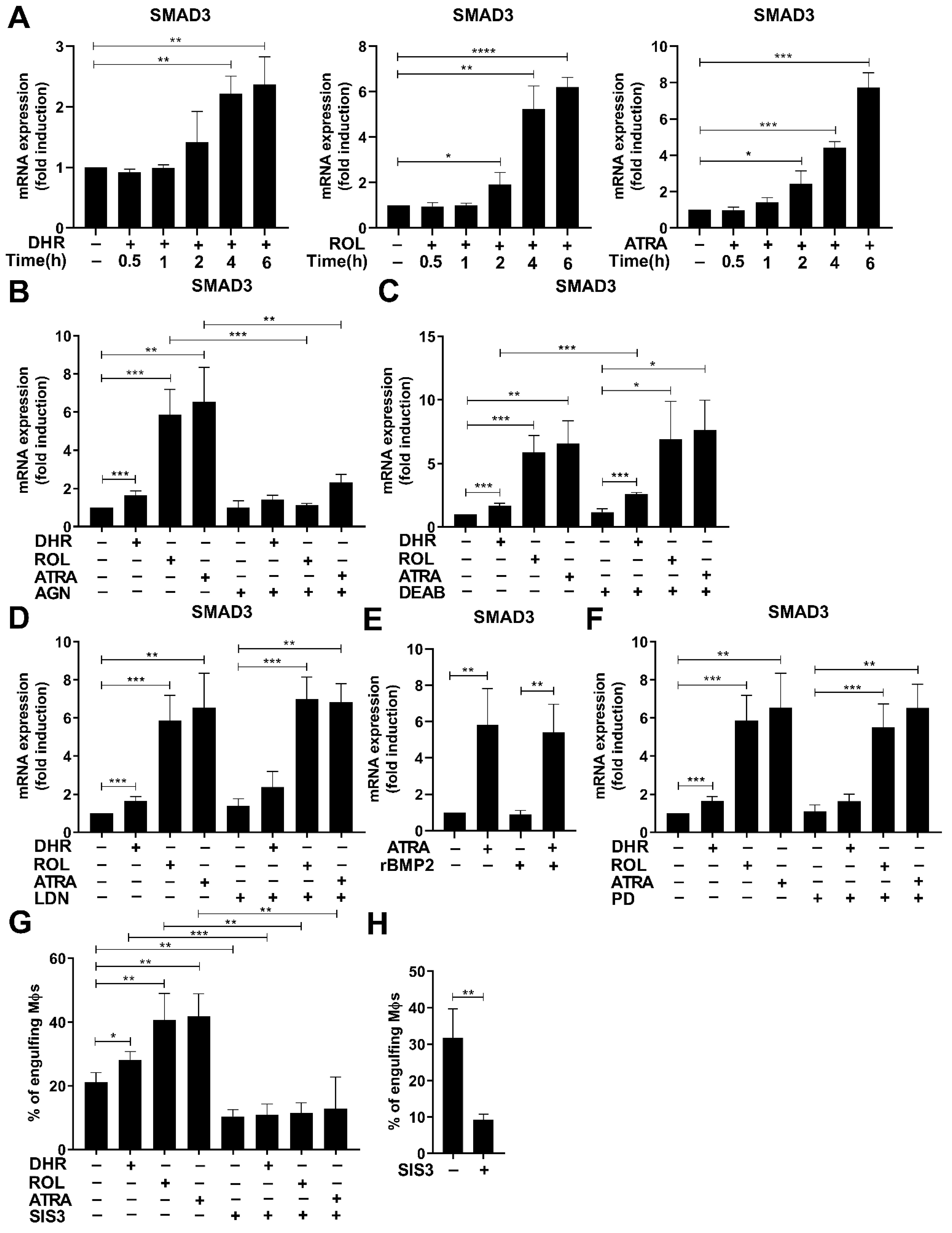
3.5. Smad3 Is also Induced by Retinoids
3.6. Smad3 also Contributes to the Retinoid-Induced Efferocytosis during Monocyte Differentiation
3.7. Retinoids also Upregulate the Expression of Vascular Endothelial Growth Factor (VEGF) A
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arandjelovic, S.; Ravichandran, K.S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 2015, 16, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Voelker, D.R.; Campbell, P.A.; Cohen, J.J.; Bratton, D.L.; Henson, P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992, 48, 2207–2216. [Google Scholar]
- Park, D.; Tosello-Trampont, A.C.; Elliott, M.R.; Lu, M.; Haney, L.B.; Ma, Z.; Klibanov, A.L.; Mandell, J.W.; Ravichandran, K.S. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007, 450, 430–434. [Google Scholar] [CrossRef]
- Park, S.Y.; Jung, M.Y.; Kim, H.J.; Lee, S.J.; Kim, S.Y.; Lee, B.H.; Kwon, T.H.; Park, R.W.; Kim, I.S. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008, 15, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Miyanishi, M.; Tada, K.; Koike, M.; Uchiyama, Y.; Kitamura, T.; Nagata, S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Hanayama, R.; Tanaka, M.; Miwa, K.; Shinohara, A.; Iwamatsu, A.; Nagata, S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002, 417, 182–187. [Google Scholar] [CrossRef]
- Savill, J.S.; Hogg, N.; Ren, Y.; Haslett, C. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Investig. 1992, 90, 1513–1522. [Google Scholar] [CrossRef]
- Stitt, T.N.; Conn, G.; Gore, M.; Lai, C.; Bruno, J.; Radziejewski, C.; Mattsson, K.; Fisher, J.; Gies, D.R.; Jones, P.F.; et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 1995, 80, 661–670. [Google Scholar] [CrossRef]
- Botto, M.; Dell’Agnola, C.; Bygrave, A.E.; Thompson, E.M.; Cook, H.T.; Petry, F.; Loos, M.; Pandolfi, P.P.; Walport, M.J. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 1998, 19, 56–59. [Google Scholar] [CrossRef]
- Park, D.; Hochreiter-Hufford, A.; Ravichandran, K.S. The phosphatidylserine receptor TIM-4 does not mediate direct signaling. Curr. Biol. 2009, 19, 346–351. [Google Scholar] [CrossRef]
- Devitt, A.; Parker, K.G.; Ogden, C.A.; Oldreive, C.; Clay, M.F.; Melville, L.A.; Bellamy, C.O.; Lacy-Hulbert, A.; Gangloff, S.C.; Goyert, S.M.; et al. Persistence of apoptotic cells without autoimmune disease or inflammation in CD14-/- mice. J. Cell. Biol. 2004, 167, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.E.; Sun, M.; Zhang, R.; Febbraio, M.; Silverstein, R.; Hazen, S.L. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J. Exp. Med. 2006, 203, 2613–2625. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.L.; Caricchio, R.; Abraham, V.; Camenisch, T.D.; Jennette, J.C.; Roubey, R.A.; Earp, H.S.; Matsushima, G.; Reap, E.A. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-Mertk membrane tyrosine kinase. J. Exp. Med. 2002, 196, 135–140. [Google Scholar] [CrossRef]
