Computational Insights into the Sequence-Activity Relationships of the NGF(1–14) Peptide by Molecular Dynamics Simulations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Preparation and Molecular Dynamics Simulation
2.2. Binding Free Energy Calculations and Per-Residue Energy Decomposition Analysis
3. Results and Discussion
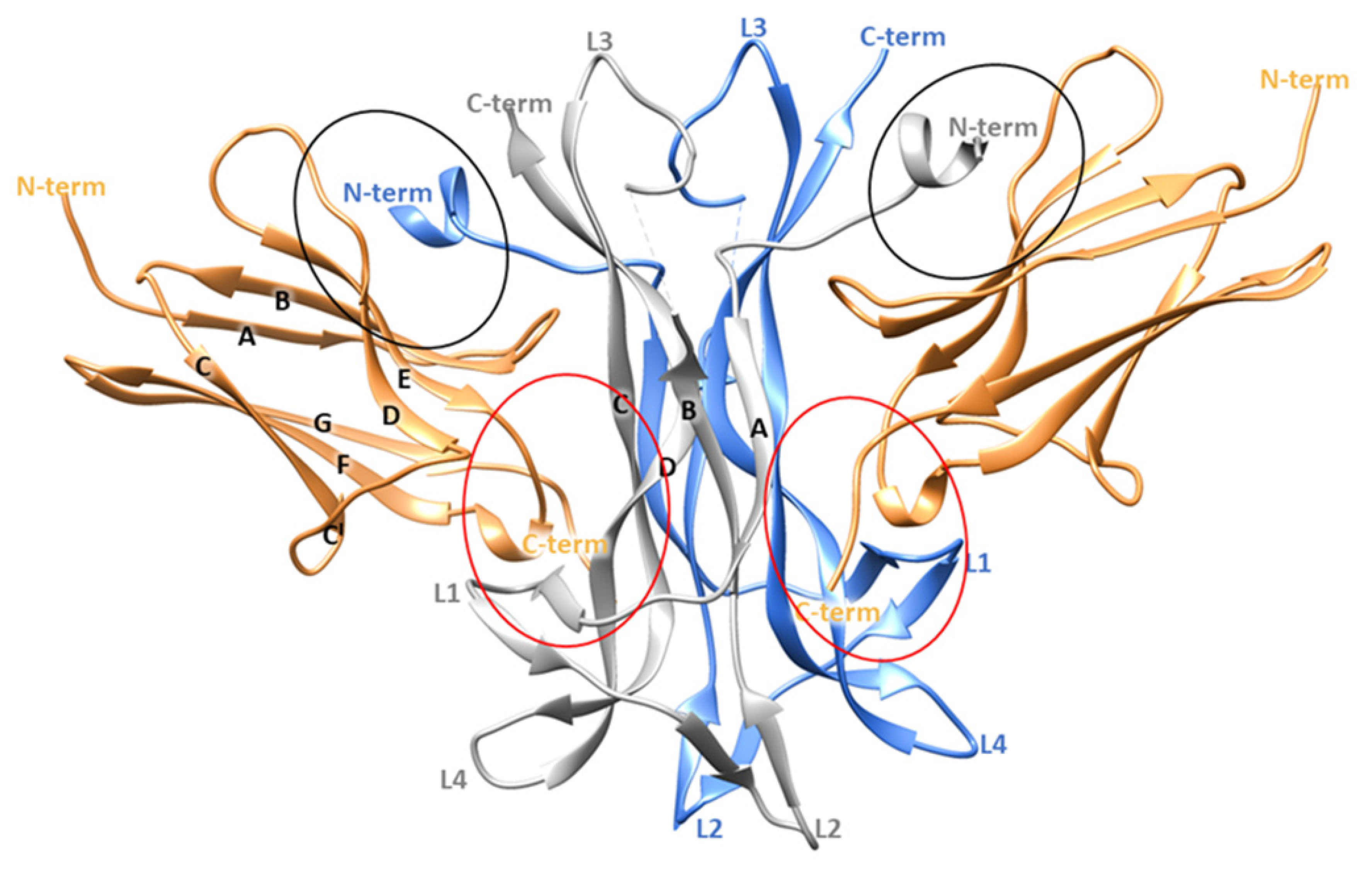
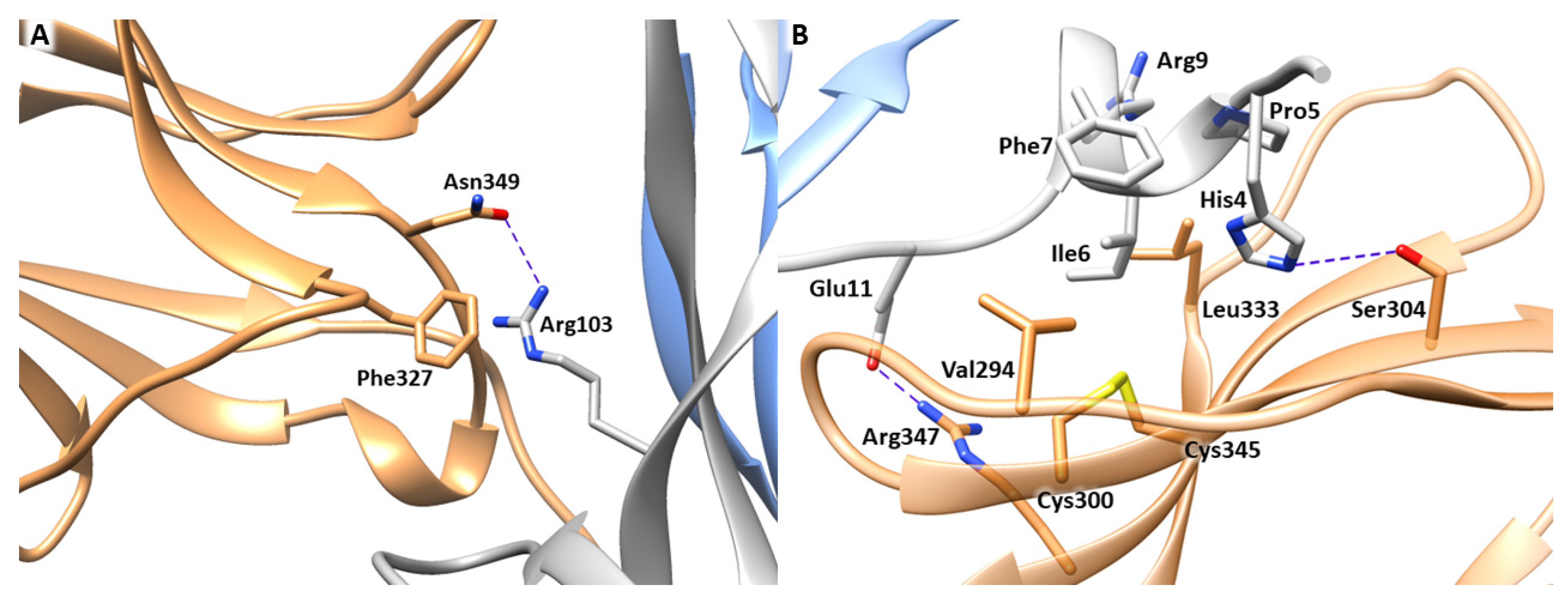
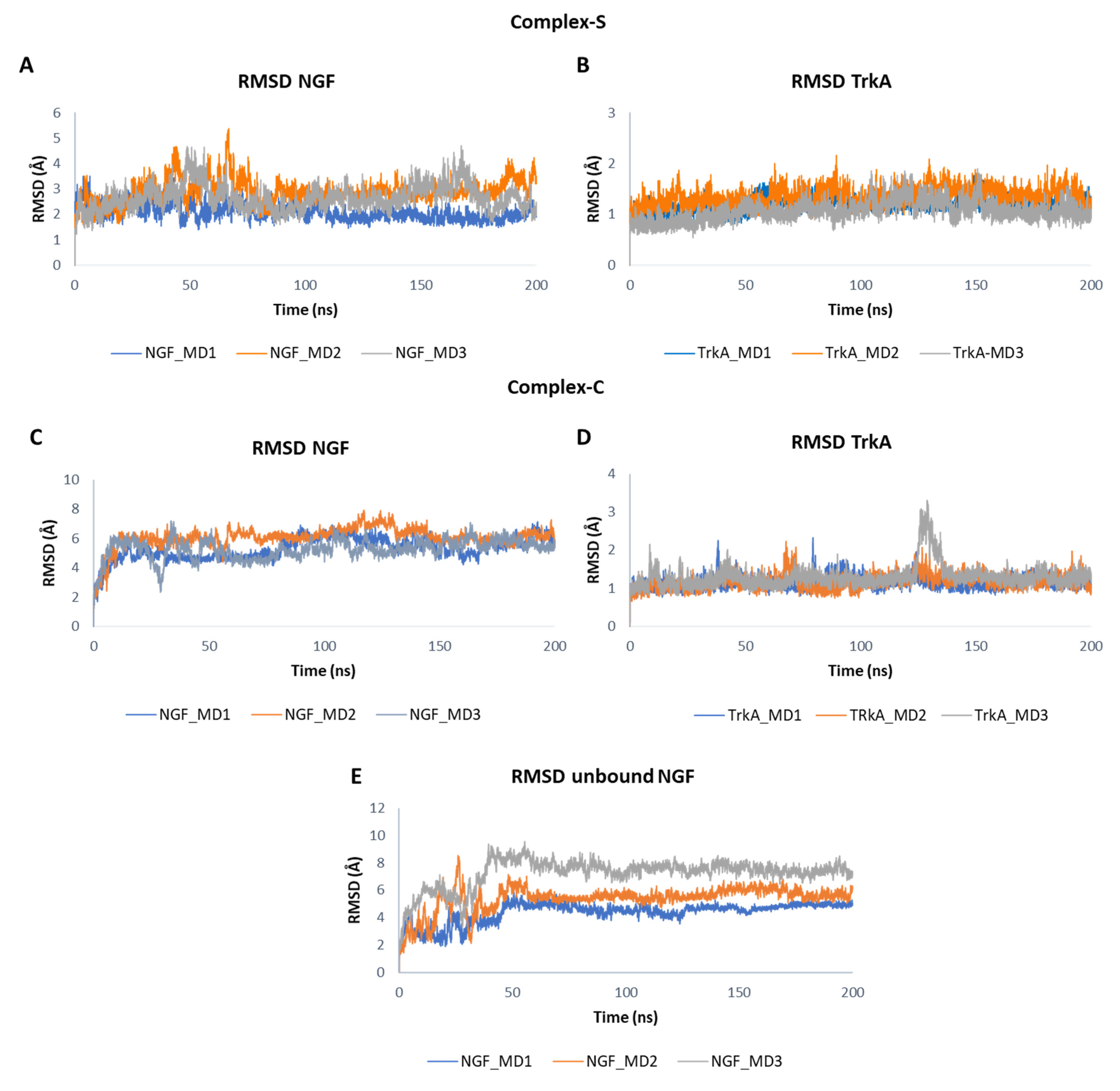
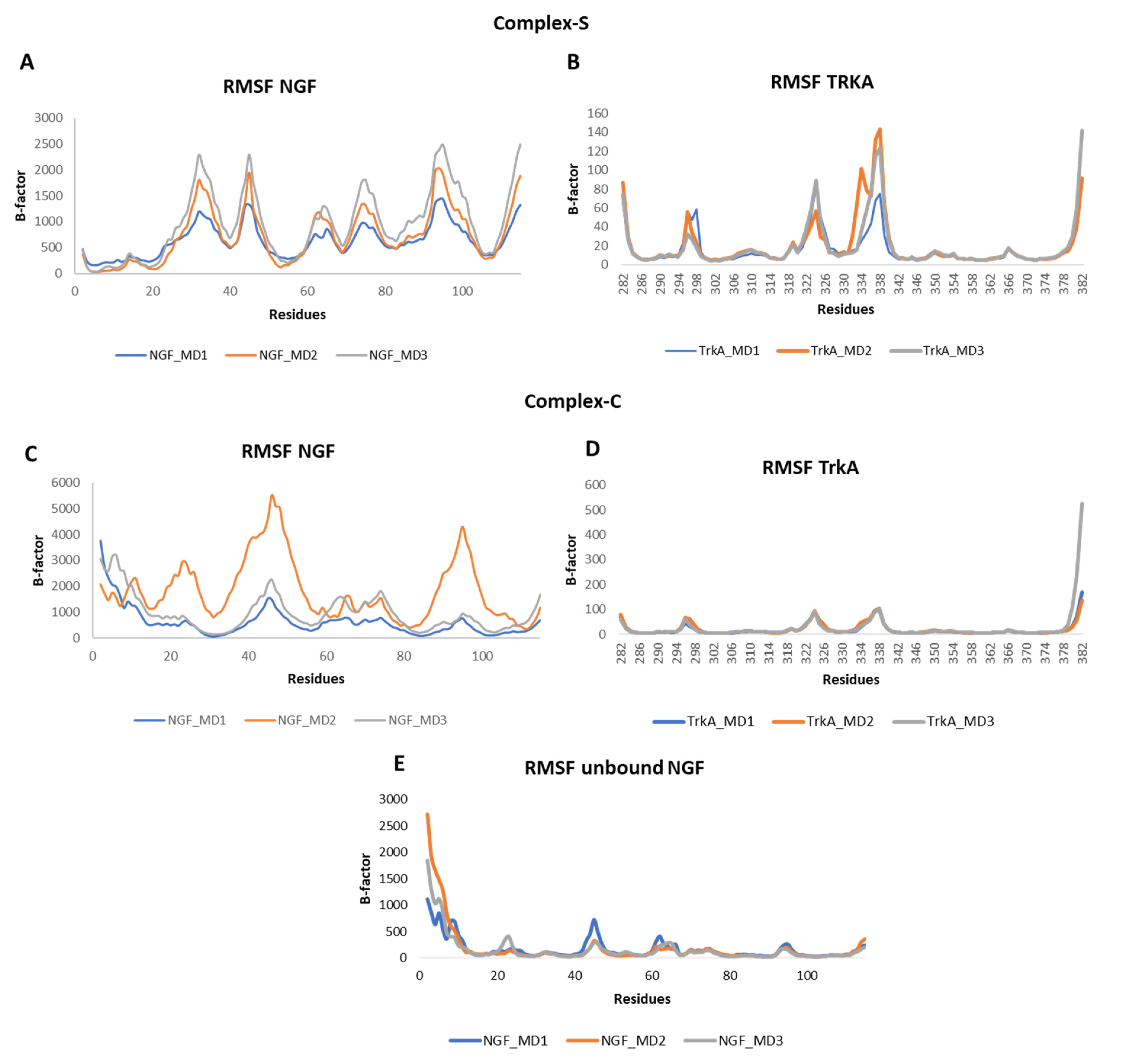
3.1. MD Simulation of NGF-TrkA Complex
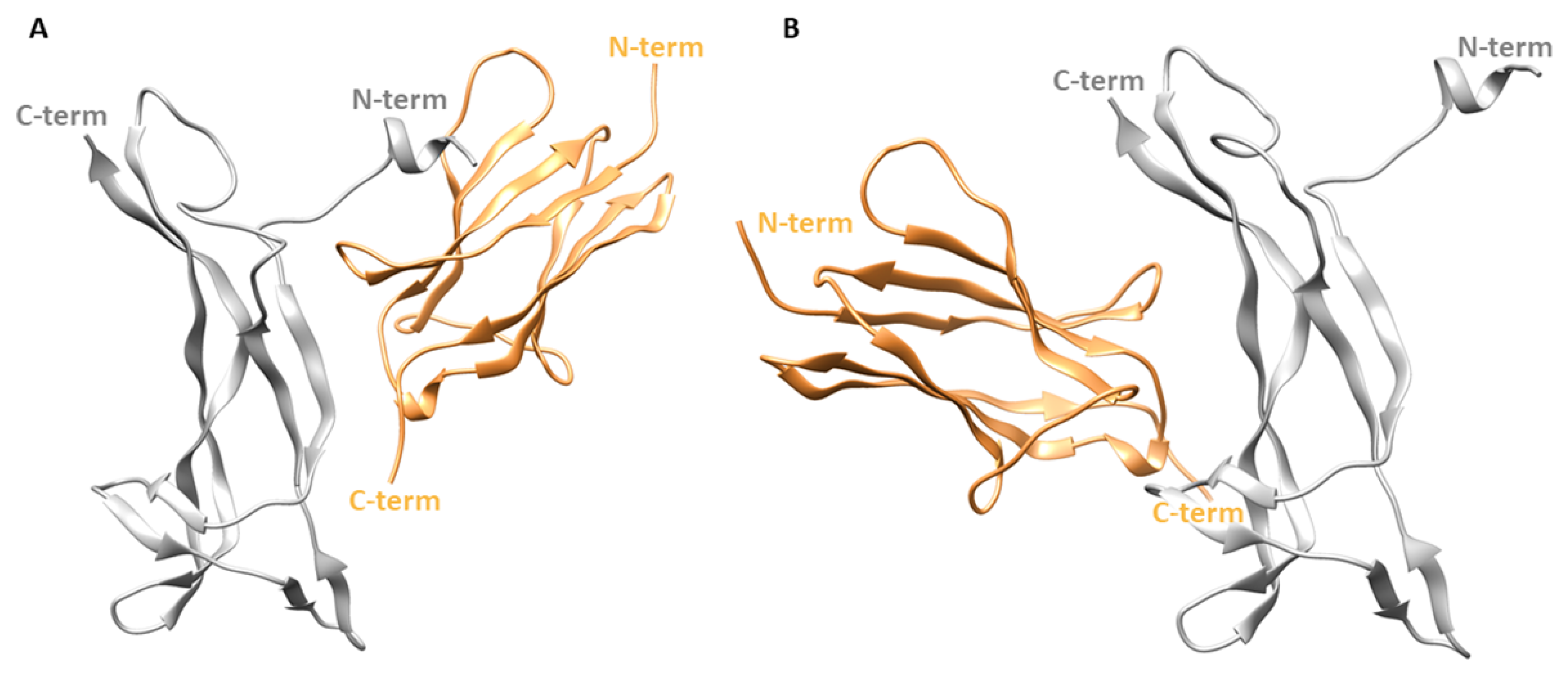
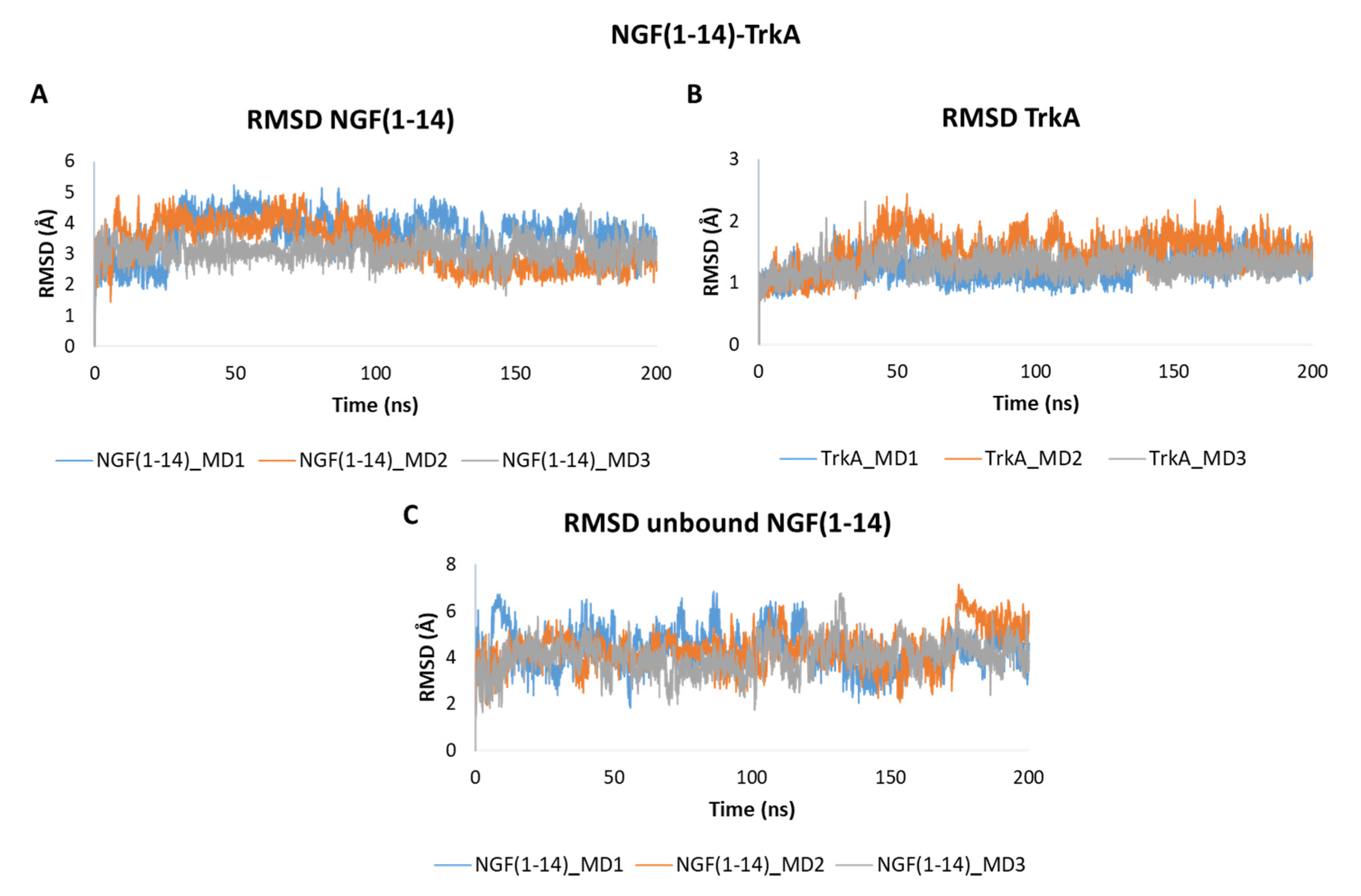
3.2. MD Simulation of NGF(1–14)-TrkA Complex
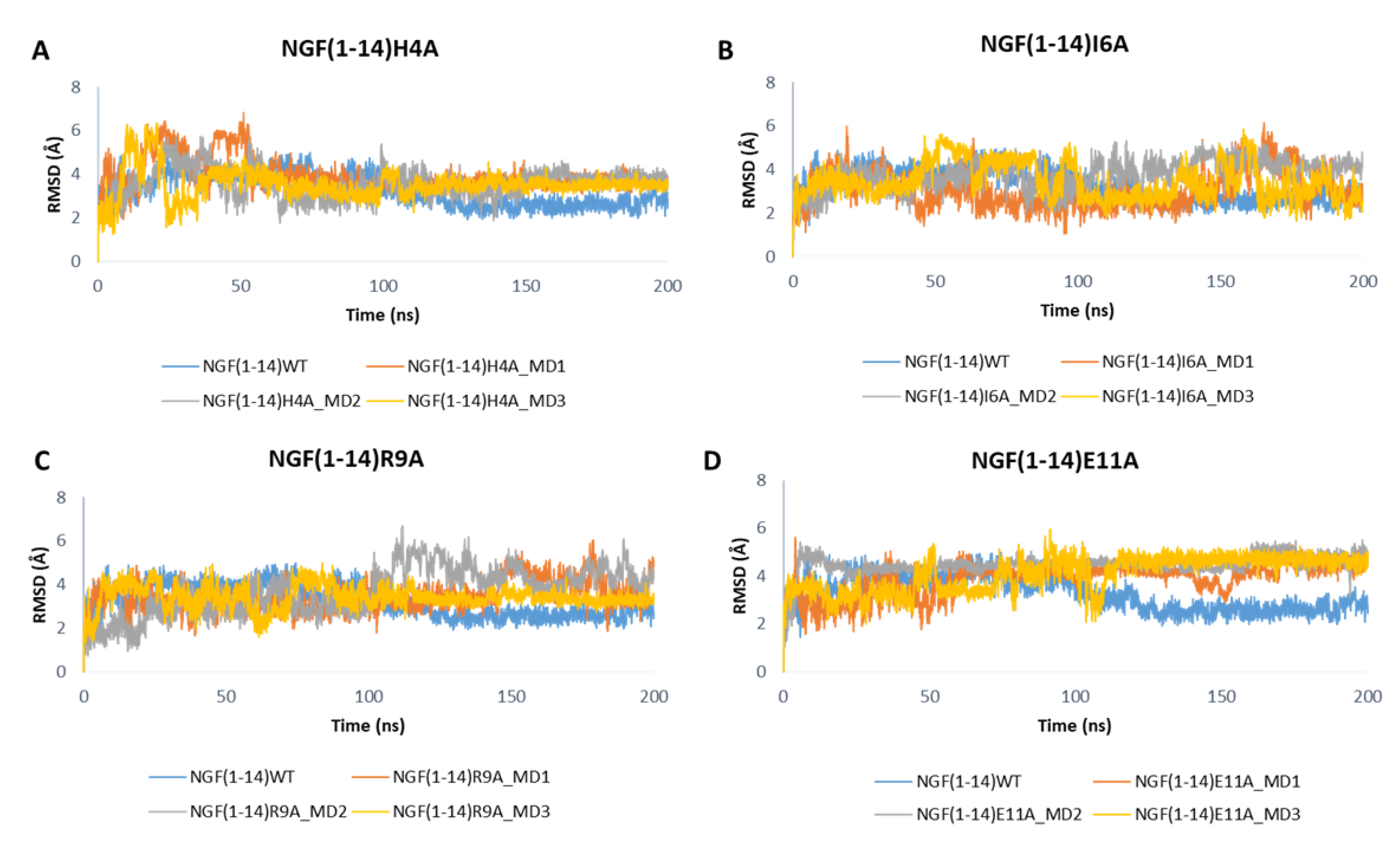
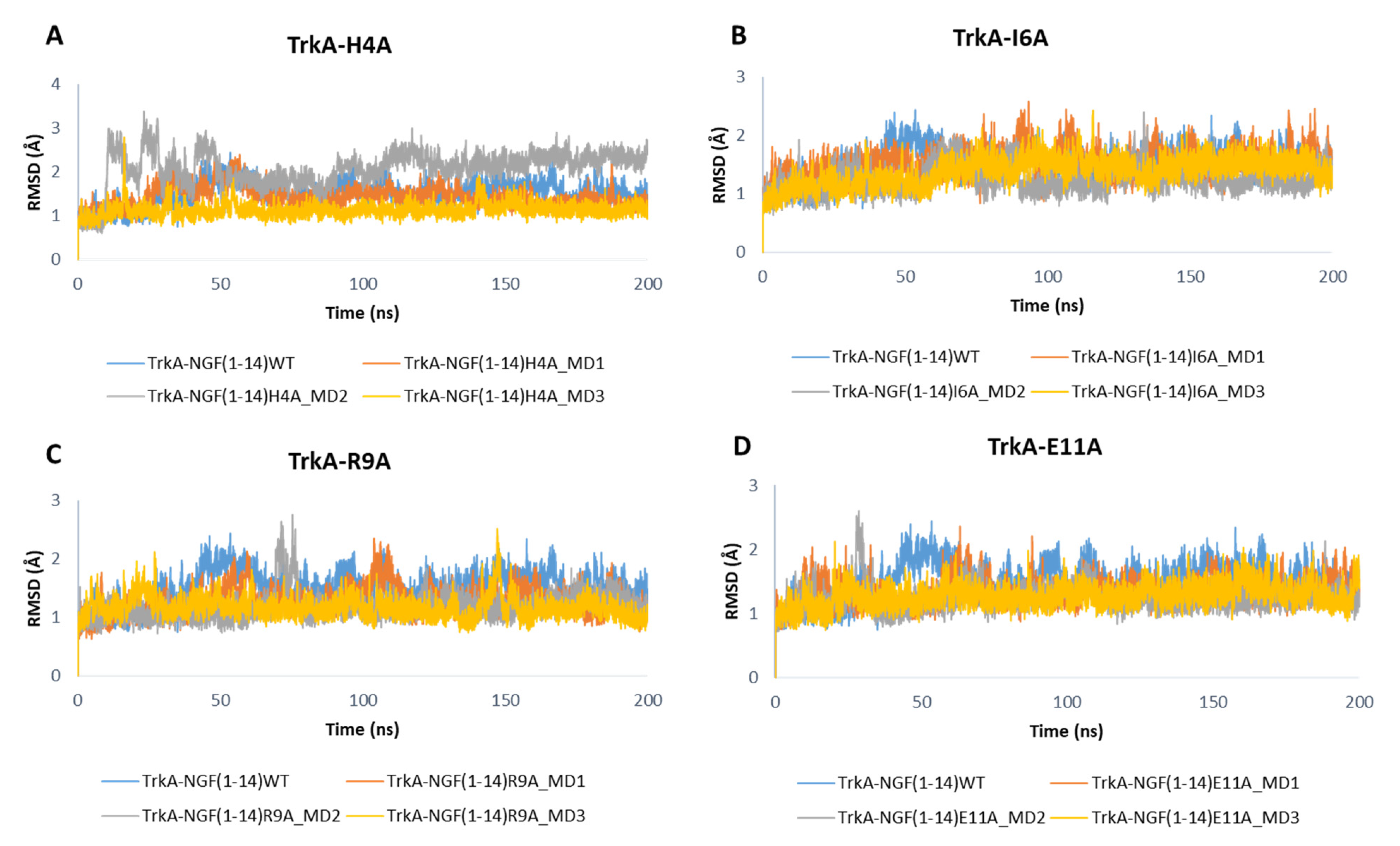
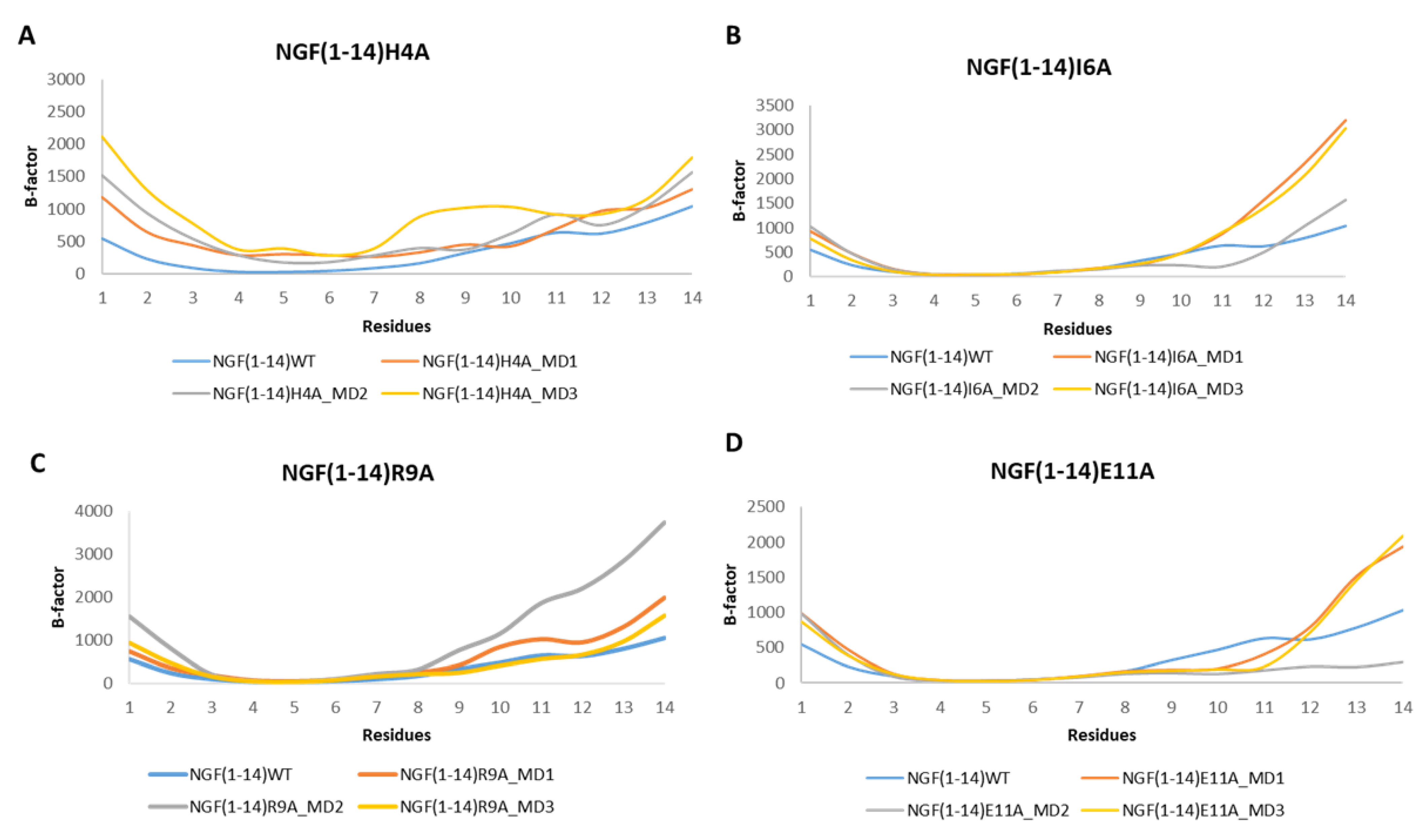
3.3. MD Simulations of TrkA in Complex with NGF(1–14) Mutants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hirose, M.; Kuroda, Y.; Murata, E. NGF/TrkA Signaling as a Therapeutic Target for Pain. Pain Pract. 2016, 16, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.V. Neurotrophins and Their Receptors: A Convergence Point for Many Signalling Pathways. Nat. Rev. Neurosci. 