Mitochondrial Genome Variants as a Cause of Mitochondrial Cardiomyopathy
Abstract
1. Introduction
1.1. Mitochondria: “The Powerhouse of the Cell”
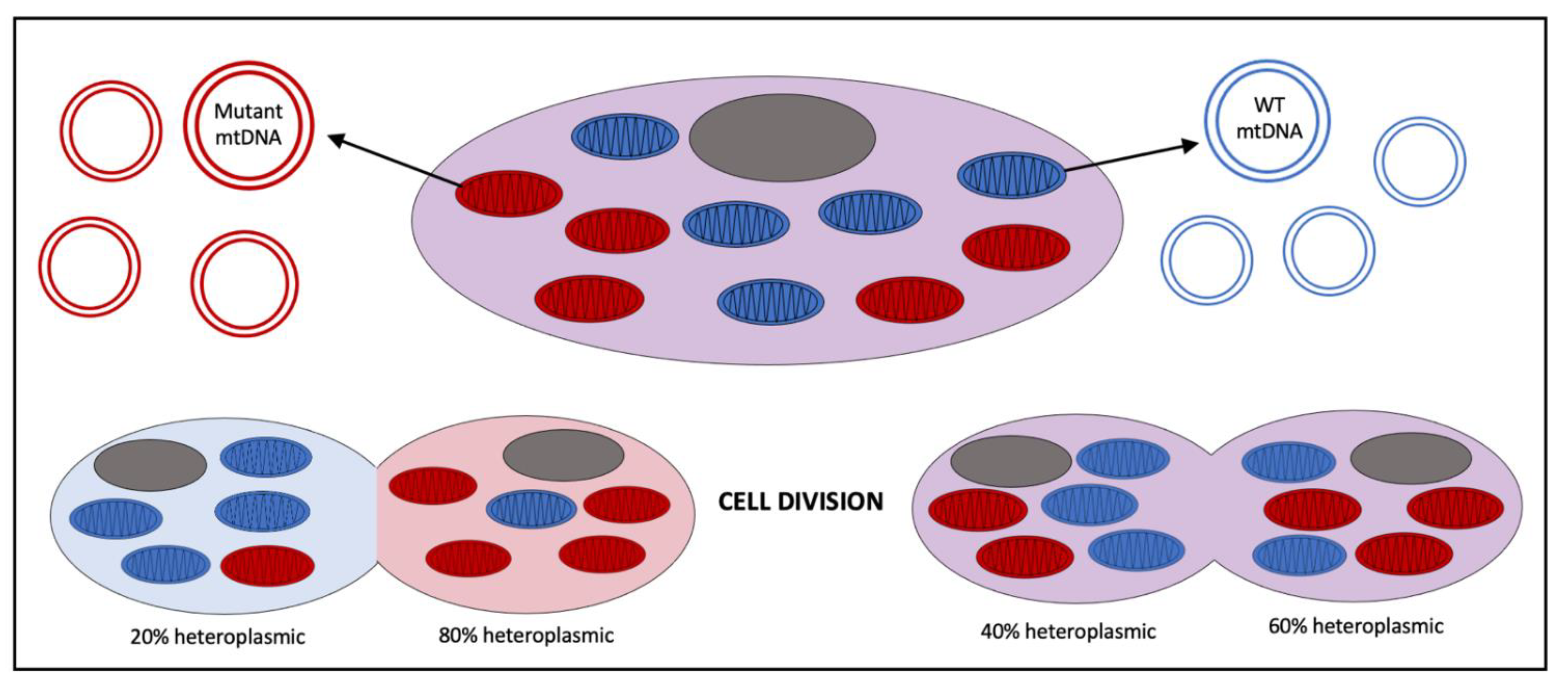
1.2. Mitochondrial Genetics
1.3. Mitochondrial Biodynamics
2. Mitochondrial Function in the Heart
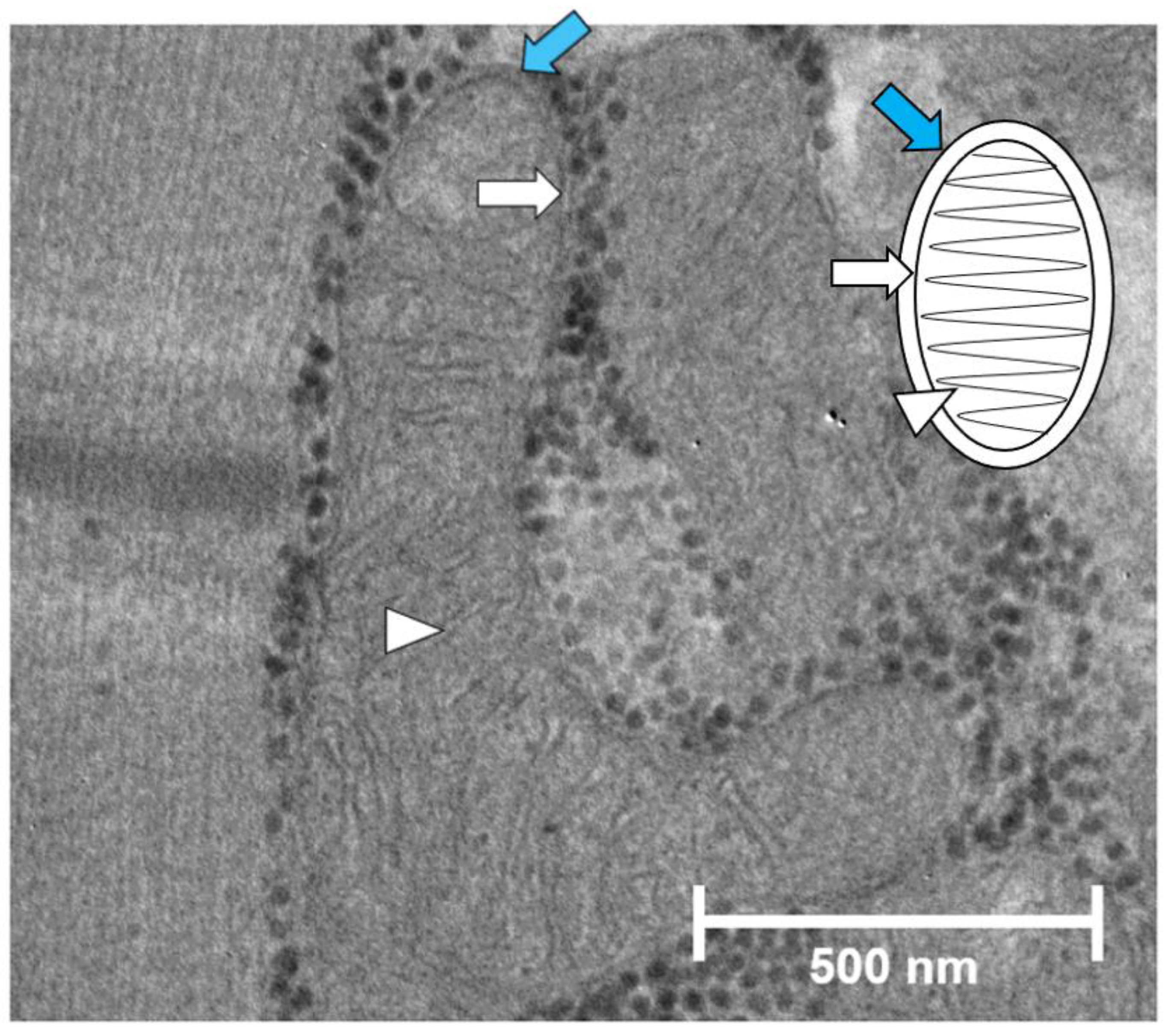
2.1. Mitochondrial Organization in the Cardiomyocytes
2.2. Ca2+ Homeostasis in the Cardiomyocytes
2.3. Iron Regulation and the Mitochondria
3. Mitochondrial Cardiomyopathy
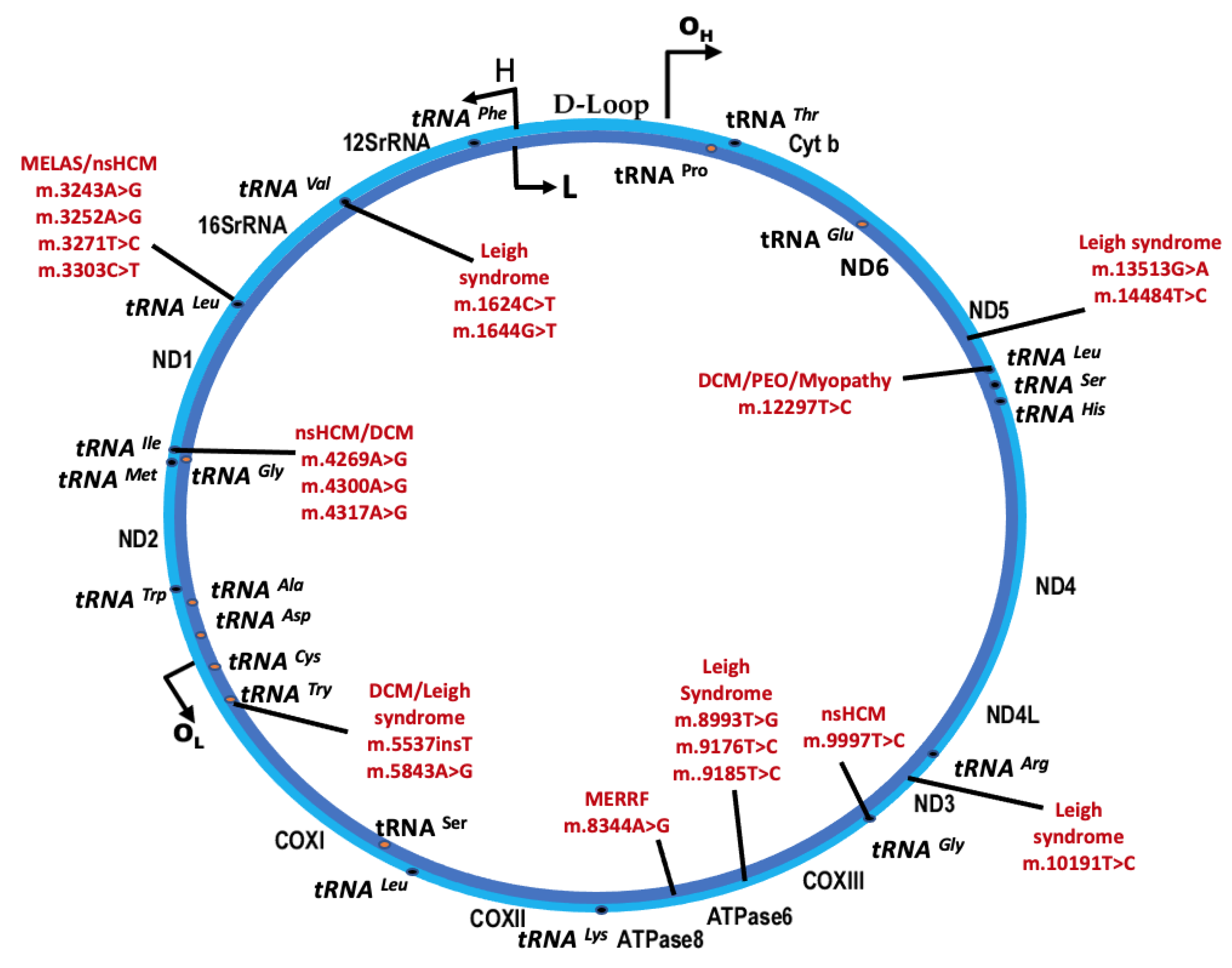
3.1. Syndromic mtDNA-Related PMD and Cardiomyopathy
3.1.1. Mitochondrial Myopathy, Encephalopathy, Lactic Acidosis, and Stroke-Like Episodes (MELAS)
3.1.2. Myoclonic Epilepsy with Ragged Red Fibers (MERRF)
3.1.3. Leigh Syndrome
3.1.4. Mitochondrial DNA Deletion Syndromes (MDDS)
3.2. Nonsyndromic mtDNA-Related Cardiomyopathy
3.2.1. Maternally Inherited Cardiomyopathy
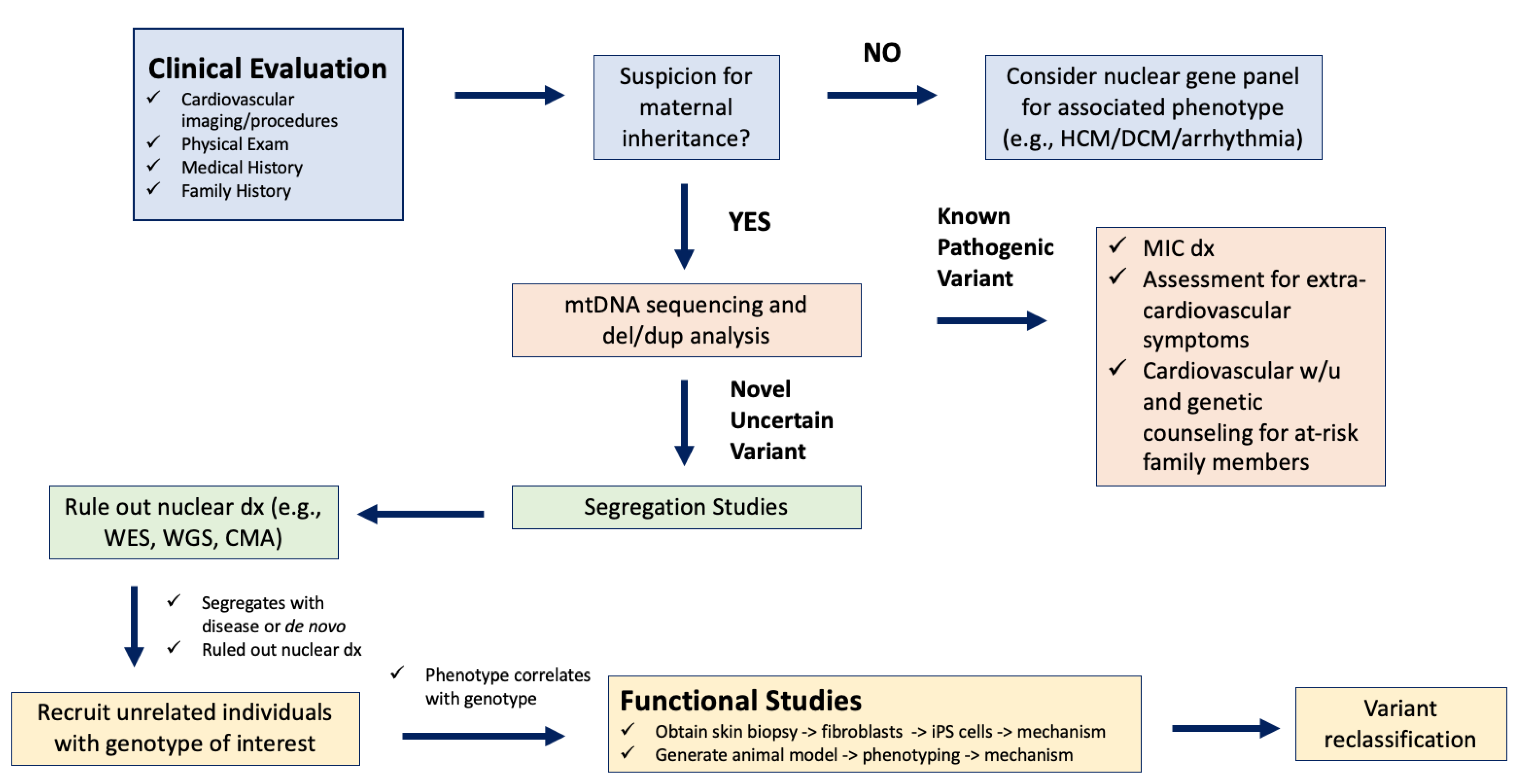
3.2.2. Clinical Assessment for MIC
3.2.3. Discovery and Classification of Novel mtDNA Variants in Association with MIC
3.3. mtDNA Variants as a Risk Factor for Multifactorial Cardiovascular Disease
4. Mechanisms of Pathophysiology
4.1. Insufficient Energy Metabolism in the Cardiomyocyte
4.2. Abnormal ROS Homeostasis
4.3. Altered Mitochondrial Dynamics
4.4. Calcium Dysregulation in the Heart
4.5. Iron Overload in the Mitochondria
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kühlbrandt, W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Ribas, V.; García-Ruiz, C.; Fernández-Checa, J.C. Glutathione and mitochondria. Front. Pharmacol. 2014, 5, 151. [Google Scholar] [CrossRef]
- Gaschler, M.M.; Andia, A.A.; Liu, H.; Csuka, J.M.; Hurlocker, B.; Vaiana, C.A.; Heindel, D.W.; Zuckerman, D.S.; Bos, P.H.; Reznik, E.; et al. FINO2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol. 2018, 14, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef] [PubMed]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2021, 595, 976–1002. [Google Scholar] [CrossRef]
- Ashar, F.N.; Zhang, Y.; Longchamps, R.J.; Lane, J.; Moes, A.; Grove, M.L.; Mychaleckyj, J.C.; Taylor, K.D.; Coresh, J.; Rotter, J.I.; et al. Association of Mitochondrial DNA Copy Number with Cardiovascular Disease. JAMA Cardiol. 2017, 2, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Jing, S.; Liu, L.; Ma, F.; Zhang, Y.; Wang, C.; Duan, H.; Zhou, K.; Hua, Y.; Wu, G.; et al. Association between mitochondrial DNA copy number and cardiovascular disease: Current evidence based on a systematic review and meta-analysis. PLoS ONE 2018, 13, e0206003. [Google Scholar] [CrossRef]
- Burgstaller, J.P.; Kolbe, T.; Havlicek, V.; Hembach, S.; Poulton, J.; Piálek, J.; Steinborn, R.; Rülicke, T.; Brem, G.; Jones, N.S.; et al. Large-scale genetic analysis reveals mammalian mtDNA heteroplasmy dynamics and variance increase through lifetimes and generations. Nat. Commun. 2018, 9, 2488. [Google Scholar] [CrossRef]
- Lee, H.R.; Johnson, K.A. Fidelity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2006, 281, 36236–36240. [Google Scholar] [CrossRef]
- Li, H.; Slone, J.; Fei, L.; Huang, T. Mitochondrial DNA Variants and Common Diseases: A Mathematical Model for the Diversity of Age-Related mtDNA Mutations. Cells 2019, 8, 608. [Google Scholar] [CrossRef]
- Yu, T.; Slone, J.; Liu, W.; Barnes, R.; Opresko, P.L.; Wark, L.; Mai, S.; Horvath, S.; Huang, T. Premature aging is associated with higher levels of 8-oxoguanine and increased DNA damage in the Polg mutator mouse. Aging Cell 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Wang, X.; Tippner-Hedges, R.; Ma, H.; Folmes, C.D.; Gutierrez, N.M.; Lee, Y.; Van Dyken, C.; Ahmed, R.; Li, Y.; et al. Age-Related Accumulation of Somatic Mitochondrial DNA Mutations in Adult-Derived Human iPSCs. Cell Stem Cell 2016, 18, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Robin, E.D.; Wong, R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol. 1988, 136, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L.; Losón, O.C.; Hellberg, K.; Young, N.P.; Chen, H.; Polleux, F.; Chan, D.C.; et al. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef]
- Burman, J.L.; Pickles, S.; Wang, C.; Sekine, S.; Vargas, J.N.S.; Zhang, Z.; Youle, A.M.; Nezich, C.L.; Wu, X.; Hammer, J.A.; et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 2017, 216, 3231–3247. [Google Scholar] [CrossRef]
- Yamano, K.; Youle, R.J. PINK1 is degraded through the N-end rule pathway. Autophagy 2013, 9, 1758–1769. [Google Scholar] [CrossRef]
- Okatsu, K.; Uno, M.; Koyano, F.; Go, E.; Kimura, M.; Oka, T.; Tanaka, K.; Matsuda, N. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 2013, 288, 36372–36384. [Google Scholar] [CrossRef]
- Morciano, G.; Patergnani, S.; Bonora, M.; Pedriali, G.; Tarocco, A.; Bouhamida, E.; Marchi, S.; Ancora, G.; Anania, G.; Wieckowski, M.R.; et al. Mitophagy in Cardiovascular Diseases. J. Clin. Med. 2020, 9, 892. [Google Scholar] [CrossRef]
- Chen, H.; Vermulst, M.; Wang, Y.E.; Chomyn, A.; Prolla, T.A.; McCaffery, J.M.; Chan, D.C. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 2010, 141, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Isobe, K.; Nakada, K.; Hayashi, J.I. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 2001, 28, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Barth, E.; Stämmler, G.; Speiser, B.; Schaper, J. Ultrastructural quantitation of mitochondria and myofilaments in cardiac muscle from 10 different animal species including man. J. Mol. Cell Cardiol. 1992, 24, 669–681. [Google Scholar] [CrossRef]
- Stanley, W.C.; Recchia, F.A.; Lopaschuk, G.D. Myocardial substrate metabolism in the normal and failing heart. Physiol. Rev. 2005, 85, 1093–1129. [Google Scholar] [CrossRef]