- Tóth, B.; Garabuczi, E.; Sarang, Z.; Vereb, G.; Vámosi, G.; Aeschlimann, D.; Blaskó, B.; Bécsi, B.; Erdődi, F.; Lacy-Hulbert, A.; et al. Transglutaminase 2 is needed for the formation of an efficient phagocyte portal in macrophages engulfing apoptotic cells. J. Immunol. 2009, 182, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, J.M.; Cabello, J.; Klingle, D.; Wong, K.; Freichtinger, R.; Schnabel, H.; Schnabel, R.; Hengartner, M.O. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 2005, 43, 93–99. [Google Scholar] [CrossRef]
- Rogers, N.J.; Lees, M.J.; Gabriel, L.; Maniati, E.; Rose, S.J.; Potter, P.K.; Morley, B.J. A defect in Marco expression contributes to systemic lupus erythematosus development via failure to clear apoptotic cells. J. Immunol. 2009, 182, 1982–1990. [Google Scholar] [CrossRef]
- Kanno, S.; Hirano, S.; Sakamoto, T.; Furuyama, A.; Takase, H.; Kato, H.; Fukuta, M.; Aoki, Y. Scavenger receptor MARCO contributes to cellular internalization of exosomes by dynamin-dependent endocytosis and micropinocytosis. Sci. Rep. 2020, 10, 21795. [Google Scholar] [CrossRef]
- Morioka, S.; Maueröder, C.; Ravichandran, K.S. Living on the Edge: Efferocytosis at the Interface of Homeostasis and Pathology. Immunity 2019, 50, 1149–1162. [Google Scholar] [CrossRef]
- Chazaud, B. Inflammation and skeletal muscle regeneration: Leave it to the macrophages. Trends Immunol. 2020, 6, 481–492. [Google Scholar] [CrossRef]
- Moise, A.R.; Domínguez, M.; Alvarez, S.; Alvarez, R.; Schupp, M.; Cristancho, A.G.; Kiser, P.D.; de Lera, A.R.; Lazar, M.A.; Palczewski, K. Stereospecificity of retinol saturase: Absolute configuration, synthesis, and biological evaluation of dihydroretinoids. J. Am. Chem. Soc. 2008, 130, 1154–1155. [Google Scholar] [CrossRef]
- Sarang, Z.; Sághy, T.; Budai, Z.; Újlaky-Nagy, L.; Bedekovics, J.; Beke, L.; Méhes, G.; Nagy, G.; Moise, A.R.; Palczewski, K.; et al. Retinol Saturase Knock Out Mice are Characterized with Impaired Clearance of Apoptotic Cells and Develop Mild Autoimmunity. Biomolecules 2019, 9, 737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moise, A.R.; Alvarez, S.; Domínguez, M.; Alvarez, R.; Golczak, M.; Lobo, G.P.; von Lintig, J.; de Lera, A.R.; Palczewski, K. Activation of retinoic acid receptors by dihydroretinoids. Mol. Pharmacol. 2009, 76, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Sarang, Z.; Joós, G.; Garabuczi, É.; Rühl, R.; Gregory, C.D.; Szondy, Z. Macrophages engulfing apoptotic cells produce nonclassical retinoids to enhance their phagocytic capacity. J. Immunol. 2014, 192, 5730–5738. [Google Scholar] [CrossRef] [PubMed]
- Moise, A.R.; Lobo, G.P.; Erokwu, B.; Wilson, D.L.; Peck, D.; Alvarez, S.; Domínguez, M.; Alvarez, R.; Flask, C.A.; de Lera, A.R.; et al. Increased adiposity in the retinol saturase-knockout mouse. FASEB J. 2010, 24, 1261–1270. [Google Scholar] [CrossRef]
- Heidenreich, S.; Witte, N.; Weber, P.; Goehring, I.; Tolkachov, A.; von Loeffelholz, C.; Döcke, S.; Bauer, M.; Stockmann, M.; Pfeiffer, A.F.H.; et al. Retinol saturase coordinates liver metabolism by regulating ChREBP activity. Nat. Commun. 2017, 8, 384. [Google Scholar] [CrossRef]