2003, 4, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.J.; Allen, S.J.; Dawbarn, D. Targeting Nerve Growth Factor in Pain: What Is the Therapeutic Potential? BioDrugs 2008, 22, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Behbahani, H.; Eriksdotter, M. Innovative Therapy for Alzheimer’s Disease-with Focus on Biodelivery of NGF. Front. Neurosci. 2019, 13, 1–21. [Google Scholar] [CrossRef]
- Cattaneo, A.; Capsoni, S. Painless Nerve Growth Factor: A TrkA Biased Agonist Mediating a Broad Neuroprotection via Its Actions on Microglia Cells. Pharmacol. Res. 2019, 139, 17–25. [Google Scholar] [CrossRef]
- Pflugfelder, S.C.; Massaro-Giordano, M.; Perez, V.L.; Hamrah, P.; Deng, S.X.; Espandar, L.; Foster, C.S.; Affeldt, J.; Seedor, J.A.; Afshari, N.A.; et al. Topical Recombinant Human Nerve Growth Factor (Cenegermin) for Neurotrophic Keratopathy. Ophthalmology 2020, 127, 14–26. [Google Scholar] [CrossRef]
- Poole, A.J.; Frigotto, L.; Smith, M.E.; Baar, C.; Ivanova-Berndt, G.; Jaulent, A.; Stace, C.; Ullman, C.G.; Hine, A.V. A C-Terminal Cysteine Residue Is Required for Peptide-Based Inhibition of the NGF/TrkA Interaction at NM Concentrations: Implications for Peptide-Based Analgesics. Sci. Rep. 2019, 9, 930. [Google Scholar] [CrossRef]
