Abstract
One of the most damaging issues to cultivatable land is soil salinity. While salt stress influences plant growth and yields at low to moderate levels, severe salt stress is harmful to plant growth. Mineral shortages and toxicities frequently exacerbate the problem of salinity. The growth of many plants is quantitatively reduced by various levels of salt stress depending on the stage of development and duration of stress. Plants have developed various mechanisms to withstand salt stress. One of the key strategies is the utilization of microRNAs (miRNAs) that can influence gene regulation at the post-transcriptional stage under different environmental conditions, including salinity. Here, we have reviewed the miRNA-mediated adaptations of various plant species to salt stress and other abiotic variables. Moreover, salt responsive (SR)-miRNAs, their targets, and corresponding pathways have also been discussed. The review article concludes by suggesting that the utilization of miRNAs may be a vital strategy to generate salt tolerant crops ensuring food security in the future.
1. Introduction
Salinity is characterized by the accumulation of large levels of soluble salts in the soil, resulting in a high salt concentration that is harmful to plant growth [1]. Hyper-ionic and hyper-osmotic stress are caused by high salinity, resulting in physiological drought conditions that can harm plant species [2]. Prolonged saline conditions result in the production of reactive oxygen species (ROS), including an increase in mono-oxygen, superoxide, hydroxyl radicals, and hydrogen peroxide [2]. Salinity-induced ROS causes oxidative damage to cellular components such as proteins, lipids, and DNA and disrupts key plant biological activities [3]. A reversible slowdown in metabolism and growth or irreversible cell death could occur due to tissue injuries. Such conditions make soil salinity a crucial stress among abiotic factors.
Salinization arises because of an imbalance in the water cycle when water runs out on irrigated land and salt accumulates because of inadequate drainage [4]. With the expansion of irrigation networks, salinity is also increasing. Salinization is estimated to affect nearly 20% of total irrigated land. That percentage is likely to climb to 30% in the following two and a half decades and to around 50% by the middle of the 21st century [5]. Saline soils have caused significant agronomic problems in Asia, Africa, and South America’s dry and semi-arid regions, with around 20% of cultivated land facing salinity stress [6].
Plants’ responses to salinity have complicated physiological characteristics, embedding several changes in metabolic processes and implications of gene networks [7]. MicroRNAs (miRNAs) are 20–22 nucleotide regulatory RNAs encoded by endogenous miR genes in plants [8]. Their primary transcripts generate precursor RNAs with a partially double-stranded stem-loop structure, which Dicer-like (DCL) proteins convert into mature miRNAs. In the miRNA biogenesis pathway, RNA polymerase II (Pol II) transcribes primordial miRNAs (pri-miRNAs) from nuclear-encoded miR genes, resulting in precursor transcripts with a distinctive hairpin structure [9]. DCL1 converts pri-miRNA into pre-miRNA with the help of the HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE) proteins [10,11]. miRNA duplexes are formed by the de novo synthesis of the pre-miRNA hairpin precursor regulated by DCL1, HYL1, and SE. HASTY (HST1), an exportin protein, is sent into the cytoplasm after methylation by Hua enhancer 1 (HEN1) [12,13]. RNA-induced silencing complex’s (RISC’s) catalytic component, Argonaute (AGO) protein, binds one strand of the duplex (miRNA) in the cytoplasm and uses sequence complementarity to direct RISC to cognate target transcripts [14]. miRNAs influence gene expression by creating epigenetic changes like DNA and histone methylation and influencing post-transcriptional targets [15,16]. Several excellent reviews have recently been published on the biogenesis of miRNAs in plants [10,11,13,17,18,19,20,21].
In eukaryotes, miRNAs influence gene expression after the occurrence of transcription [22]. miRNA strands are normally destroyed, but they can also be transformed into functional guide strands that play regulatory roles in plants [23]. In addition to their role in plant growth and developmental processes, miRNAs regulate plant responses to biotic and abiotic stress conditions [24,25,26]. Many salinity-responsive miRNAs and their targets in plants have been found. Some of the miRNAs, their target genes, and their functions are given in Table 1. Keeping in view the regulatory role of miRNAs during saline conditions, this review focused on new insights about plant miRNAs responsive to salt stress and miRNA-associated regulatory networks that trigger molecular events in plants under such conditions. Correspondingly, this review also highlighted the recent findings about SR-miRNAs, their target genes, and corresponding pathways.
2. Abiotic Stress Responsive miRNAs in Plants
When confronted with environmental stresses, some miRNAs exhibit altered regulations. Nutrient imbalance [27,28], drought [29,30,31], salinity [32,33,34], cold [35,36,37], heat or high temperatures [38,39,40], ultraviolet (UV)-B/C rays [41,42] result in altered expressions of various miRNA families. miR398 was the first miRNA to be directly linked to environmental stress tolerance in plants. miR398 specifically targets two genes encoding Copper/Zinc (Cu/Zn) and superoxide dismutases (SODs) in Arabidopsis; CSD1 (cytosolic) and CSD2 (extracellular) [43]. Sunkar et al. [44] discovered that down-regulating miR398 increased oxidative stress tolerance in transgenic lines than in wild-type (WT) plants.
The expression levels of 117 miRNAs in Arabidopsis were determined using microarray technology and miRNA chips under salt and other abiotic stress conditions [45]. Examining how their promoter regions affect cis-regulation and assessing their expression patterns verified the findings of more than a dozen stress-induced miRNAs [45]. A new miRNA family targeting SOD, laccases (LACs), and ATP sulfurylases (APS) has been discovered in Arabidopsis [46]. Sulfate (S) deficiency increased the expression of miR395, demonstrating that miRNA expression could arise under environmental stress conditions [47]. Inorganic S assimilation is performed by the APS-encoding genes (APS1, APS3, and APS4), which are targeted by miR395 [48]. Cold, dryness, excessive salinity, and abscisic acid (ABA) treatments, have all been found to up-regulate miR393 [49]. A response to all stressors up-regulated miR397b and miR402 [50]; however, miR319c was only up-regulated in a cold environment [51], while responses to all stressors resulted in miR398a down-regulation [52].
Abiotic stress has been shown to differentially affect miR394 expression profiles [45]. Ren et al. [53] showed that several gene-encoding transcription factors (TFs) and many enzymes were targeted by pto-miR394, whose expression increased with drought but decreased under high water conditions. These included gene-encoding F-box domains, MYB (myeloblastosis)-like DNA-binding domains, ABC transporter transmembrane regions, DNA polymerase family B, and putative methyltransferases. Several stress conditions, including high salinity, drought, and low temperatures, were used to induce stress in Arabidopsis, which resulted in the identification of 14 stress-inducible miRNAs [45]. A real-time reverse transcription–polymerase chain reaction (RT-PCR) confirmed that these stress-inducible miRNAs were differentially expressed. Three miRNAs were up-regulated during these stressful conditions, i.e., miR168, miR171, and miR396.
Many microbial and metal stress-responsive miRNAs have been uncovered in plants [23,25,26]. According to Wang et al. [54], there were 25 known and nine novel miRNAs in Raphanus sativus that showed differential expression under lead (Pb) stress. Several stress-related signaling and secondary metabolite pathways are primarily targeted by Pb-responsive miRNAs. In response to low nitrogen stress, Wang et al. [55] observed differential expression of some soybean miRNAs via utilization of deep sequencing (DS). The authors further discovered that the potential targets of these miRNAs were involved in different biological functions.
In two tea plant cultivars that had been treated with cold stress, Zhang et al. [56] identified 106 known miRNAs and 98 potentially novel miRNAs. The authors also uncovered 238 common targets in response to cold and control treatment. In addition, 455 and 591 genes were identified as miRNA cleavage targets in cold and control treatment groups, respectively. According to gene ontology (GO) annotations, miRNA target genes were involved in transcription, stress responses, and developmental processes. These results offered valuable insight into miRNA functions associated with cold stress in tea cultivars. There are at least 40 plant miRNA families associated with abiotic stress. These families are mostly related to salt and drought stress [55,57,58]. Candar-Cakir et al. [59] applied small RNA (sRNA) and degradome sequencing (DgS) to systematically identify tissue-specific miRNAs in multiple tomato genotypes under drought stress. The key miRNAs involved in the tomato response to drought stress included miR160, miR165, miR166, miR171, miR398, miR408, miR827, miR9472, miR9476, and miR9552. Moreover, miR169, miR172, miR393, miR5641, miR5658, and miR7997 differentially regulated the genes involved in plant hormone signal transduction pathways in all tissues regardless of genotype [59]. miRNAs do not need to be involved in plant stress adaptation, even though they are differentially regulated in response to environmental stresses.