- Available online: http://www.bowdish.ca/lab/wp-content/uploads/2018/12/L929-CONDITIONED-MEDIUM-LCM-Protocol-.pdf (accessed on 9 August 2022).
- Köröskényi, K.; Duró, E.; Pallai, A.; Sarang, Z.; Kloor, D.; Ucker, D.S.; Beceiro, S.; Castrillo, A.; Chawla, A.; Ledent, C.A.; et al. Involvement of Adenosine A2A Receptors in Engulfment-Dependent Apoptotic Cell Suppression of Inflammation. J. Immunol. 2011, 186, 7144–7155. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Taefehshokr, N.; Yin, C.; Heit, B. Rab GTPases in the differential processing of phagocytosed pathogens versus efferocytosed apoptotic cells. Histol. Histopathol. 2021, 36, 123–135. [Google Scholar]
- Richter, E.; Ventz, K.; Harms, M.; Mostertz, J.; Hochgräfe, F. Induction of Macrophage Function in Human THP-1 Cells Is Associated with Rewiring of MAPK Signaling and Activation of MAP3K7 (TAK1) Protein Kinase. Front. Cell. Dev. Biol. 2016, 4, 21. [Google Scholar] [CrossRef]
- Woods, A.; Dickerson, K.; Heath, R.; Hong, S.P.; Momcilovic, M.; Johnstone, S.R.; Carlson, M.; Carling, D. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005, 2, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.B.; Zmijewski, J.W.; Deshane, J.S.; Tadie, J.M.; Chapli, D.D.; Takashima, S.; Abraham, E. AMP-activated protein kinase enhances the phagocytic ability of macrophages and neutrophils. FASEB J. 2011, 25, 4358–4368. [Google Scholar] [CrossRef] [PubMed]
- Marques-da-Silva, C.; Burnstock, G.; Ojcius, D.M.; Coutinho-Silva, R. Purinergic receptor agonists modulate phagocytosis and clearance of apoptotic cells in macrophages. Immunobiology 2011, 216, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Huang, S.; Su, Y.; Wu, Y.J.; Hanna, A.; Brickshawana, A.; Graff, J.; Frangogiannis, H.G. Macrophage Smad3 protects the infarcted heart, stimulating phagocytosis and regulating inflammation. Circ. Res. 2019, 125, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Napoli, J.L. Retinoic acid synthesis and metabolism. Prog. Nucleic Acid Res. Mol. Biol. 1999, 63, 139–188. [Google Scholar]
- Chen, J.Y.; Penco, S.; Ostrowski, J.; Balaguer, P.; Pons, M.; Starret, J.E.; Reczek, P.; Chambon, P.; Gronemeyer, H. RAR-specific agonist/antagonists which dissociate transactivation and AP1 transrepression inhibit anchorage-independent cell proliferation. EMBO J. 1995, 14, 1187–1197. [Google Scholar] [CrossRef]
- Tóth, K.; Sarang, Z.; Scholtz, B.; Brázda, P.; Ghyselinck, N.; Chambon, P.; Fésüs, L.; Szondy, Z. Retinoids enhance glucocorticoid-induced apoptosis of T cells by facilitating glucocorticoid receptor-mediated transcription. Cell Death Differ. 2011, 18, 783–792. [Google Scholar] [CrossRef]
- Repa, J.J.; Hanson, K.K.; Clagett-Dame, M. All-trans-retinol is a ligand for the retinoic acid receptors. Proc. Natl. Acad. Sci. USA 1993, 90, 7293–7297. [Google Scholar] [CrossRef]
- Heller, L.C.; Li, Y.; Abrams, K.L.; Rogers, M.B. Transcriptional regulation of the Bmp2 gene. Retinoic acid induction in F9 embryonal carcinoma cells and Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 1394–1400. [Google Scholar] [CrossRef]