- Wang, W.; Chen, J.; Guo, X. The Role of Nerve Growth Factor and Its Receptors in Tumorigenesis and Cancer Pain. BioSci. Trends 2014, 8, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.W.; Li, R.; Geetha, T.; Tao, Y.X.; Babu, J.R. Nerve Growth Factor in Metabolic Complications and Alzheimer’s Disease: Physiology and Therapeutic Potential. Biochim. Et Biophys. Act Mol. Basis Dis. 2020, 1866, 165858. [Google Scholar] [CrossRef]
- Wiesmann, C.; Ultsch, M.H.; Bass, S.H.; De Vos, A.M. Crystal Structure of Nerve Growth Factor in Complex with the Ligand-Binding Domain of the TrkA Receptor. Nature 1999, 401, 184–188. [Google Scholar] [CrossRef]
- Wiesmann, C.; De Vos, A.M. Nerve Growth Factor: Structure and Function. Cell. Mol. Life Sci. 2001, 58, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.; Laramee, G.R.; Schmelzer, C.H.; Burton, L.E.; Winslow, J.W. Mutagenesis Identifies Amino-Terminal Residues of Nerve Growth Factor Necessary for Trk Receptor Binding and Biological Activity. J. Biol. Chem. 1994, 269, 27679–27686. [Google Scholar] [CrossRef]