- Cluntun, A.A.; Badolia, R.; Lettlova, S.; Parnell, K.M.; Shankar, T.S.; Diakos, N.A.; Olson, K.A.; Taleb, I.; Tatum, S.M.; Berg, J.A.; et al. The pyruvate-lactate axis modulates cardiac hypertrophy and heart failure. Cell Metab. 2021, 33, 629–648.e610. [Google Scholar] [CrossRef]
- Gertz, E.W.; Wisneski, J.A.; Stanley, W.C.; Neese, R.A. Myocardial substrate utilization during exercise in humans. Dual carbon-labeled carbohydrate isotope experiments. J. Clin. Investig. 1988, 82, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Karwi, Q.G.; Tian, R.; Wende, A.R.; Abel, E.D. Cardiac Energy Metabolism in Heart Failure. Circ. Res. 2021, 128, 1487–1513. [Google Scholar] [CrossRef]
- Hollander, J.M.; Thapa, D.; Shepherd, D.L. Physiological and structural differences in spatially distinct subpopulations of cardiac mitochondria: Influence of cardiac pathologies. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H1–H14. [Google Scholar] [CrossRef]
- Ikonomidis, J.S.; Salerno, T.A.; Wittnich, C. Calcium and the heart: An essential partnership. Can. J. Cardiol. 1990, 6, 305–316. [Google Scholar]
- Denton, R.M. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 2009, 1787, 1309–1316. [Google Scholar] [CrossRef]
- Territo, P.R.; Mootha, V.K.; French, S.A.; Balaban, R.S. Ca2+ activation of heart mitochondrial oxidative phosphorylation: Role of the F0/F1-ATPase. Am. J. Physiol. Cell Physiol. 2000, 278, C423–C435. [Google Scholar] [CrossRef]
- Wescott, A.P.; Kao, J.P.Y.; Lederer, W.J.; Boyman, L. Voltage-energized Calcium-sensitive ATP Production by Mitochondria. Nat. Metab. 2019, 1, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Hansford, R.G.; Castro, F. Intramitochondrial and extramitochondrial free calcium ion concentrations of suspensions of heart mitochondria with very low, plausibly physiological, contents of total calcium. J. Bioenerg. Biomembr. 1982, 14, 361–376. [Google Scholar] [CrossRef]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L.; et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef]
- Palty, R.; Silverman, W.F.; Hershfinkel, M.; Caporale, T.; Sensi, S.L.; Parnis, J.; Nolte, C.; Fishman, D.; Shoshan-Barmatz, V.; Herrmann, S.; et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. USA 2010, 107, 436–441. [Google Scholar] [CrossRef]
- Bonora, M.; Giorgi, C.; Pinton, P. Molecular mechanisms and consequences of mitochondrial permeability transition. Nat. Rev. Mol. Cell Biol. 2022, 23, 266–285. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Li, J.; Zhang, Y.; Chang, Y.Z. Cellular Iron Metabolism and Regulation. Adv. Exp. Med. Biol. 2019, 1173, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Chapa-Dubocq, X.R.; Tyurina, Y.Y.; St Croix, C.M.; Kapralov, A.A.; Tyurin, V.A.; Bayır, H.; Kagan, V.E.; Javadov, S. Elucidating the contribution of mitochondrial glutathione to ferroptosis in cardiomyocytes. Redox Biol. 2021, 45, 102021. [Google Scholar] [CrossRef]
- Fang, X.; Ardehali, H.; Min, J.; Wang, F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat. Rev. Cardiol. 2022. [Google Scholar] [CrossRef]
- Lakhal-Littleton, S.; Wolna, M.; Carr, C.A.; Miller, J.J.; Christian, H.C.; Ball, V.; Santos, A.; Diaz, R.; Biggs, D.; Stillion, R.; et al. Cardiac ferroportin regulates cellular iron homeostasis and is important for cardiac function. Proc. Natl. Acad. Sci. USA 2015, 112, 3164–3169. [Google Scholar] [CrossRef]
- Lane, D.J.; Merlot, A.M.; Huang, M.L.; Bae, D.H.; Jansson, P.J.; Sahni, S.; Kalinowski, D.S.; Richardson, D.R. Cellular iron uptake, trafficking and metabolism: Key molecules and mechanisms and their roles in disease. Biochim. Biophys. Acta 2015, 1853, 1130–1144. [Google Scholar] [CrossRef]
- Rauen, U.; Springer, A.; Weisheit, D.; Petrat, F.; Korth, H.G.; de Groot, H.; Sustmann, R. Assessment of chelatable mitochondrial iron by using mitochondrion-selective fluorescent iron indicators with different iron-binding affinities. ChemBioChem 2007, 8, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Shvartsman, M.; Kikkeri, R.; Shanzer, A.; Cabantchik, Z.I. Non-transferrin-bound iron reaches mitochondria by a chelator-inaccessible mechanism: Biological and clinical implications. Am. J. Physiol. Cell Physiol. 2007, 293, C1383–C1394. [Google Scholar] [CrossRef] [PubMed]
- Wolff, N.A.; Ghio, A.J.; Garrick, L.M.; Garrick, M.D.; Zhao, L.; Fenton, R.A.; Thévenod, F. Evidence for mitochondrial localization of divalent metal transporter 1 (DMT1). FASEB J. 2014, 28, 2134–2145. [Google Scholar] [CrossRef]
- Paradkar, P.N.; Zumbrennen, K.B.; Paw, B.H.; Ward, D.M.; Kaplan, J. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol. 2009, 29, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Sripetchwandee, J.; Sanit, J.; Chattipakorn, N.; Chattipakorn, S.C. Mitochondrial calcium uniporter blocker effectively prevents brain mitochondrial dysfunction caused by iron overload. Life Sci. 2013, 92, 298–304. [Google Scholar] [CrossRef]
- Braymer, J.J.; Lill, R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017, 292, 12754–12763. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhao, Y.; Guo, J.; Zhao, S.; Fei, C.; Xiao, C.; Wu, D.; Wu, L.; Li, X.; Chang, C. Iron overload promotes mitochondrial fragmentation in mesenchymal stromal cells from myelodysplastic syndrome patients through activation of the AMPK/MFF/Drp1 pathway. Cell Death Dis. 2018, 9, 515. [Google Scholar] [CrossRef]
- Hoes, M.F.; Grote Beverborg, N.; Kijlstra, J.D.; Kuipers, J.; Swinkels, D.W.; Giepmans, B.N.G.; Rodenburg, R.J.; van Veldhuisen, D.J.; de Boer, R.A.; van der Meer, P. Iron deficiency impairs contractility of human cardiomyocytes through decreased mitochondrial function. Eur. J. Heart Fail. 2018, 20, 910–919. [Google Scholar] [CrossRef]
- Chen, X.; Comish, P.B.; Tang, D.; Kang, R. Characteristics and Biomarkers of Ferroptosis. Front. Cell Dev. Biol. 2021, 9, 637162. [Google Scholar] [CrossRef]