- Gonzalez-Junca, A.; Driscoll, K.E.; Pellicciotta, I.; Du, S.; Lo, C.H.; Roy, R.; Parry, R.; Tenvooren, I.; Marquez, D.M.; Spitzer, M.H.; et al. Autocrine TGFβ is a Survival Factor for Monocytes and Drives Immunosuppressive Lineage Commitment. Cancer Immunol. Res. 2019, 7, 306–320. [Google Scholar] [CrossRef]
- Ross, K.R.; Corey, D.A.; Dunn, J.M.; Kelley, T.J. SMAD3 expression is regulated by mitogen-activated protein kinase kinase-1 in epithelial and smooth muscle cells. Cell Signal. 2007, 19, 923–931. [Google Scholar] [CrossRef]
- Gunasekar, P.; Swier, V.J.; Fleegel, J.P.; Boosani, C.S.; Radwan, M.M.; Agrawal, D.K. Vitamin D and Macrophage Polarization in Epicardial Adipose Tissue of Atherosclerotic Swine. PLoS ONE 2018, 13, e0199411. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Zajd, C.M.; Ziemba, A.M.; Miralles, G.M.; Nguyen, T.; Feustel, P.J.; Dunn, S.M.; Gilbert, R.J.; Lennartz, M.R. Bone Marrow-Derived and Elicited Peritoneal Macrophages Are Not Created Equal: The Questions Asked Dictate the Cell Type Used. Front. Immunol. 2020, 11, 269. [Google Scholar] [CrossRef] [Green Version]
- Loi, F.; Córdova, L.A.; Pajarinen, J.; Lin, T.H.; Yao, Z.; Goodman, S.B. Inflammation, fracture and bone repair. Bone 2016, 86, 119. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiang, H.; Zheng, C.; Gu, M.; Zhen, X. Secretion of BMP-2 by tumor-associated macrophages (TAM) promotes microcalcifications in breast cancer. BMC Cancer 2022, 22, 34. [Google Scholar] [CrossRef]
- Unsworth, A.J.; Flora, G.D.; Gibbins, J.M. Non-genomic effects of nuclear receptors: Insights from the anucleate platelet. Cardiovasc. Res. 2018, 114, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, É.; Lala-Tabbert, N.; Gunanayagam, A.; St-Louis, C.; Wiper-Bergeron, N. Retinoic acid promotes myogenesis in myoblasts by antagonizing transforming growth factor-beta signaling via C/EBPβ. Skelet. Muscle 2015, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.L.; Chen, Z.H.; Teng, Y.Y.; Liu, S.Y.; Jia, Y.; Zhang, K.W.; Sun, Z.L.; Wu, J.J.; Yuan, Z.D.; Feng, Y.; et al. The Smad Dependent TGF-β and BMP Signaling Pathway in Bone Remodeling and Therapies. Front. Mol. Biosci. 2021, 8, 593310. [Google Scholar] [CrossRef]
- Holtzhausen, A.; Golzio, C.; How, T.; Lee, Y.H.; Schiemann, W.P.; Katsanis, N.; Blobe, G.C. Novel bone morphogenetic protein signaling through Smad2 and Smad3 to regulate cancer progression and development. FASEB J. 2014, 28, 1248–1267. [Google Scholar] [CrossRef]
- Patsalos, A.; Halász, L.; Medina-Serpas, M.A.; Berger, W.K.; Dániel, B.; Tzerpos, P.; Kiss, M.; Nagy, G.; Fischer, C.; Simándi, Z.; et al. A growth factor expressing macrophage subpopulation orchestrates regenerative inflammation via GDF-15. J. Exp. Med. 2021, 219, e20210420. [Google Scholar] [CrossRef]
- Li, L.; Song, J.; Chuquisana, O.; Hannocks, M.J.; Loismann, S.; Vogl, T.; Roth, J.; Hallmann, R.; Sorokin, L. Endothelial Basement Membrane Laminins as an Environmental Cue in Monocyte Differentiation to Macrophages. Front. Immunol. 2020, 11, 584229. [Google Scholar] [CrossRef] [PubMed]