- Woo, S.B.; Timm, D.E.; Neet, K.E. Alteration of NH2-Terminal Residues of Nerve Growth Factor Affects Activity and Trk Binding without Affecting Stability or Conformation. J. Biol. Chem. 1995, 270, 6278–6285. [Google Scholar] [CrossRef]
- Woo, S.B.; Neet, K.E. Characterization of Histidine Residues Essential for Receptor Binding and Activity of Nerve Growth Factor. J. Biol. Chem. 1996, 271, 24433–24441. [Google Scholar] [CrossRef] [PubMed]
- Berrera, M.; Cattaneo, A.; Carloni, P. Molecular Simulation of the Binding of Nerve Growth Factor Peptide Mimics to the Receptor Tyrosine Kinase A. Biophys. J. 2006, 91, 2063–2071. [Google Scholar] [CrossRef]
- Travaglia, A.; Pietropaolo, A.; Di Martino, R.; Nicoletti, V.G.; La Mendola, D.; Calissano, P.; Rizzarelli, E. A Small Linear Peptide Encompassing the NGF N-Terminus Partly Mimics the Biological Activities of the Entire Neurotrophin in PC12 Cells. ACS Chem. Neurosci. 2015, 6, 1379–1392. [Google Scholar] [CrossRef]
- Pandini, G.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Travaglia, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The Inorganic Side of NGF: Copper(II) and Zinc(II) Affect the NGF Mimicking Signaling of the N-Terminus Peptides Encompassing the Recognition Domain of TrkA Receptor. Front. Neurosci. 2016, 10, 569. [Google Scholar] [CrossRef]