It has been observed that some miRNAs are responsible for increasing plant tolerance to multiple environmental stresses. For example, in Bermuda grass grown under combined cold and salt stresses, out of five miRNAs (miR827-5p, PC-3p-49895, PC-5p-104176, PC-5p-56353, and PC-5p-67388), the down-regulation of three miRNAs (osa-miR160, osa-miR160a-5p, and PC-5p-131796) was observed [60]. It has been found that Glycine max miR169l-3p, miR5036, miR862a, and miR398a/b targeted ethylene-responsive TF4, protein phosphatase 2C, and Cu/Zn-SOD as a response to single and double stress phosphate starvation and salt [61]. Salinity and alkalinity treatments increased the sensitivity of rice seedlings overexpressing miR393 [62], while salt stress tolerance increased in plants overexpressing a miR393-resistant transport inhibitor response protein 1 (TIR1). A higher germination rate, increased water-use efficiency, delayed senescence, and stabilized chlorophyll levels were observed in A. thaliana plants under saline conditions [63]. Other studies found that exogenous pri-osa-miR393a conferred enhanced drought (heat) and salt tolerance in transgenic Agrostis stolonifera (creeping bentgrass) plants [64]. Plants expressing the transgene had fewer but longer tillers, reduced stomatal density, denser cuticles, increased potassium uptake, and enhanced expression of small heat-shock proteins, exhibiting the plant’s ability to withstand multiple stresses can be improved via genetic incorporation of miR393.
As a result of overexpression of miR408 in Arabidopsis, salinity, cold, and oxidative stress tolerance were enhanced, but drought sensitivity and osmotic stress sensitivity were also increased [65]. Nicotiana benthamiana responded similarly when Salvia miltiorrhiza miR408 was heterologously expressed. There was reduced ROS accumulation in the plants and higher tolerance to salt stress [66]. A study showed that osa-miR408 overexpression improved drought tolerance in perennial ryegrass [67] and chickpea [68]. It is possible that decreased leaf water loss is associated with morphological changes in transgenic plants, such as curled leaves and sunken stomata. It has been demonstrated that TaemiR408, a miRNA family member in wheat (Triticum aestivum), also exhibited induced expression patterns when exposed to salt stress and phosphate starvation and that its induced expression was gradually repressed when the plants reverted to normal conditions [69]. It appeared that miR408 played a crucial role in improving plant tolerance to multiple stresses caused by several abiotic factors.
Multiple stress conditions typically up-regulate miR319 via targeting Teosinte branched 1, Cycloidea, and proliferating cell nuclear antigen binding factor (TCP/PCF) [70]. Increasing miR319 expression increased creeping bentgrass’s tolerance for salt and drought stress [71]. Across transgenic potato plants, water retention and membrane integrity were increased under salt stress conditions, and Na+ accumulation was reduced [72]. osa-miR319 overexpression reduced OsPCF5 and OsPCF8 expression levels in rice, resulting in cold stress tolerance [73]. When considered together, the overexpression of miR319 in plants improved their tolerance to various environmental stresses. Genetically engineered tobacco plants expressing zmmiR156 were more resistant to drought and salt without compromising their architecture because their transcript levels of senescence-associated genes were reduced [74]. According to these findings, miRNAs could be useful in developing crop varieties that are more resistant to multiple stresses, thereby enhancing the productivity of agriculture.
3. Plant miRNAs and Salt Stress
During salt stress, numerous gene transcripts are variably regulated by miRNAs, indicating that transcription in stressed plants is tightly modulated; hence, salt stress interaction is heavily influenced by post-translational gene regulations (Table 1). There was a discrepancy between leaf and root miR398 levels of salt-stressed Populus euphratica as miR398 levels increased in the former while decreased in the latter. The expression of three miRNAs (miR164, miR166, and miR169) significantly altered P. euphratica when exposed to salt [75]. Numerous environmental factors, including salt stress, affected the expression of a diverse set of miRNAs in P. trichocarpa [76]. Furthermore, Phaseolus vulgaris harbored increased levels of miRNA-targeting genes implicated in Calmodulin-binding transcription activators (CAMTAs) [77]. During salt stress, the differential expression of two miR169 family members was also observed, i.e., miR169g and miR169n [78]. Upstream of miR169n, a cis-acting, ABA-responsive region was discovered, suggesting that this stress-responsive hormone regulates miR169n. The wheat leaves miR169 down-regulated via salinity expressed NF-YA (nuclear factor Y subunit A) [79]. Researchers found that salt-sensitive (SS) and salt-tolerant (ST) Zea mays cultivars showed down-regulation of miR156, miR164, miR171, miR167, and miR396, and up-regulation of miR162, miR168, miR395, and miR474 [80].

Table 1.
Salt-responsive microRNA, their corresponding targets, and regulations in various plant species.
Table 1.
Salt-responsive microRNA, their corresponding targets, and regulations in various plant species.
| microRNA | Target | Plant Species | miRNA/Target Module Function | Regulations | References | |
|---|---|---|---|---|---|---|
| Up | Down | |||||
| miR156 | Unknown | Gossypium raimondii | Abiotic stress tolerance | [81] | ||
| SPLs | Raphanus sativus | Delays flowering; regulates leaf development, fruit ripening, vegetative and reproductive stage transitions; tillering and branching; plays key roles in embryogenesis, morphogenesis, life cycle stage transformation, and flower formation. | [82] | |||
| SPLs | Panicum virgatum | [83] | ||||
| SPLs | Arabidopsis thaliana | [45] | ||||
| SPLs | Malus domestica | [84] | ||||
| POPTR_0007s01030 | Populus trichocarpa | Unknown | [85] | |||
| Unknown | Medicago truncatula | Abiotic stress tolerance | [86] | |||
| Unknown | Solanum lycopersicum | [87] | ||||
| UGTs | Hordeum spontaneum | Increases anthocyanin synthesis, leading to enhanced antioxidative capacity. | [88] | |||
| miR157 | SPLs | P. virgatum | Modulate leaf initiation rate | [83] | ||
| Unknown | G. raimondii | Regulation of biological processes | [81] | |||
| miR159 | MYBs | Oryza sativa | Growth and flowering, role in fruit development. | [89] | ||
| MYBs | P. virgatum | [83] | ||||
| MYBs | Nicotiana tabacum | [90] | ||||
| MYBs | M. truncatula | [91] | ||||
| miR160 | ARFs | G. raimondii | Regulating plant growth and development through auxin signaling pathways | [81] | ||
| ARFs | R. sativus | [82] | ||||
| ARFs | O. sativa | [89] | ||||
| ARFs | Setaria italica | [92] | ||||
| ARFs | Triticum aestivum | [93] | ||||
| miR161 | AGO | A. thaliana | Vital in salinity stress response | [94] | ||
| miR162 | DCLs | S. italica | miRNA biogenesis plays a vital role in saline and drought conditions | [92] | ||
| DCLs | P. virgatum | [83] | ||||
| miR164 | NAC | R. sativus | Critical role in regulating the response to salt and drought stress | [82] | ||
| NAC | A. thaliana | [95] | ||||
| NAC | P.euphratica | [96] | ||||
| Pavirv00056088m | P.virgatum | Despite regulating salt stress, involvement in any other regulatory mechanisms is still unknown. | [81] | |||
| POPTR_0007s08420 | P.trichocarpa | [97] | ||||
| GRMZM2G114850 | Zea mays | [98] | ||||
| miR165 | unknown | T. aestivum | Determining the positional fate of leaf tissues (adaxial or abaxial) and xylem differentiation in root stele tissues | [99] | ||
| HD-ZIP | A. thaliana | [45] | ||||
| miR166 | Unknown | G. raimondii | Plant development processes and abiotic stresses resistance | [81] | ||
| SPB-like | A. thaliana | [95] | ||||
| SPB-like | Glycine max | [100] | ||||
| SPB-like | Z. mays | [101] | ||||
| miR167 | Unknown | G. raimondii | Regulates some reproductive development processes, such as anther dehiscence, and ovule, embryonic, and seed development. | [81] | ||
| ARF | A. thaliana | [45] | ||||
| ARF | N. tabacum | [90] | ||||
| ARF | T. aestivum | [93] | ||||
| ARF | Z. mays | [102] | ||||