- Francke, A.; Herold, J.; Weinert, S.; Strasser, R.H.; Braun-Dullaeus, R.C. Generation of mature murine monocytes from heterogeneous bone marrow and description of their properties. J. Histochem. Cytochem. 2011, 59, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Szondy, Z.; Al-Zaeed, N.; Tarban, N.; Fige, E.; Garabuczi, E.; Sarang, Z. Involvement of phosphatidylserine receptors in skeletal muscle regeneration. Therapeutic implications. J. Cachexia Sarcopenia Muscle 2022, 3, 1961–1973. [Google Scholar] [CrossRef]
- Cinti, S.; Mitchell, G.; Barbatelli, G.; Murano, I.; Ceresi, E.; Faloia, E.; Wang, S.; Fortier, M.; Greenberg, A.S.; Obin, M.S. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid. Res. 2005, 46, 2347–2355. [Google Scholar] [CrossRef] [PubMed]
- Szondy, Z.; Garabuczi, E.; Joós, G.; Tsay, G.J.; Sarang, Z. Impaired clearance of apoptotic cells in chronic inflammatory diseases: Therapeutic implications. Front. Immunol. 2014, 5, 354. [Google Scholar] [CrossRef] [PubMed]
- Werner, F.; Jain, M.K.; Feinberg, M.W.; Sibinga, N.; Pellacani, A.; Wiesel, P.; Chin, M.T.; Topper, J.N.; Perrella, M.A.; Lee, M.E. Transforming Growth Factor-β1 Inhibition of Macrophage Activation Is Mediated via Smad3. J. Biol. Chem. 2000, 275, 36653–36658. [Google Scholar] [CrossRef]
- Fadok, V.A.; Bratton, D.L.; Konowal, A.; Freed, P.W.; Westcott, J.Y.; Henson, P.M. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Investig. 1998, 101, 890–898. [Google Scholar] [CrossRef]
- He, L.; Jhong, J.H.; Chen, Q.; Huang, K.Y.; Strittmatter, K.; Kreuzer, J.; DeRan, M.; Wu, X.; Lee, T.Y.; Slavov, N.; et al. Global characterization of macrophage polarization mechanisms and identification of M2-type polarization inhibitors. Cell Rep. 2021, 37, 109955. [Google Scholar] [CrossRef]
- Chen, B.; Li, R.; Hernandez, S.C.; Hanna, A.; Su, K.; Shinde, A.V.; Frangogiannis, N.G. Differential effects of Smad2 and Smad3 in regulation of macrophage phenotype and function in the infarcted myocardium. J. Mol. Cell. Cardiol. 2022, 171, 1–15. [Google Scholar] [CrossRef]
- Devalaraja, S.; To, T.K.J.; Folkert, I.W.; Natesan, R.; Alam, M.Z.; Li, M.; Tada, Y.; Budagyan, K.; Dang, M.T.; Zhai, L.; et al. Tumor-Derived Retinoic Acid Regulates Intratumoral Monocyte Differentiation to Promote Immune Suppression. Cell 2020, 180, 1098–1114. [Google Scholar] [CrossRef]
- Vellozo, N.S.; Pereira-Marques, S.T.; Cabral-Piccin, M.P.; Filardy, A.A.; Ribeiro-Gomes, F.L.; Rigoni, T.S.; DosReis, G.A.; Lopes, M.F. All-Trans. Retinoic Acid Promotes an M1- to M2-Phenotype Shift and Inhibits Macrophage-Mediated Immunity to Leishmania major. Front. Immunol. 2017, 8, 1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Corr. p Val. | FC | Gene Symbol | Gene Title |
|---|---|---|---|
| 1.21 × 10−6 | 212.53 | Camkk1 | calcium/calmodulin-dependent protein kinase 1, alpha |
| 1.73 × 10−3 | 8.35 | P2rx1 | purinergic receptor P2X, ligand-gated ion channel, 1 |
| 1.04 × 10−7 | 4.1 | Smad3 | SMAD family member 3 |
| 4.38 × 10−3 | 3.65 | Marco | macrophage receptor with collagenous structure |
| 1.34 × 10−7 | 3.59 | Rab20 | RAB20, member RAS oncogene family |