- Niyazov, D.M.; Kahler, S.G.; Frye, R.E. Primary Mitochondrial Disease and Secondary Mitochondrial Dysfunction: Importance of Distinction for Diagnosis and Treatment. Mol. Syndromol. 2016, 7, 122–137. [Google Scholar] [CrossRef]
- Timpani, C.A.; Hayes, A.; Rybalka, E. Revisiting the dystrophin-ATP connection: How half a century of research still implicates mitochondrial dysfunction in Duchenne Muscular Dystrophy aetiology. Med. Hypotheses 2015, 85, 1021–1033. [Google Scholar] [CrossRef]
- Ommen, S.R.; Mital, S.; Burke, M.A.; Day, S.M.; Deswal, A.; Elliott, P.; Evanovich, L.L.; Hung, J.; Joglar, J.A.; Kantor, P.; et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients with Hypertrophic Cardiomyopathy: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2020, 142, e558–e631. [Google Scholar] [CrossRef]
- Bozkurt, B.; Colvin, M.; Cook, J.; Cooper, L.T.; Deswal, A.; Fonarow, G.C.; Francis, G.S.; Lenihan, D.; Lewis, E.F.; McNamara, D.M.; et al. Current Diagnostic and Treatment Strategies for Specific Dilated Cardiomyopathies: A Scientific Statement From the American Heart Association. Circulation 2016, 134, e579–e646. [Google Scholar] [CrossRef] [PubMed]
- Goto, D.; Kinugawa, S.; Hamaguchi, S.; Sakakibara, M.; Tsuchihashi-Makaya, M.; Yokota, T.; Yamada, S.; Yokoshiki, H.; Tsutsui, H.; Investigators, J.-C. Clinical characteristics and outcomes of dilated phase of hypertrophic cardiomyopathy: Report from the registry data in Japan. J. Cardiol. 2013, 61, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Szczepanowska, J.; Malinska, D.; Wieckowski, M.R.; Duszynski, J. Effect of mtDNA point mutations on cellular bioenergetics. Biochim. Biophys. Acta 2012, 1817, 1740–1746. [Google Scholar] [CrossRef] [PubMed]
- Scholle, L.M.; Zierz, S.; Mawrin, C.; Wickenhauser, C.; Urban, D.L. Heteroplasmy and Copy Number in the Common m.3243A>G Mutation-A Post-Mortem Genotype-Phenotype Analysis. Genes 2020, 11, 212. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic. Acids. Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Lott, M.T.; Leipzig, J.N.; Derbeneva, O.; Xie, H.M.; Chalkia, D.; Sarmady, M.; Procaccio, V.; Wallace, D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013, 44, 1.23.21–1.23.26. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University: Baltimore, MD, USA. Available online: https://omim.org/ (accessed on 1 July 2022).
- Majamaa-Voltti, K.; Peuhkurinen, K.; Kortelainen, M.L.; Hassinen, I.E.; Majamaa, K. Cardiac abnormalities in patients with mitochondrial DNA mutation 3243A>G. BMC Cardiovasc. Disord. 2002, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Uusimaa, J.; Moilanen, J.S.; Vainionpää, L.; Tapanainen, P.; Lindholm, P.; Nuutinen, M.; Löppönen, T.; Mäki-Torkko, E.; Rantala, H.; Majamaa, K. Prevalence, segregation, and phenotype of the mitochondrial DNA 3243A>G mutation in children. Ann. Neurol. 2007, 62, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, V.; Pitceathly, R.D.; Turnbull, D.M.; Taylor, R.W.; Sweeney, M.G.; Mudanohwo, E.E.; Rahman, S.; Hanna, M.G.; McFarland, R. The UK MRC Mitochondrial Disease Patient Cohort Study: Clinical phenotypes associated with the m.3243A>G mutation--implications for diagnosis and management. J. Neurol. Neurosurg. Psychiatry 2013, 84, 936–938. [Google Scholar] [CrossRef]
- Niedermayr, K.; Pölzl, G.; Scholl-Bürgi, S.; Fauth, C.; Schweigmann, U.; Haberlandt, E.; Albrecht, U.; Zlamy, M.; Sperl, W.; Mayr, J.A.; et al. Mitochondrial DNA mutation “m.3243A>G”-Heterogeneous clinical picture for cardiologists (“m.3243A>G”: A phenotypic chameleon). Congenit. Heart Dis. 2018, 13, 671–677. [Google Scholar] [CrossRef]
- Quadir, A.; Pontifex, C.S.; Lee Robertson, H.; Labos, C.; Pfeffer, G. Systematic review and meta-analysis of cardiac involvement in mitochondrial myopathy. Neurol. Genet. 2019, 5, e339. [Google Scholar] [CrossRef]
- Wahbi, K.; Bougouin, W.; Béhin, A.; Stojkovic, T.; Bécane, H.M.; Jardel, C.; Berber, N.; Mochel, F.; Lombès, A.; Eymard, B.; et al. Long-term cardiac prognosis and risk stratification in 260 adults presenting with mitochondrial diseases. Eur. Heart J. 2015, 36, 2886–2893. [Google Scholar] [CrossRef] [PubMed]
- Vydt, T.C.; de Coo, R.F.; Soliman, O.I.; Ten Cate, F.J.; van Geuns, R.J.; Vletter, W.B.; Schoonderwoerd, K.; van den Bosch, B.J.; Smeets, H.J.; Geleijnse, M.L. Cardiac involvement in adults with m.3243A>G MELAS gene mutation. Am. J. Cardiol. 2007, 99, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Okajima, Y.; Tanabe, Y.; Takayanagi, M.; Aotsuka, H. A follow up study of myocardial involvement in patients with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS). Heart 1998, 80, 292–295. [Google Scholar] [CrossRef]
- Hsu, Y.R.; Yogasundaram, H.; Parajuli, N.; Valtuille, L.; Sergi, C.; Oudit, G.Y. MELAS syndrome and cardiomyopathy: Linking mitochondrial function to heart failure pathogenesis. Heart Fail. Rev. 2016, 21, 103–116. [Google Scholar] [CrossRef]
- Sproule, D.M.; Kaufmann, P.; Engelstad, K.; Starc, T.J.; Hordof, A.J.; De Vivo, D.C. Wolff-Parkinson-White syndrome in Patients With MELAS. Arch. Neurol. 2007, 64, 1625–1627. [Google Scholar] [CrossRef]
- Finsterer, J.; Zarrouk-Mahjoub, S.; Shoffner, J.M. MERRF Classification: Implications for Diagnosis and Clinical Trials. Pediatr. Neurol. 2018, 80, 8–23. [Google Scholar] [CrossRef]
- Richter, U.; Evans, M.E.; Clark, W.C.; Marttinen, P.; Shoubridge, E.A.; Suomalainen, A.; Wredenberg, A.; Wedell, A.; Pan, T.; Battersby, B.J. RNA modification landscape of the human mitochondrial tRNA. Nat. Commun. 2018, 9, 3966. [Google Scholar] [CrossRef]
- Schaefer, A.M.; McFarland, R.; Blakely, E.L.; He, L.; Whittaker, R.G.; Taylor, R.W.; Chinnery, P.F.; Turnbull, D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008, 63, 35–39. [Google Scholar] [CrossRef]