- Triaca, V.; Fico, E.; Sposato, V.; Caioli, S.; Ciotti, M.T.; Zona, C.; Mercanti, D.; La Mendola, D.; Satriano, C.; Rizzarelli, E.; et al. HNGF Peptides Elicit the NGF-TrkA Signalling Pathway in Cholinergic Neurons and Retain Full Neurotrophic Activity in the DRG Assay. Biomolecules 2020, 10, 216. [Google Scholar] [CrossRef]
- Naletova, I.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Pandini, G.; Triaca, V.; Amadoro, G.; Latina, V.; Calissano, P.; Travaglia, A.; et al. The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides. Cells 2019, 8, 301. [Google Scholar] [CrossRef] [Green Version]
- Gascon, S.; Jann, J.; Langlois-Blais, C.; Plourde, M.; Lavoie, C.; Faucheux, N. Peptides Derived from Growth Factors to Treat Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 6071. [Google Scholar] [CrossRef]
- Dege, N.; Gökce, H.; Doğan, O.E.; Alpaslan, G.; Ağar, T.; Muthu, S.; Sert, Y. Quantum Computational, Spectroscopic Investigations on N-(2-((2-Chloro-4,5-Dicyanophenyl)Amino)Ethyl)-4-Methylbenzenesulfonamide by DFT/TD-DFT with Different Solvents, Molecular Docking and Drug-Likeness Researches. Colloids Surf. A Physicochem. Eng. Asp. 2022, 638, 128311. [Google Scholar] [CrossRef]