| 1.07 × 10−3 | 3.21 | Stab2 | stabilin 2 |
| 4.88 × 10−4 | 1.99 | THBS-1 | thrombospondin 1 |
| 6.24 × 10−8 | 1.58 | Tgm2 | transglutaminase 2, C polypeptide |
| 5.23 × 10−3 | 1.56 | Axl | AXL receptor tyrosine kinase |
| 1.43 × 10−4 | 1.53 | CD36 | CD36 antigen |
| Upregulated Transcripts | |||
|---|---|---|---|
| Corr. p Val. | FC | Gene Symbol | Gene Title |
| 1.8 × 10−8 | 30.3 | Hic1 | hypermethylated in cancer 1 |
| 1.2 × 10−6 | 25.0 | Camkk1 | calcium/calmodulin-dependent protein kinase 1, alpha |
| 2.4 × 10−3 | 10.8 | Gm15927 | predicted gene 15927 |
| 8.9 × 10−8 | 7.6 | Bmp2 | bone morphogenetic protein 2 |
| 2.1 × 10−5 | 7.4 | B230378P21Rik | RIKEN cDNA B230378P21 gene |
| 3.2 × 10−6 | 6.1 | Art2a-ps | ADP-ribosyltransferase 2a, pseudogene |
| 2.4 × 10−3 | 5.1 | Dll1 | delta-like 1 (Drosophila) |
| 2.8 × 10−5 | 5.0 | Tox3 | TOX high mobility group box family member 3 |
| 1.6 × 10−3 | 4.8 | Fam20a | family with sequence similarity 20, member A |
| 4.3 × 10−5 | 4.5 | Kcnip3 | Kv channel interacting protein 3, calsenilin |
| 6.1 × 10−3 | 4.2 | AI848285 | expressed sequence AI848285 |
| 3.4 × 10−4 | 4.1 | Kcng1 | potassium voltage-gated channel, subfamily G, member 1 |
| 2.9 × 10−4 | 3.9 | Il2rb | interleukin 2 receptor, beta chain |
| 1.5 × 10−2 | 3.3 | Vash1 | vasohibin 1 |
| 2.0 × 10−7 | 3.2 | Gm13431 | predicted gene 13431 |
| 5.4 × 10−3 | 3.1 | Bfsp1 | beaded filament structural protein 1, in lens-CP94 |
| 8.6 × 10−5 | 3.1 | Hbegf | heparin-binding EGF-like growth factor |
| 4.9 × 10−8 | 3.1 | Pram1 | PML-RAR alpha-regulated adaptor molecule 1 |
| 9.6 × 10−3 | 2.9 | Fam124a | family with sequence similarity 124, member A |
| 5.9 × 10−5 | 2.8 | Shcbp1l | Shc SH2-domain binding protein 1-like |
| 1.2 × 10−6 | 2.8 | Nppa | natriuretic peptide type A |
| 2.1 × 10−7 | 2.8 | Hivep2 | human immunodeficiency virus type I enhancer binding protein 2 |
| 3.8 × 10−5 | 2.7 | Robo3 | roundabout homolog 3 (Drosophila) |
| 9.1 × 10−7 | 2.7 | Vegfa | vascular endothelial growth factor A |
| 3.2 × 10−3 | 2.7 | Gm9733 | predicted gene 9733 |
| 1.0 × 10−7 | 2.7 | Smad3 | SMAD family member 3 |
| 8.3 × 10−4 | 2.6 | Tubb3 | tubulin, beta 3 class III |
| 5.4 × 10−3 | 2.5 | Gm7148 | predicted gene 7148 |
| 1.7 × 10−4 | 2.3 | Btnl4 | butyrophilin-like 4 |
| 3.4 × 10−3 | 2.3 | Rpsa-ps3 | ribosomal protein SA, pseudogene 3 |
| 6.5 × 10−8 | 2.3 | Osgin1 | oxidative stress induced growth inhibitor 1 |
| 8.8 × 10−9 | 2.2 | Dtx4 | deltex 4 homolog (Drosophila) |
| 1.3 × 10−10 | 2.2 | Ptgs1 | prostaglandin-endoperoxide synthase 1 |
| 1.3 × 10−7 | 2.2 | Rab20 | RAB20, member RAS oncogene family |
| 4.8 × 10−3 | 2.2 | Gm11870 | predicted gene 11870 |
| 6.9 × 10−7 | 2.2 | Il21r | interleukin 21 receptor |
| 4.4 × 10−4 | 2.1 | Map6d1 | MAP6 domain containing 1 |
| 1.9 × 10−3 | 2.1 | Corin | corin |
| 2.6 × 10−9 | 2.1 | 2510009E07Rik | RIKEN cDNA 2510009E07 gene |
| 2.0 × 10−6 | 2.1 | Socs2 | suppressor of cytokine signaling 2 |
| 1.1 × 10−9 | 2.0 | Mcart1 | mitochondrial carrier triple repeat 1 |
| 2.2 × 10−7 | 2.0 | Asb10 | ankyrin repeat and SOCS box-containing 10 |