| miR168 | AGOs | Saccharum spp. | Facilitates plant adaptation to K+-deficiency stress, influences phase transition, leaf epinasty, and fruit development | [103] | ||
| AGOs | A. thaliana | [45] | ||||
| MYBs | P. euphratica | [84] | ||||
| AGOs | Z. mays | [102] | ||||
| Unknown | G. raimondii | [81] | ||||
| Unknown | Vigna unguiculata | [104] | ||||
| miR169 | NY-FA | Z. mays | Regulates tolerance to abiotic stresses in both monocots and dicots; plays a key role in nutrient uptake. | [78] | ||
| CCAAT-binding | A. thaliana | [45] | ||||
| CBF HAP2-like factor | N. tabacum | [90] | ||||
| CCAAT-binding TF | P. euphratica | [84] | ||||
| CBF HAP2-like factor | G. max | [105] | ||||
| CCAAT-binding TF | V. unguiculata | [104] | ||||
| miR171 | Scarecrow-like TFs | A. thaliana | Plant growth and development | [45] | ||
| AP2 | P. trichocarpa | [84] | ||||
| AP2 | S. italica | [92] | ||||
| Unknown | S. lycopersicum | [86] | ||||
| miR172 | AP2 | G. raimondii | Regulates the transitions between developmental stages and specifies floral organ identity | [81] | ||
| AP2 | N. tabacum | [90] | ||||
| AGOs | A. thaliana | [94] | ||||
| AP2 | H. spontaneum | [88] | ||||
| NNC1 | G. max | [106] | ||||
| MYBs | S. lycopersicum | [107] | ||||
| miR319 | TCPs | A. thaliana | Cooperatively regulates downstream genes, such as CUC genes, for cotyledon boundary, leaf serration formation, and other physiological responses. | [45] | ||
| MTR_3g011610 | M. truncatula | [108] | ||||
| PvPCF5 | A.s thaliana | [109] | ||||
| miR390 | ARFs | Populus spp. | Directs the production of tasiRNAs from Trans-acting siRNA3 (TAS3) transcripts to regulated ARF genes | [110] | ||
| TAS | Helianthus tuberosus | [111] | ||||
| miR393 | F-box | A. thaliana | Regulates the expression of different sets of TAAR genes following pathogen infection or nitrate treatment and regulates expression of the TIR1/AFB2 auxin receptor clade and auxin-related development | [45] | ||
| F-box | G. raimondii | [81] | ||||
| AFB2 | H. spontaneum | [88] | ||||
| AsTIR1 | Agrostis stolonifera | [64] | ||||
| miR394 | F-box | G. raimondii | Participates in the regulation of plant development and stress responses | [81] | ||
| F-box | A. thaliana | [45] | ||||
| F-box | G. max | [105] | ||||
| miR395 | Unknown | S. lycopersicum | An important regulator involved in sulfate transport and assimilation and a high-affinity sulfate transporter | [86] | ||
| ATP sulfurylase | P. virgatum | [83] | ||||
| miR396 | Unknown | G. raimondii | Control cell proliferation, margin, and vein pattern formation | [81] | ||
| GRFs | A. thaliana | [45] | ||||
| GRFs | N. tabacum | [90] | ||||
| GRFs | P. virgatum | [83] | ||||
| bHLH74 | R. sativus | [82] | ||||
| GRFs | A. stolonifera | [112] | ||||
| GRFs | A. thaliana | [113] | ||||
| miR397 | LACs | S. linnaeanum | Functioning in lignin synthesis and are involved in the development of plants under various conditions | [45] | ||
| cDNA l-ascorbate oxidase precursor | P. virgatum | [83] | ||||
| miR398 | Cu/Zn Superoxide dismutase | A. thaliana | Regulates plant responses to oxidative stress, water deficit, salt stress, abscisic acid stress, ultraviolet stress, copper and phosphate deficiency | [45] | ||
| miR399 | ATP-dependent RNA helicase | M. truncatula | Regulates phosphate homeostasis | [108] | ||
| ATP-dependent RNA helicase | T. aestivum | [93] | ||||
| miR402 | DEMETER-LIKE protein 3 | A. thaliana | Regulator of seed germination and seedling growth | [109] | ||
| miR408 | DEAD-box helicases | O. sativa | Provide an important cross-link between plant growth, development, and stress response. | [114] | ||
| SnRK2 | T. aestivum | [69] | ||||
| Cu-binding proteins | N. benthamiana | [66] | ||||
| miR414 | GhFSD1 | A. thaliana | Critical role in regulating the growth and development of plants’ cell development and cell differentiation | [115] | ||
| miR474 | PPR | Populus cathayana | Plant nutrient homeostasis | [116] | ||
| miR482 | TIR-NBS-LRR | P. trichocarpa | Regulates defense mechanisms | [117] | ||
| GRAS | S. lycopersicum | [107] | ||||
| miR530 | F-box | P. trichocarpa | Plant resistance against multiple pathogens and nutrient homeostasis | [117] | ||
| miR1444 | POPTR_0001s39950 | P. trichocarpa | Regulates copper homeostasis | [117] | ||
| miR1445 | Unknown | P. trichocarpa | Unknown | [117] | ||
| miR1446 | GRM-like protein | P. euphratica | Nutrient homeostasis | [84] | ||
| miR1447 | ABC transport protein | P. euphratica | Abiotic stress tolerance | [84] | ||
| miR1448 | unknown | P. euphratica | Disease resistance against fungal pathogens | [84] | ||
| miR1507 | NBS-LRR | G. max | Activators of plant defense | [105] | ||
| miR1711 | unknown | P. trichocarpa | Unknown | [117] | ||
| miR2118 | APS-reductase | Phaseolus vulgaris | Involved in the production of 21-nt phasiRNAs | [118,119] | ||
| miRNVL5 | GhCHR | G. hirsutum | Vital in plant response to salinity | [120] | ||
Squamosa promoter-binding protein-like (SPB-Like); Auxin response factors (ARFs); Dicer-like (DCL); Argonaute (AGO); Nucleotide-binding site–leucine-rich repeat (NBS-LRR); ATP-binding cassette (ABC); Gibberellin response modulator (GRM); Toll/interleukin receptor (TIR); Pentatricopeptide repeat (PPR); SNF1-related protein kinase 2 (SnRK2); Auxin signaling F-BOX 2 (AFB2); Teosinte branched1-cycloidea-proliferating cell factor (TCP); Apetala 2, (AP2); NACs (NAM, no apical meristem, petunia, ATAF1–2, Arabidopsis thaliana activating factor, and CUC2, cup-shaped cotyledon); Homeodomain-leucine zipper (HD-Zip); Squamosa promoter-binding-like (SPL).
miR393 is a conserved miRNA found in several plant species. The rice miR393 family consists of two members: osa-miR393 and osa-miR393b [62]. The expression of osa-miR393 significantly changed under salt and alkaline stress, whereas the expression of osa-miR393b remained stable. According to the authors, over-expression of athmiR395c detained seed germination in Arabidopsis under conditions of excessive salt or dehydration, but over-expression of athmiR395e improved seed germination under saline conditions [121]. As a result, miRNAs from the same family may play a variety of roles. Next-generation sequencing (NGS) techniques were used to explore soybean stress-related miRNAs [61,122]. The researchers discovered that under salt stress circumstances, soybeans generated 133 conserved miRNAs from 95 distinct miRNA families, differentially expressing 50 miRNAs [122]. miR159 and miR319 expression increased in artichoke tissues following a saline solution treatment [123]. Medicago truncatula and M. sativa had different expression patterns for several miRNAs, including miR156 and miR166, indicating that these two plant species had different levels of salt tolerance [124]. According to DS of transcripts and miRNAs, salt stress inhibited the expression of most miRNAs in banana roots. Moreover, other stress-related functions, such as cellular homeostasis, metabolism, and cellular stress responses, were also inhibited [125]. Salt-stress-sensitive miRNAs have been identified in nursery seedlings of Eutrema salsugineum using Solexa sequencing, which was vital to analyzing the direct and indirect responses to salinity [126]. The authors proposed that salt-responsive (SR) precursor miRNA genes contain numerous stress- and phytohormone-regulatory cis-regulatory elements [126].
Salt-responsive miRNAs of Solanum lycopersicum and S. pimpinellifolium were characterized by generating libraries of miRNAs from NaCl-treated and untreated seedlings. miRNAs were found to belong to 45 different families, with 95 conserved or known and 254 unknown. In response to salt stress, 109 novel and 14 conserved miRNAs were significantly regulated—specifically, the interaction of the miR156e-5p with miR23b and miR50a in S. pimpinellifolium [107]. In wild emmer, researchers have also identified salt-induced miRNAs. They discovered a total of 212 miRNAs, 50 of which were salinity-sensitive, with 32 significantly up-regulated and 18 down-regulated. miR172b and miR1120a, as well as miR393a, were the most significantly differentially expressed. Based on these results, wild wheat miRNAs can be explored in terms of their biological functions and evolution [127].