- Mancuso, M.; Orsucci, D.; Angelini, C.; Bertini, E.; Carelli, V.; Comi, G.P.; Minetti, C.; Moggio, M.; Mongini, T.; Servidei, S.; et al. Phenotypic heterogeneity of the 8344A>G mtDNA “MERRF” mutation. Neurology 2013, 80, 2049–2054. [Google Scholar] [CrossRef] [PubMed]
- Darin, N.; Oldfors, A.; Moslemi, A.R.; Holme, E.; Tulinius, M. The incidence of mitochondrial encephalomyopathies in childhood: Clinical features and morphological, biochemical, and DNA abnormalities. Ann. Neurol. 2001, 49, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, J.B.; Harding, B.N. Pathogenic factors underlying the lesions in Leigh’s disease. Tissue responses to cellular energy deprivation and their clinico-pathological consequences. Brain 1994, 117 Pt 6, 1357–1376. [Google Scholar] [CrossRef]
- Schubert Baldo, M.; Vilarinho, L. Molecular basis of Leigh syndrome: A current look. Orphanet. J. Rare Dis. 2020, 15, 31. [Google Scholar] [CrossRef]
- Sofou, K.; de Coo, I.F.M.; Ostergaard, E.; Isohanni, P.; Naess, K.; De Meirleir, L.; Tzoulis, C.; Uusimaa, J.; Lönnqvist, T.; Bindoff, L.A.; et al. Phenotype-genotype correlations in Leigh syndrome: New insights from a multicentre study of 96 patients. J. Med. Genet. 2018, 55, 21–27. [Google Scholar] [CrossRef]
- Yaplito-Lee, J.; Weintraub, R.; Jamsen, K.; Chow, C.W.; Thorburn, D.R.; Boneh, A. Cardiac manifestations in oxidative phosphorylation disorders of childhood. J. Pediatr. 2007, 150, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Ruiter, E.M.; Siers, M.H.; van den Elzen, C.; van Engelen, B.G.; Smeitink, J.A.; Rodenburg, R.J.; Hol, F.A. The mitochondrial 13513G > A mutation is most frequent in Leigh syndrome combined with reduced complex I activity, optic atrophy and/or Wolff-Parkinson-White. Eur. J. Hum. Genet. 2007, 15, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.B.; Weng, W.C.; Lee, N.C.; Hwu, W.L.; Fan, P.C.; Lee, W.T. Mutation of mitochondrial DNA G13513A presenting with Leigh syndrome, Wolff-Parkinson-White syndrome and cardiomyopathy. Pediatr. Neonatol. 2008, 49, 145–149. [Google Scholar] [CrossRef]
- Monlleo-Neila, L.; Toro, M.D.; Bornstein, B.; Garcia-Arumi, E.; Sarrias, A.; Roig-Quilis, M.; Munell, F. Leigh Syndrome and the Mitochondrial m.13513G>A Mutation: Expanding the Clinical Spectrum. J. Child Neurol. 2013, 28, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Khambatta, S.; Nguyen, D.L.; Beckman, T.J.; Wittich, C.M. Kearns-Sayre syndrome: A case series of 35 adults and children. Int. J. Gen. Med. 2014, 7, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Farruggia, P.; Di Cataldo, A.; Pinto, R.M.; Palmisani, E.; Macaluso, A.; Valvo, L.L.; Cantarini, M.E.; Tornesello, A.; Corti, P.; Fioredda, F.; et al. Pearson Syndrome: A Retrospective Cohort Study from the Marrow Failure Study Group of A.I.E.O.P. (Associazione Italiana Emato-Oncologia Pediatrica). JIMD Rep. 2016, 26, 37–43. [Google Scholar] [CrossRef]
- Maceluch, J.A.; Niedziela, M. The clinical diagnosis and molecular genetics of kearns-sayre syndrome: A complex mitochondrial encephalomyopathy. Pediatr. Endocrinol. Rev. 2006, 4, 117–137. [Google Scholar]
- Holt, I.J.; Harding, A.E.; Morgan-Hughes, J.A. Deletions of muscle mitochondrial DNA in mitochondrial myopathies: Sequence analysis and possible mechanisms. Nucleic. Acids. Res. 1989, 17, 4465–4469. [Google Scholar] [CrossRef]
- Bris, C.; Goudenège, D.; Desquiret-Dumas, V.; Gueguen, N.; Bannwarth, S.; Gaignard, P.; Rucheton, B.; Trimouille, A.; Allouche, S.; Rouzier, C.; et al. Improved detection of mitochondrial DNA instability in mitochondrial genome maintenance disorders. Genet. Med. 2021, 23, 1769–1778. [Google Scholar] [CrossRef]
- Imamura, T.; Sumitomo, N.; Muraji, S.; Mori, H.; Osada, Y.; Oyanagi, T.; Kojima, T.; Yoshiba, S.; Kobayashi, T.; Ono, K. The necessity of implantable cardioverter defibrillators in patients with Kearns-Sayre syndrome—Systematic review of the articles. Int. J. Cardiol. 2019, 279, 105–111. [Google Scholar] [CrossRef]
- van Beynum, I.; Morava, E.; Taher, M.; Rodenburg, R.J.; Karteszi, J.; Toth, K.; Szabados, E. Cardiac arrest in kearns-sayre syndrome. JIMD Rep. 2012, 2, 7–10. [Google Scholar] [CrossRef]
- Kabunga, P.; Lau, A.K.; Phan, K.; Puranik, R.; Liang, C.; Davis, R.L.; Sue, C.M.; Sy, R.W. Systematic review of cardiac electrical disease in Kearns-Sayre syndrome and mitochondrial cytopathy. Int. J. Cardiol. 2015, 181, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Di Mambro, C.; Tamborrino, P.P.; Silvetti, M.S.; Yammine, M.L.; Marcolin, C.; Righi, D.; Baban, A.; Martinelli, D.; Dionisi Vici, C.; Drago, F. Progressive involvement of cardiac conduction system in paediatric patients with Kearns-Sayre syndrome: How to predict occurrence of complete heart block and sudden cardiac death? Europace 2021, 23, 948–957. [Google Scholar] [CrossRef]
- Schwartzkopff, B.; Frenzel, H.; Breithardt, G.; Deckert, M.; Lösse, B.; Toyka, K.V.; Borggrefe, M.; Hort, W. Ultrastructural findings in endomyocardial biopsy of patients with Kearns-Sayre syndrome. J. Am. Coll. Cardiol. 1988, 12, 1522–1528. [Google Scholar] [CrossRef]
- Govindaraj, P.; Khan, N.A.; Rani, B.; Rani, D.S.; Selvaraj, P.; Jyothi, V.; Bahl, A.; Narasimhan, C.; Rakshak, D.; Premkumar, K.; et al. Mitochondrial DNA variations associated with hypertrophic cardiomyopathy. Mitochondrion 2014, 16, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Merante, F.; Myint, T.; Tein, I.; Benson, L.; Robinson, B.H. An additional mitochondrial tRNA(Ile) point mutation (A-to-G at nucleotide 4295) causing hypertrophic cardiomyopathy. Hum. Mutat. 1996, 8, 216–222. [Google Scholar] [CrossRef]