| 6.9 × 10−8 | 2.0 | Neurl3 | neuralized homolog 3 homolog (Drosophila) |
| 1.4 × 10−4 | 1.9 | Mex3b | mex3 homolog B (C. elegans) |
| 2.0 × 10−5 | 1.9 | Hs3st3b1 | heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| 1.3 × 10−3 | 1.9 | Gm15708 | predicted gene 15708 |
| 1.1 × 10−2 | 1.9 | Pih1d2 | PIH1 domain containing 2 |
| 1.4 × 10−5 | 1.9 | Hrh1 | histamine receptor H1 |
| 4.5 × 10−3 | 1.9 | Elmo3 | engulfment and cell motility 3 |
| 1.3 × 10−5 | 1.9 | AA467197 | expressed sequence AA467197 |
| 2.5 × 10−6 | 1.8 | Gm22 | predicted gene 22 |
| 4.5 × 10−8 | 1.8 | Fam117a | family with sequence similarity 117, member A |
| 2.1 × 10−6 | 1.8 | Dchs1 | dachsous 1 (Drosophila) |
| 9.8 × 10−7 | 1.8 | Klhl12 | kelch-like 12 (Drosophila) |
| 1.8 × 10−8 | 1.8 | Dusp5 | dual specificity phosphatase 5 |
| 6.7 × 10−3 | 1.8 | Sema3d | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| 9.6 × 10−3 | 1.7 | 1700042O10Rik | RIKEN cDNA 1700042O10 gene |
| 1.4 × 10−6 | 1.7 | Cmah | cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| 1.4 × 10−5 | 1.7 | Pilrb1 | paired immunoglobin-like type 2 receptor beta 1 |
| 1.2 × 10−4 | 1.7 | Vangl2 | vang-like 2 (van gogh, Drosophila) |
| 2.1 × 10−7 | 1.7 | Ikbke | inhibitor of kappaB kinase epsilon |
| 4.9 × 10−8 | 1.7 | Fam20c | family with sequence similarity 20, member C |
| 8.9 × 10−8 | 1.7 | Tagap | T cell activation Rho GTPase activating protein |
| 2.5 × 10−7 | 1.6 | Mafb | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| 3.9 × 10−4 | 1.6 | Vdr | vitamin D receptor |
| 5.4 × 10−7 | 1.6 | Gda | guanine deaminase |
| 5.3 × 10−5 | 1.6 | Fbxo32 | F-box protein 32 |
| 1.2 × 10−5 | 1.6 | Gm16010 | predicted gene 16010 |
| 2.0 × 10−7 | 1.6 | Bcl3 | B cell leukemia/lymphoma 3 |
| 1.9 × 10−7 | 1.6 | Hcfc2 | host cell factor C2 |
| 1.4 × 10−6 | 1.6 | Cd97 | CD97 antigen |
| 1.6 × 10−7 | 1.6 | Lfng | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| 4.2 × 10−6 | 1.5 | Aifm2 | apoptosis-inducing factor, mitochondrion-associated 2 |
| 1.8 × 10−6 | 1.5 | Spsb4 | splA/ryanodine receptor domain and SOCS box containing 4 |
| Downregulated Transcripts | |||
| Corr. P val. | FC | Gene Symbol | Gene Title |
| 2.3 × 10−3 | −4.1 | Klf5 | Kruppel-like factor 5 |
| 1.3 × 10−3 | −3.9 | Zfp831 | zinc finger protein 831 |
| 6.8 × 10−3 | −3.6 | St8sia6 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| 3.6 × 10−4 | −3.2 | Gm1564 | predicted gene 1564 |
| 9.3 × 10−5 | −3.1 | Efna1 | ephrin A1 |
| 1.2 × 10−3 | −3.0 | Plat | plasminogen activator, tissue |
| 1.0 × 10−2 | −2.9 | Ucp3 | uncoupling protein 3 (mitochondrial, proton carrier) |
| 2.6 × 10−3 | −2.7 | Havcr1 | hepatitis A virus cellular receptor 1 |
| 5.4 × 10−3 | −2.7 | Heyl | hairy/enhancer-of-split related with YRPW motif-like |
| 1.7 × 10−3 | −2.7 | Akr1b7 | aldo-keto reductase family 1, member B7 |
| 5.6 × 10−3 | −2.5 | Mir425 | microRNA 425 |
| 2.5 × 10−4 | −2.4 | Ighv6-3 | immunoglobulin heavy variable 6-3 |
| 2.5 × 10−3 | −2.2 | Cyp4f41-ps | cytochrome P450, family 4, subfamily f, polypeptide 41 pseudogene |