One-hundred-fifty conserved miRNAs and 348 new miRNAs were discovered in salt-treated Oryza glaberrima (African rice). Salinity stress differentially regulated 29 known and 32 new miRNAs. The GO and Kyoto encyclopedia of genes and genomes (KEGG) analyses showed that salinity stress tolerance was mediated by several targets. Based on the analysis of RT-PCR data, it appeared that some miRNAs were expressed in the adipose tissue in the same manner as indicated by Illumina sequencing data. An inverse correlation was found between the target gene and miRNA expression [128]. osa-miR396c was overexpressed in transgenic creeping bentgrass (Agrostis stolonifera) in perennial monocots under saline conditions. The leaves in mutants were smaller, the internodes were shorter, the leaves had fewer leaf veins, and there were fewer epidermal cells per square inch as compared with their wild counterparts. When exposed to high salinity levels, transgenic plants showed improved water retaining capacity, chlorophyll content, and cell membrane integrity. This provided insights into miRNA-mediated regulatory networks by establishing molecular links between upstream regulatory elements and downstream functional elements in the miR396 pathway [112]. The overexpression of a particular miRNA, miR1841, in rice (Oryza sativa) plants significantly alleviated salt stress [129]. Originally discovered in Dongxiang wild rice (Oryza rufipogon Griff.) [130], miR1861 plays a crucial role in salt stress in rice (Oryza sativa) plants.
The researchers investigated the role of 40 miRNAs belonging to 19 different families in the superfruit guava (Psidium guajava L.). In response to salinity stress, seven guava miRNAs (miR156f, miR160c, miR162, miR164b, miR166t, miR167a, and miR390b) showed differential regulation [33]. In another study, under salt conditions, the Hassawi-3 (a faba bean SS genotype) underwent a comparison with the ST-ILB4347 genotype. It was found that Hassawi-3 and ILB4347 differentially expressed 527 and 693 miRNAs, respectively. There was also a significant increase in 284 miRNAs in Hassawi-3 under control and a reduction in 243 miRNAs in ILB4347 plants during stress conditions [131].
The NGS and qRT-PCR validations of miRNAs associated with salinity stress responses in Niger (Guizotia abyssinica Cass.) were recently conducted [132]. The research findings identified 212 conserved miRNAs in oil-producing plants (300 mM NaCl) and 203 miRNAs in control libraries. From these libraries, 6 and 16 new miRNAs were predicted from stressed and control Niger, respectively. Based on qRT-PCR evaluations, it was found that six miRNAs were up-regulated (miR166, miR169, miR156, miR6173, miR6478, and miR166), while four miRNAs were down-regulated (miR166e, miR156a, miR159b, and miR169h) [132]. Setaria viridis, which is an emerging monocotyledonous grass model species, has similarly been shown to accumulate miR397 from Arabidopsis transformant lines [133]. A range of developmental phenotypes was observed in the transformed lines, termed Sv-MIR397 plants, exhibiting mild to severe dwarfism. By using qRT-PCR, the authors determined that miR397 overabundance repressed expression of the LAC target gene and reduced lignin contents in the Sv-MIR397 transformant plant population. Sv-MIR397 transformants were also more sensitive to salt stress than WT Arabidopsis plants were after exposure to a seven-day salt stress regime [133].
Different cultivars of the same plant species may exhibit variable responses to salt stress conditions resulting in differential expression of miRNAs. Among two rice cultivars, IR26 (sub. Xian) and JCQ and Jiucaiqing (sub. Geng), 73 mature SR-miRNAs were identified through DS. In addition to transcriptional regulation, miRNAs targeting these genes were also involved in responding to stimuli [67]. Moreover, to determine whether alfalfa and WT plants have different SR-miRNAs, researchers constructed sRNA libraries. Alfalfa plants grown under normal or saline conditions contained 128 miRNAs. Accordingly, 29 and 23 different miRNAs were differentially expressed between the Alfalfa-CK and WT-CK salt-supplemented plants, respectively [134]. Correspondingly, SR-miRNAs in sweet potatoes were uncovered through DS using libraries constructed from leaves and roots of sweet potatoes treated with NaCl (Na-150) and NaCl-free (CK) [135]. The results exhibited the existence of 175 novel and 66 conserved miRNAs. Salinity stress increased 51 miRNAs (22 known and 29 novel miRNAs) and significantly reduced 76 (61 known and 15 novel miRNAs) in sweet potato leaves. In roots, 13 miRNAs were significantly up-regulated (12 known and 1 novel miRNA), and 9 were significantly down-regulated (seven known miRNAs and two novel miRNAs) [135]. Furthermore, miRNA profiling showed that miR169, miR395, miR396, miR397, miR398, and miR408 played major roles in shoot and root tissue of two S. viridis accessions (A10 and ME-034V) when exposed to salinity [136]. The sRNA and DgS of barley, which is among the most salt-tolerant cereal crops, revealed 40 and 51 SR-miRNAs in the roots and shoots of ST-XZ16 and Golden Promise (GP), respectively [88]. There were several miRNAs involved in salt tolerance in roots, such as miR156d, miR164a, miR393a, miR319a, and miR172b, which targeted Uridine 5′-diphospho-glucuronosyltransferase (UDP-glucuronosyltransferase, UGTs), NACs (NAM no apical meristem, petunia, ATAF1–2 Arabidopsis thaliana activating factor, and CUC2 cup-shaped cotyledon), TIR1, TCP4, and APETALA2 (AP2). It has been suggested that miR159a, miR169i, miR319a/miR396e modules, and miR172b, which regulate MYB33, TCP4, growth regulating factors (GRFs), and AP2, contributed to salt tolerance in shoots [88].
Using Illumina high throughput sequencing and comprehensive in-silico analysis to obtain insight into salinity tolerance in the roots of two contrasting wheat cvv, namely Suntop (ST) and Sunmate (SS), 110 conserved and 81 novel miRNAs were identified. There were 191 miRNAs identified in both cultivars. Among them, 181 miRNAs were shared between the two cultivars. In total, these miRNAs belonged to 35 known families, of which 23 were conserved, and 12 were unique families. Saline conditions induced 43 and 75 miRNAs in Suntop and Sunmate, respectively [137]. These findings improved our understanding of how miRNAs participate in the cellular process of salt tolerance, and this knowledge may assist in genetically improving wheat cultivars. SR-miRNAs are being discovered in great numbers as genomic technology and procedures have evolved, leading toward our better understanding of their targets and gene expression.
4. The Target Genes and Related Pathways of Salt-Responsive miRNAs
Multiple miRNAs that target genes from the same family are frequently found in various plant species. Evidence suggests that miRNAs can selectively operate on certain target genes in a variety of contexts [138]. Based on maize miRNA sequencing, TFs are often targeted by miRNAs that are critical for the growth of plants and the formation of their organs. Earlier model plant research confirms these results. Targets of zma-miR164a/b/c/d are known to be MYBs, NAC1, and HD-ZIP (Homeodomain-leucine zipper proteins) [98].
Salt stress responses in plants involve several regulatory proteins, including MADS-box and zinc-finger proteins [139], that can be miRNA targets. In addition to TFs, various metabolic pathways and physiological functions are regulated by miRNA-targeted genes. The genes encoding salt-stress-responsive enzymes and proteins in plants include cytochrome oxidase and NADP-dependent malic enzyme. Both are predicted miRNA targets [140]. For example, in salt-induced soybeans treated with sulfate deficiency, miR395 regulates the sulfurylase and ASP1 genes. Moreover, miR395 is important in energy supply maintenance [141,142]. In saline conditions, miR394a, which regulates the F-box proteins, was drastically up-regulated in the leaves and roots of P. euphratica [143]. A similar trend was seen in Arabidopsis ath-miR394 during saline conditions [53]. However, a negative expression pattern was observed in rice cultivar IR26 for osa-miR394:LOC_Os01g6940 [67].
It has been found that genes from the LAC family, which are implicated in salinity response [144], are homologous to cca-miR397 and cca-miR399. With Arabidopsis as a heterologous system, S. viridis miR397 could reduce lignin content and increase salt stress sensitivity by repressing three Arabidopsis LAC genes. Furthermore, in salt-stressed Col-0, MIM399, and MIR399 plants with an elevated miR399 abundance and altered expression of the PHOSPHATE2 (PHO2) target genes, significant changes were observed in the expression levels of the PO4 transporter genes (PHT1;4 and PHT1;9. PHT1;4 and PHT1;9). PO4 transporters were elevated in salt-stressed Arabidopsis and could enhance PO4 translocation from the roots to the shoots, which would increase the amount of this precious cellular resource available. To maintain essential biological processes or to mount an adaptive response to salt stress, Arabidopsis aerial tissues could use salt stress to maintain essential biological processes [145]. In this model, it was clearly demonstrated that the anthocyanin biosynthesis pathway was accelerated to a higher degree when induced by stress in the MIR399 transformant lines compared to the degree to which this antioxidant pigment production pathway was stimulated in Col-0 plants or MIM399 plants [145]. Using the above molecular manipulation model, the same authors’ group showed that Arabidopsis responded to salt stress by altering miR396/GRF expression [113].