- Campbell, T.; Lou, X.; Slone, J.; Brown, J.; Bromwell, M.; Liu, J.; Bai, R.; Haude, K.; Balog, A.; Cui, H.; et al. Mitochondrial genome variant m.3250T>C as a possible risk factor for mitochondrial cardiomyopathy. Hum. Mutat. 2021, 42, 177–188. [Google Scholar] [CrossRef]
- Li, D.; Sun, Y.; Zhuang, Q.; Song, Y.; Wu, B.; Jia, Z.; Pan, H.; Zhou, H.; Hu, S.; Zhang, B.; et al. Mitochondrial dysfunction caused by m.2336T>C mutation with hypertrophic cardiomyopathy in cybrid cell lines. Mitochondrion 2019, 46, 313–320. [Google Scholar] [CrossRef]
- Finsterer, J.; Kothari, S. Cardiac manifestations of primary mitochondrial disorders. Int. J. Cardiol. 2014, 177, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wu, Z.; Bai, Y.; Jiao, Y.; Li, P. Screening for Mitochondrial tRNA Mutations in 318 Patients with Dilated Cardiomyopathy. Hum. Hered. 2022, 87, 1–11. [Google Scholar] [CrossRef]
- Silvestri, G.; Santorelli, F.M.; Shanske, S.; Whitley, C.B.; Schimmenti, L.A.; Smith, S.A.; DiMauro, S. A new mtDNA mutation in the tRNA(Leu(UUR)) gene associated with maternally inherited cardiomyopathy. Hum. Mutat. 1994, 3, 37–43. [Google Scholar] [CrossRef]
- Tang, S.; Batra, A.; Zhang, Y.; Ebenroth, E.S.; Huang, T. Left ventricular noncompaction is associated with mutations in the mitochondrial genome. Mitochondrion 2010, 10, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bai, Y.; Huang, J.; Zhao, H.; Zhang, X.; Hu, S.; Wei, Y. Do mitochondria contribute to left ventricular non-compaction cardiomyopathy? New findings from myocardium of patients with left ventricular non-compaction cardiomyopathy. Mol. Genet. Metab. 2013, 109, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Finsterer, J.; Josef, F.; Stöllberger, C.; Claudia, S.; Gelpi, E.; Ellen, G. Successful heart failure therapy in mitochondrial disorder with noncompaction cardiomyopathy. Int. J. Cardiovasc. Imaging 2006, 22, 393–398. [Google Scholar] [CrossRef][Green Version]
- Bregel, L.V.; Belozerov, I.M.; Ogloblina, M.L.; Golubev, S.S.; Zemchenko, O.A.; Pavlenok, K.N.; Antoshkina, E.P.; Bochkareva, A.K.; Popod’ko, T.N. Histiocytoid cardiomyopathy in an infant. Kardiologiia 2012, 52, 93–96. [Google Scholar]
- Papadimitriou, A.; Neustein, H.B.; Dimauro, S.; Stanton, R.; Bresolin, N. Histiocytoid cardiomyopathy of infancy: Deficiency of reducible cytochrome b in heart mitochondria. Pediatr. Res. 1984, 18, 1023–1028. [Google Scholar] [CrossRef]
- Vallance, H.D.; Jeven, G.; Wallace, D.C.; Brown, M.D. A case of sporadic infantile histiocytoid cardiomyopathy caused by the A8344G (MERRF) mitochondrial DNA mutation. Pediatr. Cardiol. 2004, 25, 538–540. [Google Scholar] [CrossRef]
- Ware, S.M.; El-Hassan, N.; Kahler, S.G.; Zhang, Q.; Ma, Y.W.; Miller, E.; Wong, B.; Spicer, R.L.; Craigen, W.J.; Kozel, B.A.; et al. Infantile cardiomyopathy caused by a mutation in the overlapping region of mitochondrial ATPase 6 and 8 genes. J. Med. Genet. 2009, 46, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Ino, H.; Ohno, K.; Hattori, K.; Sato, W.; Ozawa, T.; Tanaka, T.; Itoyama, S. Mitochondrial mutation in fatal infantile cardiomyopathy. Lancet 1990, 336, 1452. [Google Scholar] [CrossRef]
- Finsterer, J.; Zarrouk-Mahjoub, S. The heart in m.3243A>G carriers. Herz 2020, 45, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Chau, E.M.C.; Ma, E.S.K.; Chan, A.O.O.; Tsoi, T.H.; Law, W.L. Mitochondrial cardiomyopathy due to m.3243A>G mitochondrial DNA mutation presenting in late adulthood: A case report. Hong Kong Med. J. 2020, 26, 240–242. [Google Scholar] [CrossRef]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation 2021, 144, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Stafford, F.; Thomson, K.; Butters, A.; Ingles, J. Hypertrophic Cardiomyopathy: Genetic Testing and Risk Stratification. Curr. Cardiol. Rep. 2021, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Hagen, C.M.; Aidt, F.H.; Hedley, P.L.; Jensen, M.K.; Havndrup, O.; Kanters, J.K.; Moolman-Smook, J.C.; Larsen, S.O.; Bundgaard, H.; Christiansen, M. Mitochondrial haplogroups modify the risk of developing hypertrophic cardiomyopathy in a Danish population. PLoS ONE 2013, 8, e71904. [Google Scholar] [CrossRef] [PubMed]
- van der Westhuizen, F.H.; Sinxadi, P.Z.; Dandara, C.; Smuts, I.; Riordan, G.; Meldau, S.; Malik, A.N.; Sweeney, M.G.; Tsai, Y.; Towers, G.W.; et al. Understanding the Implications of Mitochondrial DNA Variation in the Health of Black Southern African Populations: The 2014 Workshop. Hum. Mutat. 2015, 36, 569–571. [Google Scholar] [CrossRef]
- McCormick, E.M.; Lott, M.T.; Dulik, M.C.; Shen, L.; Attimonelli, M.; Vitale, O.; Karaa, A.; Bai, R.; Pineda-Alvarez, D.E.; Singh, L.N.; et al. Specifications of the ACMG/AMP standards and guidelines for mitochondrial DNA variant interpretation. Hum. Mutat. 2020, 41, 2028–2057. [Google Scholar] [CrossRef]
- Falk, M.J.; Shen, L.; Gai, X. From case studies to community knowledge base: MSeqDR provides a platform for the curation and genomic analysis of mitochondrial diseases. Cold Spring Harb. Mol. Case Stud. 2016, 2, a001065. [Google Scholar] [CrossRef][Green Version]
- Umbria, M.; Ramos, A.; Aluja, M.P.; Santos, C. The role of control region mitochondrial DNA mutations in cardiovascular disease: Stroke and myocardial infarction. Sci. Rep. 2020, 10, 2766. [Google Scholar] [CrossRef]
- Piotrowska-Nowak, A.; Elson, J.L.; Sobczyk-Kopciol, A.; Piwonska, A.; Puch-Walczak, A.; Drygas, W.; Ploski, R.; Bartnik, E.; Tonska, K. New mtDNA Association Model, MutPred Variant Load, Suggests Individuals with Multiple Mildly Deleterious mtDNA Variants Are More Likely to Suffer From Atherosclerosis. Front. Genet. 2018, 9, 702. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Gao, B.B.; Huang, J.Y. The role of mitochondrial DNA mutations in coronary heart disease. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8502–8509. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y. Mitochondrial tRNA Mutations Associated With Essential Hypertension: From Molecular Genetics to Function. Front. Cell Dev. Biol. 2020, 8, 634137. [Google Scholar] [CrossRef]