| 4.7 × 10−5 | −2.2 | Emr4 | EGF-like module containing, mucin-like, hormone receptor-like sequence 4 |
| 9.4 × 10−5 | −2.1 | Styk1 | serine/threonine/tyrosine kinase 1 |
| 2.0 × 10−4 | −2.1 | Gpr182 | G protein-coupled receptor 182 |
| 1.2 × 10−5 | −2.1 | U6 | U6 spliceosomal RNA |
| 1.8 × 10−4 | −2.0 | Ch25h | cholesterol 25-hydroxylase |
| 8.2 × 10−9 | −1.9 | Rnd3 | Rho family GTPase 3 |
| 1.7 × 10−2 | −1.8 | Gm6776 | predicted pseudogene 6776 |
| 1.1 × 10−2 | −1.8 | Pxdc1 | PX domain containing 1 |
| 1.2 × 10−4 | −1.8 | Socs3 | suppressor of cytokine signaling 3 |
| 1.7 × 10−6 | −1.8 | Tnf | tumor necrosis factor |
| 1.8 × 10−10 | −1.8 | Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| 3.1 × 10−6 | −1.6 | Spata13 | spermatogenesis associated 13 |
| 1.8 × 10−8 | −1.6 | Rasgef1b | RasGEF domain family, member 1B |
| 7.3 × 10−6 | −1.6 | Gpr85 | G protein-coupled receptor 85 |
| 1.5 × 10−4 | −1.6 | Rgs7bp | regulator of G-protein signaling 7 binding protein |
| 1.3 × 10−7 | −1.5 | Dusp1 | dual specificity phosphatase 1 |
| 1.2 × 10−3 | −1.5 | Tnfaip3 | tumor necrosis factor, alpha-induced protein 3 |
| 1.9 × 10−3 | −1.5 | Gm16541 | predicted gene 16541 |
| 4.7 × 10−6 | −1.5 | Tmem178 | transmembrane protein 178 |
| Corr. p Val. | FC | Gene Symbol | Gene Title |
|---|---|---|---|
| 8.93 × 10−8 | 249.6 | Bmp2 | bone morphogenetic protein 2 |
| 1.83 × 10−4 | 14.8 | Cyp26b1 | cytochrome P450, family 26, subfamily b, polypeptide 1 |
| 9.13 × 10−7 | 4.4 | Vegfa | vascular endothelial growth factor A |
| 3.76 × 10−4 | 4.1 | Aldh1a2 | aldehyde dehydrogenase family 1, subfamily A2 |
| 4.38 × 10−3 | 3.6 | Marco | macrophage receptor with collagenous structure |
| 4.85 × 10−7 | 2.8 | Clec7a | C-type lectin domain family 7, member a |
| 2.01 × 10−4 | 1.9 | Irf4 | interferon regulatory factor 4 |
| 5.56 × 10−5 | 1.7 | Siglec1 | sialic acid binding Ig-like lectin 1, sialoadhesin |
| 6.24 × 10−8 | 1.6 | Tgm2 | transglutaminase 2, C polypeptide |
| 1.43 × 10−4 | 1.5 | Cd36 | CD36 antigen |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fige, É.; Sarang, Z.; Sós, L.; Szondy, Z. Retinoids Promote Mouse Bone Marrow-Derived Macrophage Differentiation and Efferocytosis via Upregulating Bone Morphogenetic Protein-2 and Smad3. Cells 2022, 11, 2928. https://doi.org/10.3390/cells11182928
Fige É, Sarang Z, Sós L, Szondy Z. Retinoids Promote Mouse Bone Marrow-Derived Macrophage Differentiation and Efferocytosis via Upregulating Bone Morphogenetic Protein-2 and Smad3. Cells. 2022; 11(18):2928. https://doi.org/10.3390/cells11182928
Chicago/Turabian StyleFige, Éva, Zsolt Sarang, László Sós, and Zsuzsa Szondy. 2022. "Retinoids Promote Mouse Bone Marrow-Derived Macrophage Differentiation and Efferocytosis via Upregulating Bone Morphogenetic Protein-2 and Smad3" Cells 11, no. 18: 2928. https://doi.org/10.3390/cells11182928
APA StyleFige, É., Sarang, Z., Sós, L., & Szondy, Z. (2022). Retinoids Promote Mouse Bone Marrow-Derived Macrophage Differentiation and Efferocytosis via Upregulating Bone Morphogenetic Protein-2 and Smad3. Cells, 11(18), 2928. https://doi.org/10.3390/cells11182928