B3 DNA-binding domain proteins and auxin response factors (ARFs) are bound by miR160 [146]. Growth, development, and response to environmental conditions are influenced by several ARFs [147,148]. In RNA-seq analyses, 29 and 30 SR-ARF members from A. duranensis and A. ipaensis were identified, respectively [149]. There was a prediction that miR160 might target Arahy.7DXUOK, an ARF gene. Plants transgenic with miR160OX may exhibit increased salt tolerance due to overexpression of miR160. The TATA-box is essential for the expression of ARF genes, as most members of the ARF genes participate in related signaling pathways. P-box elements [150], tricarboxylic acid (TCA) elements [151], ABRE (ABA-responsive elements) motifs [152], Gibberellic acid responsive elements (GARE) motifs [153], TGACG and CGTCA motifs have all been identified as hormone-responsive elements in ARF gene members. The two wild peanuts showed functional diversity and involvement in various biological processes, including responses to phytohormones, abiotic stresses, and even tissue development, according to a cis-element analysis [149].
Based on GO analysis, many important physiological players (genes) have recently been targeted by SR-miRNAs, such as ARFs, AA/IAA-ARF-dimerization, Cytochrome P450, Chlorophyll A-B binding protein, NADPH-cytochrome P450 reductase, Homeobox leucine-zipper proteins, NF-YA, and MYBs [132]. miR166a is a prominent SR-miRNA that is associated with an increased rooting rate, DNA glycosylase, and phosphotransferase activity in Larix leptolepis, promoting lateral root formation and carbohydrate metabolism [154]. This particular miRNA targets HD-Zipsi which is essential for root development in Niger plants during salt stress [132].
Auxins (AA) are vital hormones in the development of lateral roots and apical dominance in plants by influencing cell division, elongation, and differentiation [155]. In Vigna unguiculata, miR160a expression increased in saline conditions. miR160a and miR160b expression, on the other hand, increased after five hours of salt treatment but declined 24 h later [104]. The target gene for miR398b encodes Cu/Zn-SOD; however, miR398b and miR395 are generally expressed in opposite directions [156] (Figure 1). APS and Kelch motif proteins are molecular targets of miRNAs. Plants produce ATP using APS and the pyrophosphate anion (Ul96) [157]. Plant development was considerably impacted by miR396 inhibition via targeting GRFs [158] (Figure 1). Specifically targeting the cytochrome oxidase subunit I gene, the Z. mays miR396 family was down-regulated in plants growing in saline soil [102]. P. cathayana had a diminished expression of miR396f and activated target genes that produce GRF when salt stress was applied [116].
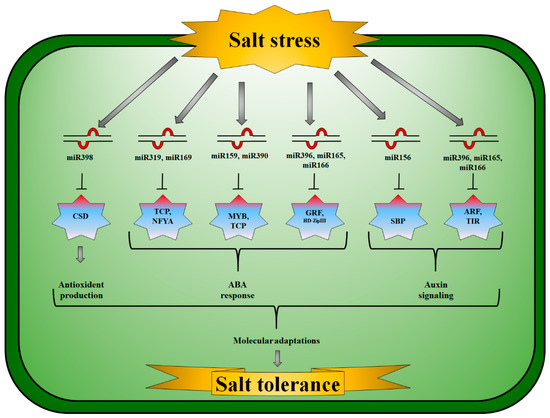
Figure 1.
Salt-responsive microRNA and their corresponding targets and signaling pathways in plants. miR398/CSD (Copper superoxide dismutase) module produces antioxidants, miR319/TCPs (Teosinte branched1/Cycloidea/Proliferating cell factor), miR169/NFYA (Nuclear factor Y subunit A), miR159/MYBs (Myeloblastosis), miR390/TCPs, miR165/miR166/HD-Zip (Homeodomain-leucine zipper proteins), and miR396/GRF (Growth-regulating factors) module are key ABA-responsive elements. miR156/SBPs (Squamosa promoter binding protein), miR165/miR166/TIR (Toll/Interleukin receptor), and miR396/ARF (Auxin response factors) modules play vital roles in auxin signaling to initiate molecular response to tolerate salinity in different plant species.
AGO1 gene, which encodes the enzyme that creates RNA from miRNAs, is controlled by miR168 [159]. miR168 and AGO1 affect miRNA target genes in the same manner. Salt-stressed maize also produces miR168, which performs a regulatory function [101] miR161 and miR173 expression increased, while pri-miR161 and pri-miR173 expression decreased [94]. In the cytoplasm, these miRNAs stabilized, whereas their expression in the nucleus was negatively regulated by AGO1. It was concluded that AGO1 took part in interacting with chromatin at the loci of the above miRNAs, causing the transcriptional complex to disassemble and short, unpolyadenylated transcripts to be released [160].
Target genes are implicated in a range of critical pathways, including AA and ethylene (ET) signaling, RNA-mediated silencing, and DNA methylation [161]. According to researchers, miRNVL5 and GhCHR are two critical components involved in salinity response. Further, salt stress increased the expression of GhCHR (a gene of miRNA ovary line 5) while reducing the expression of corresponding miRNA [120]. It was concluded that the constitutive expression of miRNVL5 made transgenic plants susceptible to salt stress, whereas GhCHR expression enhanced salt tolerance [162]. In NaCl-treated and untreated seedlings of S. lycopersicum and S. pimpinellifolium, SR-miRNA’s target genes were predicted via GO and miRNA involvement in salt-stress-related biochemical pathways, including photosynthetic pathways, hormonal signaling pathways, phospholipid signaling pathways, and calcium signaling pathways [107]. Through the use of GO and KEGG analyses, targets for miR172b and miR1120a were predicted in wild emmer. Target proteins such as TFs and stress-related proteins were enriched in salinity-responsive miRNAs [127]. The GO classification and KEGG pathway analysis of the potential target genes in WT and M. alfalfa plants under normal and saline conditions predicted that the majority of target genes were related to plant growth and development and showed significant differences between WT and M. alfalfa plants. Moreover, miR172-CNGC (cyclic nucleotide-gated channel), miR319-CAX2 (CATION EXCHANGER 2), miR408-NHX (NA+/H+ exchanger), and miR2590-CHX14/15 (cation/H+ exchanger) were significantly up-regulated in M. alfalfa plants compared with WT plants, suggesting that M. alfalfa plants have higher ion transport levels [134].
As specified by GO and KEGG, SR-O. glaberrima miRNA targets several genes that participate in salinity stress resistance pathways [128]. Four potential miR396 targets were additionally identified in creeping bentgrass, and the levels of these targets were elevated when the plants were stressed by salt. In stressful conditions, miR396 was found to be required for salt stress tolerance through both functional and regulatory proteins (TFs and protein kinases) [112]. Based on results from the psRNATarget tool, P. guajava miRNA target transcripts were characterized to have 49 potential targets, mostly involved in metabolism, cellular development, and stress responses [33]. Faba bean genotypes Hassawi-3 (SS) and ST-ILB4347 have specific miRNA targets involved in regulating specific SR genes, primarily TFs, LACs, SODs, plantacyanins, and F-box proteins.
Salinity-induced miRNAs and their targets are involved in corresponding biological networks and associated pathways such as ABC transportation, MAPK (mitogen-activated protein kinase) signaling networks, and plant hormone networks, indicating that miRNAs play a role in salt stress tolerance in the ILB4347 genotype [131]. Similarly, in-silico analysis of two contrasting SR-wheat cultivars (Suntop and Sunmate) identified more than 800 targets for the 75 known miRNAs. Signaling activities of miR156, miR160, miR171, miR319, miR159, miR9657, and miR59 were linked to ARFs, SPLs (Squamosa-promoter binding protein-like), and Scarecrow-like 6 (SCL6). It was predicted that the proteins PCF5 (binds to the core sequence of the promoter), R2R3-MYB, and CBL-CIPK (CBL-interacting protein kinases) were involved in salt tolerance [137]. Similar to this, a miRNA-DgS of sweet potato samples treated with and without salt revealed that the SPL/miR156 and miR169/TOP1 modules, as well as GC4, HSP90, UBXN1, miR393/AFB2, and miR162/DCL modules, played vital roles during saline conditions [135].
miRNAs have been found to regulate hormone pathways in plants under salt stress. Plant stress responses may be linked to AA signaling through miR160, miR167, and miR393 [163]. As a result of their low expression levels, miR393, miR160, and miR167 are slightly inhibited by the expression of ARFs under non-stressed conditions. Up-regulated miR393 repressed AA signaling by lowering TIR1, which increased AA/IAA-ARF heterodimerization under stress conditions [164]. ARF levels were also directly reduced by miR160 and miR167 up-regulation [164] and miR162 down-regulation [32]. Salt treatment of Kentucky bluegrass (Poa pratensis L.) led to changes in the expression levels of miRNAs [165]. Salt treatment increased the expression of miR162, miR173, miR391, miR408, miR773, and miR857 by 70%, then declined to levels similar to those of the control after 24 h. A 20% decrease in miR775 and miR827 expression levels was observed after 24 h, followed by an 80% decrease after 144 h. miR841 expression increased by 50% after 24 h of salt treatment but stabilized after 144 h. Salt treatment significantly increased the expression of ARFs between 12 and 144 h, respectively. When salinity stress was applied to the callus, miRNAs were found to regulate SR-gene families [165].