- Çapan, İ.; Shehu, A.; Sert, Y.; Çelik, İ.; Erol, M.; Koca, İ.; Servi, S. Synthesis, Molecular Docking, Molecular Dynamics and Evaluation of Drug-Likeness Properties of the Fused N -Formyl Pyrazoline Substituted New Dehydroepiandrosterone Derivatives. J. Biomol. Struct. Dyn. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pedretti, A.; Mazzolari, A.; Gervasoni, S.; Fumagalli, L.; Vistoli, G. The VEGA Suite of Programs: An Versatile Platform for Cheminformatics and Drug Design Projects. Bioinformatics 2021, 37, 1174–1175. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A Server for Estimating PKas and Adding Missing Hydrogens to Macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Paterlini, M.G.; Ferguson, D.M. Constant Temperature Simulations Using the Langevin Equation with Velocity Verlet Integration. Chem. Phys. 1998, 236, 243–252. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Kräutler, V.; Van Gunsteren, W.F.; Hünenberger, P.H. A Fast SHAKE Algorithm to Solve Distance Constraint Equations for Small Molecules in Molecular Dynamics Simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Vittorio, S.; Ielo, L.; Mirabile, S.; Gitto, R.; Fais, A.; Floris, S.; Rapisarda, A.; Germanò, M.P.; De