- Stewart, J.B. Current progress with mammalian models of mitochondrial DNA disease. J. Inherit. Metab. Dis. 2021, 44, 325–342. [Google Scholar] [CrossRef]
- Richter, U.; McFarland, R.; Taylor, R.W.; Pickett, S.J. The molecular pathology of pathogenic mitochondrial tRNA variants. FEBS Lett. 2021, 595, 1003–1024. [Google Scholar] [CrossRef] [PubMed]
- Zeviani, M.; Gellera, C.; Antozzi, C.; Rimoldi, M.; Morandi, L.; Villani, F.; Tiranti, V.; DiDonato, S. Maternally inherited myopathy and cardiomyopathy: Association with mutation in mitochondrial DNA tRNA(Leu)(UUR). Lancet 1991, 338, 143–147. [Google Scholar] [CrossRef]
- Merante, F.; Tein, I.; Benson, L.; Robinson, B.H. Maternally inherited hypertrophic cardiomyopathy due to a novel T-to-C transition at nucleotide 9997 in the mitochondrial tRNA(glycine) gene. Am. J. Hum. Genet. 1994, 55, 437–446. [Google Scholar]
- Picard, M.; Zhang, J.; Hancock, S.; Derbeneva, O.; Golhar, R.; Golik, P.; O’Hearn, S.; Levy, S.; Potluri, P.; Lvova, M.; et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl. Acad. Sci. USA 2014, 111, E4033–E4042. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, S.; Horn, M.; Cramer, M.; Harre, K.; Newell, J.B.; Peters, W.; Pabst, T.; Ertl, G.; Hahn, D.; Ingwall, J.S.; et al. Myocardial phosphocreatine-to-ATP ratio is a predictor of mortality in patients with dilated cardiomyopathy. Circulation 1997, 96, 2190–2196. [Google Scholar] [CrossRef] [PubMed]
- Desquiret-Dumas, V.; Gueguen, N.; Barth, M.; Chevrollier, A.; Hancock, S.; Wallace, D.C.; Amati-Bonneau, P.; Henrion, D.; Bonneau, D.; Reynier, P.; et al. Metabolically induced heteroplasmy shifting and l-arginine treatment reduce the energetic defect in a neuronal-like model of MELAS. Biochim. Biophys. Acta 2012, 1822, 1019–1029. [Google Scholar] [CrossRef]
- Fillmore, N.; Levasseur, J.L.; Fukushima, A.; Wagg, C.S.; Wang, W.; Dyck, J.R.B.; Lopaschuk, G.D. Uncoupling of glycolysis from glucose oxidation accompanies the development of heart failure with preserved ejection fraction. Mol. Med. 2018, 24, 3. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Zuryn, S. Mitochondrial Genome (mtDNA) Mutations that Generate Reactive Oxygen Species. Antioxidants 2019, 8, 392. [Google Scholar] [CrossRef]
- Ganetzky, R.D.; Stendel, C.; McCormick, E.M.; Zolkipli-Cunningham, Z.; Goldstein, A.C.; Klopstock, T.; Falk, M.J. MT-ATP6 mitochondrial disease variants: Phenotypic and biochemical features analysis in 218 published cases and cohort of 14 new cases. Hum. Mutat. 2019, 40, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Nadworny, A.S.; Guruju, M.R.; Poor, D.; Doran, R.M.; Sharma, R.V.; Kotlikoff, M.I.; Davisson, R.L. Nox2 and Nox4 influence neonatal c-kit(+) cardiac precursor cell status and differentiation. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H829–H842. [Google Scholar] [CrossRef]
- Amberg, G.C.; Earley, S.; Glapa, S.A. Local regulation of arterial L-type calcium channels by reactive oxygen species. Circ. Res. 2010, 107, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Zima, A.V.; Blatter, L.A. Redox regulation of cardiac calcium channels and transporters. Cardiovasc. Res. 2006, 71, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Filburn, C.R.; Klotz, L.O.; Zweier, J.L.; Sollott, S.J. Reactive oxygen species (ROS)-induced ROS release: A new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J. Exp. Med. 2000, 192, 1001–1014. [Google Scholar] [CrossRef]
- Kwong, J.Q.; Molkentin, J.D. Physiological and pathological roles of the mitochondrial permeability transition pore in the heart. Cell Metab. 2015, 21, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Brady, N.R.; Elmore, S.P.; van Beek, J.J.; Krab, K.; Courtoy, P.J.; Hue, L.; Westerhoff, H.V. Coordinated behavior of mitochondria in both space and time: A reactive oxygen species-activated wave of mitochondrial depolarization. Biophys. J. 2004, 87, 2022–2034. [Google Scholar] [CrossRef]
- Cadenas, S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic. Biol. Med. 2018, 117, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.; Terentyeva, R.; Perger, F.; Hernández Orengo, B.; Martin, B.; Gorr, M.W.; Belevych, A.E.; Clements, R.T.; Györke, S.; Terentyev, D. MCU overexpression evokes disparate dose-dependent effects on mito-ROS and spontaneous Ca. Am. J. Physiol. Heart Circ. Physiol. 2021, 321, H615–H632. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Q.; Watson, L.J.; Jones, S.P.; Epstein, P.N. Cardiac overexpression of 8-oxoguanine DNA glycosylase 1 protects mitochondrial DNA and reduces cardiac fibrosis following transaortic constriction. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H2073–H2080. [Google Scholar] [CrossRef]
- Vendelin, M.; Béraud, N.; Guerrero, K.; Andrienko, T.; Kuznetsov, A.V.; Olivares, J.; Kay, L.; Saks, V.A. Mitochondrial regular arrangement in muscle cells: A “crystal-like” pattern. Am. J. Physiol. Cell Physiol. 2005, 288, C757–C767. [Google Scholar] [CrossRef]
- Song, M.; Mihara, K.; Chen, Y.; Scorrano, L.; Dorn, G.W. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015, 21, 273–286. [Google Scholar] [CrossRef]
- Kanzaki, Y.; Terasaki, F.; Okabe, M.; Otsuka, K.; Katashima, T.; Fujita, S.; Ito, T.; Kitaura, Y. Giant mitochondria in the myocardium of a patient with mitochondrial cardiomyopathy: Transmission and 3-dimensional scanning electron microscopy. Circulation 2010, 121, 831–832. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Murayama, K.; Okazaki, Y.; Imai-Okazaki, A.; Ohtake, A.; Takakuwa, E.; Yamazawa, H.; Izumi, G.; Abe, J.; Nagai, A.; et al. Advanced pathological study for definite diagnosis of mitochondrial cardiomyopathy. J. Clin. Pathol. 2020, 74, 365–371. [Google Scholar] [CrossRef]