Several studies have established that miRNAs target the TIR1 or ARF genes to contribute to salt stress responses [64,166]. He et al. [110] found that salt-induced miR390 expression stimulated tasiARF production for the degradation of ARF4 transcripts that influence ARFs. Inhibition of salt-induced AA signaling was facilitated by the decreased expression of ARF4. Nodulation and salt stress are regulated by miR390 in dual ways. By overexpressing miR390 in M. truncatula, lateral root growth is stimulated. Nodule organogenesis and rhizobial infection are prevented, and nodulation genes are inhibited, while miR390/TAS3 (trans-acting-small interfering RNA3) inactivation leads to more nodulation and rhizobial infections [167]. Using NaCl concentrations of 100 mM and 300 mM, the authors found that in Helianthus tuberosus, miR390 expression is induced by 100 mM, whereas miR390 expression is inhibited by 300 mM [111]. A miR390-TAS3-tasiARFs module pathway plays distinct functions in regulating salt stress, and nodulation is also complex. Furthermore, a recent study revealed that miR167 genes were constitutively diminished during organogenesis under target-mimicry-based conditions [166]. Under stress, miR167 mimic lines exhibited a greater magnitude of organogenesis compared to the parent line (cultured in NaCl concentrations of 12.5 and 25 mM). As a consequence of miR167 reduction paired with salt stress (up to 12.5 mM), AA transporter genes AUX1 (Auxin influx transporter 1), PIN1 (Peptidylprolyl Cis/Trans Isomerase, NIMA-Interacting 1), and PIN2 showed synergistic effects resulting in enhanced callogenesis and reduced organogenesis. miR167 reduction-associated signaling pathways were reflected in the increased relative water content, chlorophyll, and antioxidant activity of in-vitro grown miR167 mimic shoot initials [166].
Additionally, miRNAs modulate the ABA and ET metabolic pathways to provide resistance to salt toxicity. There are several miRNAs involved in the ABA metabolic pathway during salt stress. i.e., miR156, miR172, miR393, miR394, and miR399 [168,169,170,171,172]. Moreover, salt stress regulates miR319 and the ET metabolism pathway [173]. ABA-mediated pathways are negatively regulated by scaffold protein receptors for activated C kinase 1 (RACK1) [174], e.g., in A. thaliana through miR393s [175]. Several studies have demonstrated that miR6478 controls the ET signaling pathway in Niger plants under salt stress, targeting AGO/DCL protein, PAZ (Piwi, Ago, and Zwille), and CTR1 (CONSTITUTIVE TRIPLE RESPONSE 1), thus promoting miRNA turnover [132]. There is evidence that the CTR1 gene participates in the ET signal transduction pathway. Plant ET responses are negatively regulated by the amino terminus of CTR1, which was previously reported to form a complex with ET [176]. Furthermore, OsNAC2 overexpressing lines (ZUOErN3 and ZUOErN4) were more salt tolerant than WT rice seeds since their levels of ABA were higher in comparison with WT seeds [177]. RT-PCR revealed that OsNAC2-overexpressing plants expressed significantly more ABA biosynthesis genes OsNCED1 (9-cis-epoxycarotenoid dioxygenases) and OsNCED3, as well as higher levels of expression of stress-responsive genes OsP5CS1 (pyrroline-5-carboxylate synthase 1), OsLEA3 (late embryogenesis abundant), and OsRab16 (responsive to ABA) [177].
ROS accumulates in plants as a result of oxidative stress during salt stress conditions [178]. SOD converts superoxide radicals into molecular oxygen and hydrogen peroxide, which provide the first cellular defense against oxidative stress. By introducing miR397, miR398, miR408, and miR528 in the cell, Cu/Zn-SOD, and LACs are suppressed, thereby controlling ROS accumulation and the availability of Cu ions. Additionally, miR398, miR408, and miR528 are involved in heavy metal transport regulation [26]. Through its effects on iron SOD (Fe-SOD, FSD) in cotton, miRNA414c affects salinity tolerance [115]. L-ascorbate oxidase (LAO) is an enzyme that breaks down ascorbate, a molecule that detoxifies H2O2. miR12477 plays a vital role in the inhibition of LAO. Salt stress results in low ROS accumulation when miR12477 expression is high [34]. The AP2/ERF (Ethylene response factors) domain-containing TF gene INDETERMINATE SPIKELET1 (IDS1) plays a crucial role in interacting with ROS produced by salt-treated plants via miR172a/b [179].
5. Conclusions and Future Prospects
Gene expression patterns affect plants’ responses to salinity stress. miRNAs, which are active post-transcriptional regulators, are known to regulate stress-related genes. Understanding how miRNAs affect genetic expressions will enable us to understand miRNA’s role in saline conditions in many plant species. The new availability of whole plant genomes and high-throughput sequencing technology has fueled research into miRNA control under salt stress. Because miRNAs are so crucial in gene regulation networks, learning more about them should help us better understand plant salt tolerance responses. Understanding miRNA-mediated regulation networks could lead to new strategies to improve plant salt tolerance genetically. Although we are still learning about miRNA evolution, studies on salinity-responsive miRNAs and their corresponding target networks in different cell types may uncover how miRNA target networks operate in these different cell types. Identifying miRNAs that change expression levels in response to salt stress in various agricultural plants and their target genes remains a slow progress. The solution reveals new elements in plant stress tolerance and helps unravel the salt stress response’s complex regulatory network. If we can better understand miRNA activity under salt stress, we may be able to use miRNA-mediated gene regulation to improve plant stress resistance, particularly for economically important crops, to ensure future food security.
Author Contributions
Contributions from all authors were considered in preparing this manuscript. W.I. gathered information, drafted the manuscript, and prepared figures and tables; H.N. and A.W. edited, helped in the preparation of the table and data collection; F.Z. supervised, and W.I. finalized and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Program of Joint Funds of the National Natural Science Foundation of China and Xinjiang Uygur Autonomous Region of China (No. U1903102); and the National Natural Science Foundation of China (No. 41977050).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are thankful to our friend (Susan Sen, a UK national and native English speaker) from Rongqiao International School, Fuzhou, for editing the English language of our article and improving the English standards.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R. A review: Impact of salinity on plant growth. Nat. Sci. 2019, 17, 34–40. [Google Scholar]
- Goyal, V.; Jhanghel, D.; Mehrotra, S. Emerging warriors against salinity in plants: Nitric oxide and hydrogen sulphide. Physiol. Plant. 2021, 171, 896–908. [Google Scholar] [CrossRef]
- Kamran, M.; Parveen, A.; Ahmar, S.; Malik, Z.; Hussain, S.; Chattha, M.S.; Saleem, M.H.; Adil, M.; Heidari, P.; Chen, J.T. An overview of hazardous impacts of soil salinity in crops, tolerance mechanisms, and amelioration through selenium supplementation. Int. J. Mol. Sci. 2020, 21, 148. [Google Scholar] [CrossRef]
- Minhas, P.S.; Ramos, T.B.; Ben-Gal, A.; Pereira, L.S. Coping with salinity in irrigated agriculture: Crop evapotranspiration and water management issues. Agric. Water Manag. 2020, 227, 105832. [Google Scholar] [CrossRef]
- Mustafa, G.; Akhtar, M.S.; Abdullah, R. Global concern for salinity on various agro-ecosystems. In Salt Stress, Microbes, and Plant Interactions: Causes and Solution: Volume 1; Springer: Singapore, 2019; pp. 1–19. ISBN 9789811388019. [Google Scholar]