Luca, L. 4-Fluorobenzylpiperazine-Containing Derivatives as Efficient Inhibitors of Mushroom Tyrosinase. Chem. Med. Chem. 2020, 15, 1757–1764. [Google Scholar] [CrossRef] [PubMed]


| System | ΔG (Kcal/mol) | Mean | ||
|---|---|---|---|---|
| MD1 | MD2 | MD3 | ||
| Complex-S | −53.07 ± 9.01 | −55.56 ± 8.99 | −54.68 ± 10.41 | −54.44 ± 9.47 |
| Complex-C | −20.99 ± 5.95 | −22.32 ± 9.99 | −19.11 ± 7.55 | −20.81 ± 7.83 |
| NGF(1–14)-TrkA | −40.49 ± 11.35 | −38.28 ± 8.30 | −42.82 ± 8.58 | −40.53 ± 9.41 |
| Systems | ΔG (kcal/mol) | ΔΔG | |||
|---|---|---|---|---|---|
| MD1 | MD2 | MD3 | Mean | ||
| TrkA-NGF(1–14)WT | −40.49 ± 11.35 | −38.28 ± 8.30 | −42.82 ± 8.58 | −40.53 ± 9.41 | - |
| TrkA-NGF(1–14)H4A | −34.74 ± 11.82 | −34.70 ± 11.01 | −31.22 ± 7.05 | −33.55 ± 9.96 | 6.98 ± 4.40 |
| TrkA-NGF(1–14)I6A | −40.34 ± 8.68 | −40.95 ± 7.78 | −38.66 ± 9.98 | −39.98 ± 8.81 | 0.55 ± 4.27 |
| TrkA-NGF(1–14)R9A | −28.99 ± 5.72 | −31.05 ± 6.47 | −29.67 ± 5.20 | −29.90 ± 5.80 | 10.63 ± 3.90 |