- Terasaki, F.; Tanaka, M.; Kawamura, K.; Kanzaki, Y.; Okabe, M.; Hayashi, T.; Shimomura, H.; Ito, T.; Suwa, M.; Gong, J.S.; et al. A case of cardiomyopathy showing progression from the hypertrophic to the dilated form: Association of Mt8348A-->G mutation in the mitochondrial tRNA(Lys) gene with severe ultrastructural alterations of mitochondria in cardiomyocytes. Jpn. Circ. J. 2001, 65, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Suen, D.F.; Narendra, D.P.; Tanaka, A.; Manfredi, G.; Youle, R.J. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11835–11840. [Google Scholar] [CrossRef] [PubMed]
- Coronado, M.; Fajardo, G.; Nguyen, K.; Zhao, M.; Kooiker, K.; Jung, G.; Hu, D.Q.; Reddy, S.; Sandoval, E.; Stotland, A.; et al. Physiological Mitochondrial Fragmentation Is a Normal Cardiac Adaptation to Increased Energy Demand. Circ. Res. 2018, 122, 282–295. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.S.; Tanaka, M.; Suzuki, J.; Hemmi, C.; Toyo-oka, T. A novel homoplasmic mutation in mtDNA with a single evolutionary origin as a risk factor for cardiomyopathy. Am. J. Hum. Genet. 2000, 67, 1617–1620. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Yan, Z.; Zhu, Z. Mitochondria-Associated Endoplasmic Reticulum Membranes in Cardiovascular Diseases. Front. Cell Dev. Biol. 2020, 8, 604240. [Google Scholar] [CrossRef] [PubMed]
- Bravo, R.; Vicencio, J.M.; Parra, V.; Troncoso, R.; Munoz, J.P.; Bui, M.; Quiroga, C.; Rodriguez, A.E.; Verdejo, H.E.; Ferreira, J.; et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 2011, 124, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Silva, D.; Wüst, R.C.I.; Conceição, G.; Gonçalves-Rodrigues, P.; Gonçalves, N.; Gonçalves, A.; Kuster, D.W.D.; Leite-Moreira, A.F.; van der Velden, J.; de Sousa Beleza, J.M.; et al. Disturbed cardiac mitochondrial and cytosolic calcium handling in a metabolic risk-related rat model of heart failure with preserved ejection fraction. Acta Physiol. 2020, 228, e13378. [Google Scholar] [CrossRef] [PubMed]
- Givvimani, S.; Pushpakumar, S.B.; Metreveli, N.; Veeranki, S.; Kundu, S.; Tyagi, S.C. Role of mitochondrial fission and fusion in cardiomyocyte contractility. Int. J. Cardiol. 2015, 187, 325–333. [Google Scholar] [CrossRef]
- Görlach, A.; Bertram, K.; Hudecova, S.; Krizanova, O. Calcium and ROS: A mutual interplay. Redox Biol. 2015, 6, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.; Terentyeva, R.; Martin, B.; Perger, F.; Li, J.; Stepanov, A.; Bonilla, I.M.; Knollmann, B.C.; Radwański, P.B.; Györke, S.; et al. Increased RyR2 activity is exacerbated by calcium leak-induced mitochondrial ROS. Basic Res. Cardiol. 2020, 115, 38. [Google Scholar] [CrossRef] [PubMed]
- Terentyev, D.; Györke, I.; Belevych, A.E.; Terentyeva, R.; Sridhar, A.; Nishijima, Y.; de Blanco, E.C.; Khanna, S.; Sen, C.K.; Cardounel, A.J.; et al. Redox modification of ryanodine receptors contributes to sarcoplasmic reticulum Ca2+ leak in chronic heart failure. Circ. Res. 2008, 103, 1466–1472. [Google Scholar] [CrossRef] [PubMed]
- Ducamp, S.; Fleming, M.D. The molecular genetics of sideroblastic anemia. Blood 2019, 133, 59–69. [Google Scholar] [CrossRef]
- Slone, J.D.; Yang, L.; Peng, Y.; Queme, L.F.; Harris, B.; Rizzo, S.J.S.; Green, T.; Ryan, J.L.; Jankowski, M.P.; Reinholdt, L.G.; et al. Integrated analysis of the molecular pathogenesis of FDXR-associated disease. Cell Death Dis. 2020, 11, 423. [Google Scholar] [CrossRef]
- Berhe, S.; Heeney, M.M.; Campagna, D.R.; Thompson, J.F.; White, E.J.; Ross, T.; Peake, R.W.A.; Hanrahan, J.D.; Rodriguez, V.; Renaud, D.L.; et al. Recurrent heteroplasmy for the MT-ATP6 p.Ser148Asn (m.8969G>A) mutation in patients with syndromic congenital sideroblastic anemia of variable clinical severity. Haematologica 2018, 103, e561–e563. [Google Scholar] [CrossRef]
- Sripetchwandee, J.; KenKnight, S.B.; Sanit, J.; Chattipakorn, S.; Chattipakorn, N. Blockade of mitochondrial calcium uniporter prevents cardiac mitochondrial dysfunction caused by iron overload. Acta Physiol. 2014, 210, 330–341. [Google Scholar] [CrossRef]
- Pedrera, L.; Espiritu, R.A.; Ros, U.; Weber, J.; Schmitt, A.; Stroh, J.; Hailfinger, S.; von Karstedt, S.; García-Sáez, A.J. Ferroptotic pores induce Ca2+ fluxes and ESCRT-lll activation to modulate cell death kinetics. Cell Death Differ. 2021, 28, 1644–1657. [Google Scholar] [CrossRef]
| Gene |
Gene Product |
mtDNA Variants * |
OMIM ID # | Ref. |
Cardiovascular Findings Associated with Phenotype ** |
|---|---|---|---|---|---|
| MT-ND1 | Complex I subunit | m.3697G > A m.3946G > A m.3949T > C | 516000.0012 516000.0013 516000.0014 | [A] [A] | nsHCM DCM LVNC WPW Cardiac conduction abnormality |
| MT-ND5 | Complex I subunit | m.12770A > G m.13042G > A m.13045A > C m.13084A > T m.13513G > A | 516005.0004 516005.0008 516005.0005 516005.0006 516005.0007 | [A][C] [A][C] [A] [A] [A][C] | |
| MT-ND6 | Complex I subunit | m.14453G > A | 516006.0005 | [A][C] | |
| MT-TC | tRNA Cys | m.5814A > G | 590020.0001 | ||
| MT-TF | tRNA Phe | m.583G > A | 590070.0001 | [A][C] | |
| MT-TH | tRNA His | m.12147G > A m.12158A > G | 590040.0003 | [A][C] [C] | |
| MT-TK | tRNA Lys | m.8316T > C m.8356T > C | 590060.0002 | [C] [A][B][C] | |
| MT-TL1 | tRNA Leu(UUR) | m.3243A > G m.3252A > G m.3256C > T m.3258T > C m.3271T > C m. T > C | 590050.0001 590050.0005 590050.0003 590050.0002 | [A][B][C] [A] [A][C] [C] [A][C] [C] | |
| MT-TM | tRNA Met | m.4450G > A | [C] | ||
| MT-TQ | tRNA Gln | m.4332G > A | 590030.0003 | [A][B][C] | |