- Hussain, S.; Shaukat, M.; Ashraf, M.; Zhu, C.; Jin, Q.; Zhang, J. Salinity Stress in Arid and Semi-Arid Climates: Effects and Management in Field Crops. In Climate Change and Agriculture; IntechOpen Limited: London, UK, 2019. [Google Scholar]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Wang, J.; Mei, J.; Ren, G. Plant microRNAs: Biogenesis, homeostasis, and degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef]
- Tyagi, S.; Sharma, S.; Ganie, S.A.; Tahir, M.; Mir, R.R.; Pandey, R. Plant microRNAs: Biogenesis, gene silencing, web-based analysis tools and their use as molecular markers. 3 Biotech 2019, 9, 413. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Sarwat, M.; Hasan, S.; Roychodhury, N. Biogenesis, functions and fate of plant microRNAs. J. Cell. Physiol. 2012, 227, 3163–3168. [Google Scholar] [CrossRef]
- Wen-wen, K.; Hong-bo, W.; Jing, L. Biogenesis of Plant MicroRNAs. J. Northeast Agric. Univ. Engl. Ed. 2014, 21, 84–96. [Google Scholar] [CrossRef]
- Chen, X. microRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, P.J.; Park, C.M. MicroRNA biogenesis and function in higher plants. Plant Biotechnol. Rep. 2009, 3, 111–126. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Pashkovskiy, P.P.; Ryazansky, S.S. Biogenesis, evolution, and functions of plant microRNAs. Biochem. 2013, 78, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Nie, J.; Wang, H. MicroRNA biogenesis in plant. Plant Growth Regul. 2021, 93, 1–12. [Google Scholar] [CrossRef]
- Chorostecki, U.; Moro, B.; Rojas, A.M.L.; Debernardi, J.M.; Schapire, A.L.; Notredame, C.; Palatnik, J.F. Evolutionary footprints reveal insights into plant microRNA biogenesis. Plant Cell 2017, 29, 1248–1261. [Google Scholar] [CrossRef]
- Zhu, J.K. Reconstituting plant miRNA biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 9851–9852. [Google Scholar] [CrossRef]
- Islam, W.; Qasim, M.; Noman, A.; Adnan, M.; Tayyab, M.; Farooq, T.H.; Wei, H.; Wang, L. Plant microRNAs: Front line players against invading pathogens. Microb. Pathog. 2018, 118, 9–17. [Google Scholar] [CrossRef]
- Islam, W.; Noman, A.; Qasim, M.; Wang, L. Plant responses to pathogen attack: Small rnas in focus. Int. J. Mol. Sci. 2018, 19, 515. [Google Scholar] [CrossRef] [Green Version]
- Islam, W.; Adnan, M.; Huang, Z.; Lu, G.; Chen, H.Y.H. Small RNAs from seed to mature plant. CRC Crit. Rev. Plant Sci. 2019, 38, 117–139. [Google Scholar] [CrossRef]
- Islam, W.; Islam, S.U.; Qasim, M.; Wang, L. Host-Pathogen interactions modulated by small RNAs. RNA Biol. 2017, 14, 891–904. [Google Scholar] [CrossRef]
- Noman, A.; Sanaullah, T.; Khalid, N.; Islam, W.; Khan, S.; Irshad, M.K.; Aqeel, M. Crosstalk between plant miRNA and heavy metal toxicity. In Plant Metallomics and Functional Omics: A System-Wide Perspective; Springer: Cham, Denmark, 2019; pp. 145–168. ISBN 9783030191030. [Google Scholar]
- Paul, S.; Datta, S.K.; Datta, K. miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 2015, 6, 232. [Google Scholar] [CrossRef]
- Islam, W.; Tauqeer, A.; Waheed, A.; Zeng, F. MicroRNA Mediated Plant Responses to Nutrient Stress. Int. J. Mol. Sci. 2022, 23, 2562. [Google Scholar] [CrossRef]
- Liu, M.; Yu, H.; Zhao, G.; Huang, Q.; Lu, Y.; Ouyang, B. Identification of drought-responsive microRNAs in tomato using high-throughput sequencing. Funct. Integr. Genom. 2018, 18, 67–78. [Google Scholar] [CrossRef]
- Qiu, C.W.; Liu, L.; Feng, X.; Hao, P.F.; He, X.; Cao, F.; Wu, F. Genome-wide identification and characterization of drought stress responsive microRNAs in Tibetan wild barley. Int. J. Mol. Sci. 2020, 21, 2795. [Google Scholar] [CrossRef]
- Kuruvilla, L.; Sathik, M.; Luke, L.P.; Thomas, M. Identification and validation of drought-responsive microRNAs from Hevea brasiliensis. Acta Physiol. Plant. 2019, 41, 14. [Google Scholar] [CrossRef]
- Shinde, H.; Dudhate, A.; Anand, L.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Small RNA sequencing reveals the role of pearl millet miRNAs and their targets in salinity stress responses. S. Afr. J. Bot. 2020, 132, 395–402. [Google Scholar] [CrossRef]
- Sharma, A.; Ruiz-Manriquez, L.M.; Serrano-Cano, F.I.; Reyes-Pérez, P.R.; Alfaro, C.K.T.; Andrade, Y.E.B.; Aros, A.K.H.; Srivastava, A.; Paul, S. Identification of microRNAs and their expression in leaf tissues of guava (Psidium guajava L.) under salinity stress. Agronomy 2020, 10, 1920. [Google Scholar] [CrossRef]
- Parmar, S.; Gharat, S.A.; Tagirasa, R.; Chandra, T.; Behera, L.; Dash, S.K.; Shaw, B.P. Identification and expression analysis of miRNAs and elucidation of their role in salt tolerance in rice varieties susceptible and tolerant to salinity. PLoS ONE 2020, 15, e230958. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cheng, C.; Chen, F.; Ni, S.; Lin, Y.; Lai, Z. High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans). BMC Plant Biol. 2018, 18, 308. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; He, J.; Jin, G.; Chen, Z.H.; Dai, F. Identification of novel microRNAs for cold deacclimation in barley. Plant Growth Regul. 2020, 92, 389–400. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, T.; Yang, D.; Luo, B.; Wang, W.P.; Yu, D.; He, F.L.; Wang, Q.M.; Rao, L.Q. Identification and characterization of heat-responsive micrornas at the booting stage in two rice varieties, 9311 and nagina 22. Genome 2021, 64, 969–984. [Google Scholar] [CrossRef]
- Vidya, S.M.; Ravishankar, K.V.; Laxman, R.H. Genome wide analysis of heat responsive microRNAs in banana during acquired thermo tolerance. J. Hortic. Sci. 2018, 13, 61–71. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Chen, Y.; Xu, Y.; Xu, J. Profiling of heat-responsive microRNAs in creeping bentgrass (Agrostis stolonifera L.). Curr. Bioinform. 2017, 12, 319–327. [Google Scholar] [CrossRef]
- Xu, Y.; Charles, M.T.; Luo, Z.; Mimee, B.; Tong, Z.; Véronneau, P.Y.; Rolland, D.; Roussel, D. Preharvest Ultraviolet C Treatment Affected Senescence of Stored Strawberry Fruit with a Potential Role of MicroRNAs in the Activation of the Antioxidant System. J. Agric. Food Chem. 2018, 66, 12188–12197. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, J.; Cheng, J.; Jiang, Z.; Xu, N.; An, X.; Chen, Z.; Hao, J.; Yang, S.; Xu, Z.; et al. Identification of UV-B radiation responsive microRNAs and their target genes in chrysanthemum (Chrysanthemum morifolium Ramat) using high-throughput sequencing. Ind. Crops Prod. 2020, 151, 112484. [Google Scholar] [CrossRef]
- Dugas, D.V.; Bartel, B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 2008, 67, 403–417. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Cohu, C.M.; Abdel-Ghany, S.E.; Gogolin Reynolds, K.A.; Onofrio, A.M.; Bodecker, J.R.; Kimbrel, J.A.; Niyogi, K.K.; Pilon, M. Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: Regulation and unexpected phenotypes in an arabidopsis mutant. Mol. Plant 2009, 2, 1336–1350. [Google Scholar] [CrossRef]
- Yuan, N.; Yuan, S.; Li, Z.; Li, D.; Hu, Q.; Luo, H. Heterologous expression of a rice miR395 gene in Nicotiana tabacum impairs sulfate homeostasis. Sci. Rep. 2016, 6, 28791. [Google Scholar] [CrossRef] [PubMed]
- Ai, Q.; Liang, G.; Zhang, H.; Yu, D. Control of sulfate concentration by miR395-targeted APS genes in Arabidopsis thaliana. Plant Divers. 2016, 38, 92–100. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Khare, T.; Tripathi, P.; Shah, T.; Ramakrishna, C.; Aglawe, S.; Mangrauthia, S.K. miRNA applications for engineering abiotic stress tolerance in plants. Biologia 2020, 75, 1063–1081. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, Y.; Tang, R.; Qu, H.; Duan, X.; Jiang, Y. Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress 06 Biological Sciences 0604 Genetics 06 Biological Sciences 0601 Biochemistry and Cell Biology. BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Bustamante, A.; Marques, M.C.; Sanz-Carbonell, A.; Mulet, J.M.; Gomez, G. Alternative processing of its precursor is related to miR319 decreasing in melon plants exposed to cold. Sci. Rep. 2018, 8, 15538. [Google Scholar] [CrossRef]