| TrkA-NGF(1–14)E11A | −37.27 ± 6.66 | −33.10 ± 6.80 | −35.41 ± 7.61 | −35.26 ± 7.02 | 5.27 ± 4.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vittorio, S.; Manelfi, C.; Gervasoni, S.; Beccari, A.R.; Pedretti, A.; Vistoli, G.; Talarico, C. Computational Insights into the Sequence-Activity Relationships of the NGF(1–14) Peptide by Molecular Dynamics Simulations. Cells 2022, 11, 2808. https://doi.org/10.3390/cells11182808
Vittorio S, Manelfi C, Gervasoni S, Beccari AR, Pedretti A, Vistoli G, Talarico C. Computational Insights into the Sequence-Activity Relationships of the NGF(1–14) Peptide by Molecular Dynamics Simulations. Cells. 2022; 11(18):2808. https://doi.org/10.3390/cells11182808
Chicago/Turabian StyleVittorio, Serena, Candida Manelfi, Silvia Gervasoni, Andrea R. Beccari, Alessandro Pedretti, Giulio Vistoli, and Carmine Talarico. 2022. "Computational Insights into the Sequence-Activity Relationships of the NGF(1–14) Peptide by Molecular Dynamics Simulations" Cells 11, no. 18: 2808. https://doi.org/10.3390/cells11182808
APA StyleVittorio, S., Manelfi, C., Gervasoni, S., Beccari, A. R., Pedretti, A., Vistoli, G., & Talarico, C. (2022). Computational Insights into the Sequence-Activity Relationships of the NGF(1–14) Peptide by Molecular Dynamics Simulations. Cells, 11(18), 2808. https://doi.org/10.3390/cells11182808