| MT-TS1 | tRNA Ser(UCN) | m.7512T > C | 590080.0001 | [A] | |
| MT-TS2 | tRNA Ser(AGY) | m.12207G > A | 590085.0002 | [A][C] | |
| MT-TW | tRNA Trp | m.5541C > T | [C] |
| Gene | Gene Product | mtDNA Variants * | OMIM ID # | Ref. | Cardiovascular Findings Associated with Phenotype ** |
|---|---|---|---|---|---|
| MT-ND5 | Complex I subunit | m.13042G > A | 516005.0008 | [A][C] | nsHCM DCM Cardiac conduction abnormality |
| MT-TF | tRNA Phe | m.611G > A | 590070.0002 | [A] | |
| MT-TH | tRNA His | m.12147G > A | 590040.0003 | [A][C] | |
| MT-TK | tRNA Lys | m.8344A > G m.8356T > C m.8361G > A m.8363G > A | 590060.0001 590060.0002 590060.0007 590060.0003 | [A][B][C] [A][B][C] [A] [A][B][C] | |
| MT-TL1 | tRNA Leu(UUR) | m.3243A > G m.3256C > T | 590050.0001 590050.0003 | [A][B][C] [A][C] | |
| MT-TP | tRNA Pro | m.15967G > A | 590075.0003 | [A][C] | |
| MT-TS1 | tRNA Ser(UCN) | m.7512T > C | 590080.0001 | [A] | |
| MT-TS2 | tRNA Ser(AGY) | m.12207G > A | 590085.0002 | [A][C] |
| Gene | Gene Product | mtDNA Variants * | OMIM ID # | Ref. | Cardiovascular Findings Associated with Phenotype ** |
|---|---|---|---|---|---|
| MT-ND1 | Complex I subunit | m.3460G > A m.3481G > A m.3890G > A m.3946G > A | 516000.0001 516000.0013 | [A][B][C] [A] [A] [A] | nsHCM DCM WPW Cardiac conduction abnormality |
| MT-ND2 | Complex I subunit | m.4640C > A m.4681T > C | 516001.0003 516001.0006 | [A] | |
| MT-ND3 | Complex I subunit | m.10158T > C m.10191T > C m.10197G > A | 516002.0003 516002.0001 516002.0004 | [A][C] [A][C] [A][C] | |
| MT-ND4 | Complex I subunit | m.11777C > A | 516003.004 | [A][C] | |
| MT-ND5 | Complex I subunit | m.12706T > C m.13042G > A m.13063G > A m.13084A > T m.13513G > A m.13514A > G | 516005.0003 516005.0008 516005.0006 516005.0007 | [A][C] [A][C] [A][B] [A] [A][C] [A][C] | |
| MT-ND6 | Complex I subunit | m.14459G > A m.14484T > C m.14487T > C | 516006.0002 516006.0001 516006.0007 | [A][B][C] [A][B][C] [A][C] | |
| MT-CO3 | Complex IV subunit | m.9478T > C m.9537dupC | 516050.0005 | [B] [A] | |
| MT-ATP6 | Complex V subunit | m.8783G > A m.8839G > C m.8851T > C m.8993T > G m.8993T > C m.9035T > C m.9176T > G m.9176T > C m.9185T > C m.9191T > C | 516060.0001 516060.0002 516060.0011 516060.0005 516060.0008 | [A][B] [A] [A][B] [A][C] [A][B][C] [A][B][C] [A][C] [A][B][C] [A][C] [A][C] | |
| MT-TK | tRNA Lys | m.8363G > A | 590060.0003 | [A][B][C] | |
| MT-TM | tRNA Met | m.4450G > A | [C] | ||
| MT-TS2 | tRNA Ser(AGY) | m.12258C > T m.12264C > T | [A][C] [A][C] | ||
| MT-TV | tRNA Val | m.1624C > T m.1630A > G m.1659T > C | 590105.0002 | [A][B] [A][C] [A] | |
| MT-TW | tRNA Trp | m.5523T > G m.5537insT m.5540G > A m.5543T > C | 590095.0002 | [C] [A] [A] [A] |
| Gene | Gene Product | mtDNA Variants | OMIM ID # | Ref. | Cardiovascular Findings Associated with Phenotype * |
|---|---|---|---|---|---|
| MT-TL1 | tRNA Leu (UUR) | m.3249G > A m.3255G > A | 590050.0011 | [A][C] | nsHCM DCM Cardiac conduction abnormality |
| MT-TL2 | tRNA Leu (CUN) | m.12315G > A | 590055.0001 | [A][C] | |
| Deletion/ Duplications | Various | Multiple reported | [A][B][C] |
| Condition | Gene |
Gene
Product |
mtDNA Variants |
OMIM ID # | Ref. |
Other
Findings |
|---|---|---|---|---|---|---|
| nsHCM | MT-CYB | Complex III subunit | m.14849T > C | 516020.0012 | Septo-optic dysplasia, retinitis pigmentosa | |
| MT-ATP6 | Complex V subunit | m.8528T > C | 516060.0010 | [A] | Infantile | |
| MT-ATP8 | Complex V subunit | m.8528T > C m.8529G > A | 516070.0003 516070.0002 | [A] [A] | Neuropathy | |
| MT-TG | tRNAGly | m.9997T > C | 590035.0001 | [C] | Myopathy Exercise intolerance | |
| MT-TI | tRNA Ile | m.4295A > G m.4300A > G m.4317A > G | 590045.0003 590045.0006 590045.0001 | [A][C] | Infantile, SNHL Infantile | |
| MT-TK | tRNALys | m.8363G > A | 590060.0003 | [A][B][C] | SNHL | |
| MT-TL1 | tRNA Leu (UUR) | m.3243A > G m.3260A > G m.3303C > T | 590050.0001 590050.0007 590050.0004 | [A][B][C] [A][C] [A][C] | MELAS overlap Skeletal myopathy Skeletal myopathy | |
| MT-TV | tRNA Val | m.1624C > T m.1644G > A | 590105.0002 | [A][B] [A][B][C] | Leigh overlap MELAS overlap | |
| MT-TW | tRNA Trp | m.5545C > T | 590095.0001 | Multisystem | ||
| DCM | MT-TH | tRNAHis | m.12192G > A | 590040.0001 | LHON overlap | |
| MT-TI | tRNA Ile | m.4269A > G m.4300A > G m.4317A > G m.4322dupC | 590045.0002 590045.0006 590045.0001 | [A][C] [A][C] [C] | Multisystem Infantile | |
| MT-TL1 | tRNA Leu (UUR) | m.3243A > G | 590050.0001 | [A][B][C] | MELAS overlap | |
| MT-TL2 | tRNA Leu (CUN) | m.12297T > C | 590055.0003 | |||
| MT-TR | tRNA Arg | m.10415T > C | [C] | |||
| MT-TY | tRNATry | m.5843A > G | 590100.004 | Focal segmental glomerulosclerosis | ||
| LVNC | MT-TL1 | tRNA Leu (UUR) | m.3243A > G | 590050.0001 | [A][B][C] | MELAS overlap |
| MT-TV | tRNA Val | m.1612C > T | [C] | |||
| Histiocytoid cardiomyopathy | MT-CYB | Complex III subunit | m.15498G > A | 516020.0011 | [C] | Leigh overlap |
| MT-TK | tRNA Lys | m.8344A > G | 590060.0001 | [A][B][C] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campbell, T.; Slone, J.; Huang, T. Mitochondrial Genome Variants as a Cause of Mitochondrial Cardiomyopathy. Cells 2022, 11, 2835. https://doi.org/10.3390/cells11182835
Campbell T, Slone J, Huang T. Mitochondrial Genome Variants as a Cause of Mitochondrial Cardiomyopathy. Cells. 2022; 11(18):2835. https://doi.org/10.3390/cells11182835
Chicago/Turabian StyleCampbell, Teresa, Jesse Slone, and Taosheng Huang. 2022. "Mitochondrial Genome Variants as a Cause of Mitochondrial Cardiomyopathy" Cells 11, no. 18: 2835. https://doi.org/10.3390/cells11182835
APA StyleCampbell, T., Slone, J., & Huang, T. (2022). Mitochondrial Genome Variants as a Cause of Mitochondrial Cardiomyopathy. Cells, 11(18), 2835. https://doi.org/10.3390/cells11182835