- Sun, Z.; Shu, L.; Zhang, W.; Wang, Z. Cca-miR398 increases copper sulfate stress sensitivity via the regulation of CSD mRNA transcription levels in transgenic Arabidopsis thaliana. PeerJ 2020, 2020, e9105. [Google Scholar] [CrossRef]
- Ren, Y.; Chen, L.; Zhang, Y.; Kang, X.; Zhang, Z.; Wang, Y. Identification and characterization of salt-responsive microRNAs in Populus tomentosa by high-throughput sequencing. Biochimie 2013, 95, 743–750. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Shen, H.; Zhu, X.; Zhai, L.; Xu, L.; Wang, R.; Gong, Y.; Limera, C.; Liu, L. Identification of Radish (Raphanus sativus L.) miRNAs and Their Target Genes to Explore miRNA-Mediated Regulatory Networks in Lead (Pb) Stress Responses by High-Throughput Sequencing and Degradome Analysis. Plant Mol. Biol. Rep. 2015, 33, 358–376. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Hao, Q.; Sha, A.; Zhou, R.; Zhou, X.; Yuan, L. Elucidation of miRNAs-Mediated Responses to Low Nitrogen Stress by Deep Sequencing of Two Soybean Genotypes. PLoS ONE 2013, 8, e67423. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, X.; Chen, X.; Song, C.; Zou, Z.; Wang, Y.; Wang, M.; Fang, W.; Li, X. Identification and characterization of coldresponsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014, 14, 271. [Google Scholar] [CrossRef]
- Shaw, B.P. Salt stress tolerance in plants: The role of miRNAs. Adv. Plants Agric. Res. 2018, 8, 1. [Google Scholar] [CrossRef]
- Singroha, G.; Sharma, P.; Sunkur, R. Current status of microRNA-mediated regulation of drought stress responses in cereals. Physiol. Plant. 2021, 172, 1808–1821. [Google Scholar] [CrossRef] [PubMed]
- Candar-Cakir, B.; Arican, E.; Zhang, B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 2016, 14, 1727–1746. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, A.; Gitau, M.M.; Huang, X.; Chen, L.; Fu, J. Insights into the MicroRNA-regulated response of bermudagrass to cold and salt stress. Environ. Exp. Bot. 2018, 145, 64–74. [Google Scholar] [CrossRef]
- Ning, L.H.; Du, W.K.; Song, H.N.; Shao, H.B.; Qi, W.C.; Sheteiwy, M.S.A.; Yu, D.-Y. Identification of responsive miRNAs involved in combination stresses of phosphate starvation and salt stress in soybean root. Environ. Exp. Bot. 2019, 167, 103823. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. Osa-MIR393: A salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2011, 38, 237–242. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 73–83. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, S.; Zhou, M.; Yuan, N.; Li, Z.; Hu, Q.; Bethea, F.G.; Liu, H.; Li, S.; Luo, H. Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 2019, 17, 233–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Burd, S.; Lers, A. MiR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Niu, J.; Cao, X. Heterologous expression of salvia miltiorrhiza microRNA408 enhances tolerance to salt stress in nicotiana benthamiana. Int. J. Mol. Sci. 2018, 19, 3985. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Feng, J.; Wang, R.; Zhang, H.; Huang, J. Comparative analysis of microRNAs and their targets in the roots of two cultivars with contrasting salt tolerance in rice (Oryza sativa L.). Plant Growth Regul. 2019, 87, 139–148. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. MiR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef]
- Bai, Q.; Wang, X.; Chen, X.; Shi, G.; Liu, Z.; Guo, C.; Xiao, K. Wheat miRNA taemir408 acts as an essential mediator in plant tolerance to pi deprivation and salt stress via modulating stress-associated physiological processes. Front. Plant Sci. 2018, 9, 499. [Google Scholar] [CrossRef]
- Fang, Y.; Zheng, Y.; Lu, W.; Li, J.; Duan, Y.; Zhang, S.; Wang, Y. Roles of miR319-regulated TCPs in plant development and response to abiotic stress. Crop J. 2021, 9, 17–28. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef]
- Pieczynski, M.; Marczewski, W.; Hennig, J.; Dolata, J.; Bielewicz, D.; Piontek, P.; Wyrzykowska, A.; Krusiewicz, D.; Strzelczyk-Zyta, D.; Konopka-Postupolska, D.; et al. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 2013, 11, 459–469. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Mao, D.; Liu, X.; Ji, C.; Li, X.; Zhao, X.; Cheng, Z.; Chen, C.; Zhu, L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 2207–2218. [Google Scholar] [CrossRef]
- Kang, T.; Yu, C.Y.; Liu, Y.; Song, W.M.; Bao, Y.; Guo, X.T.; Li, B.; Zhang, H.X. Subtly Manipulated Expression of ZmmiR156 in Tobacco Improves Drought and Salt Tolerance Without Changing the Architecture of Transgenic Plants. Front. Plant Sci. 2020, 10, 1664. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Zhou, T.; Bo, W.; Xu, F.; Wu, R. Genome-wide analysis of salt-responsive and novel microRNAs in Populus euphratica by deep sequencing. BMC Genet. 2014, 15, S6. [Google Scholar] [CrossRef]
- Shuai, P.; Liang, D.; Zhang, Z.; Yin, W.; Xia, X. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genom. 2013, 14, 233. [Google Scholar] [CrossRef]
- Büyük, İ.; İlhan, E.; Şener, D.; Özsoy, A.U.; Aras, S. Genome-wide identification of CAMTA gene family members in Phaseolus vulgaris L. and their expression profiling during salt stress. Mol. Biol. Rep. 2019, 46, 2721–2732. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, L.; Fan, Y.; Wang, L. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 555, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ge, L.; Liang, R.; Li, W.; Ruan, K.; Lin, H.; Jin, Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009, 10, 29. [Google Scholar] [CrossRef]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015, 66, 789–804. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K.; et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 1–16. [Google Scholar] [CrossRef]
- Sun, G.; Stewart, C.N.; Xiao, P.; Zhang, B. MicroRNA expression analysis in the cellulosic biofuel crop switchgrass (panicum virgatum) under abiotic stress. PLoS ONE 2012, 7, e32017. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Duan, Z.; Xia, X.; Yin, W. Expression profiles of precursor and mature microRNAs under dehydration and high salinity shock in Populus euphratica. Plant Cell Rep. 2011, 30, 1893–1907. [Google Scholar] [CrossRef]
- Cao, C.; Long, R.; Zhang, T.; Kang, J.; Wang, Z.; Wang, P.; Sun, H.; Yu, J.; Yang, Q. Genome-wide identification of microRNAs in response to salt/alkali stress in medicago truncatula through high-throughput sequencing. Int. J. Mol. Sci. 2018, 19, 4076. [Google Scholar] [CrossRef]
- Çakır, Ö.; Arıkan, B.; Karpuz, B.; Turgut-Kara, N. Expression analysis of miRNAs and their targets related to salt stress in Solanum lycopersicum H-2274. Biotechnol. Biotechnol. Equip. 2021, 35, 283–290. [Google Scholar] [CrossRef]
- Kuang, L.; Shen, Q.; Wu, L.; Yu, J.; Fu, L.; Wu, D.; Zhang, G. Identification of microRNAs responding to salt stress in barley by high-throughput sequencing and degradome analysis. Environ. Exp. Bot. 2019, 160, 59–70. [Google Scholar] [CrossRef]
- Barrera-Figueroa, B.E.; Gao, L.; Wu, Z.; Zhou, X.; Zhu, J.; Jin, H.; Liu, R.; Zhu, J.K. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 2012, 12, 132. [Google Scholar] [CrossRef]
- Frazier, T.P.; Sun, G.; Burklew, C.E.; Zhang, B. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 2011, 49, 159–165. [Google Scholar] [CrossRef]
- Chen, L.; Wang, T.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 2012, 235, 375–386. [Google Scholar] [CrossRef]
- Khan, Y.; Yadav, A.; Bonthala, V.S.; Muthamilarasan, M.; Yadav, C.B.; Prasad, M. Comprehensive genome-wide identification and expression profiling of foxtail millet [Setaria italica (L.)] miRNAs in response to abiotic stress and development of miRNA database. Plant Cell. Tissue Organ Cult. 2014, 118, 279–292. [Google Scholar] [CrossRef]
- Lu, W.; Li, J.; Liu, F.; Gu, J.; Guo, C.; Xu, L.; Zhang, H.; Xiao, K. Expression pattern of wheat miRNAs under salinity stress and prediction of salt-inducible miRNAs targets. Front. Agric. China 2011, 5, 413–422. [Google Scholar] [CrossRef]
- Dolata, J.; Bajczyk, M.; Bielewicz, D.; Niedojadlo, K.; Niedojadlo, J.; Pietrykowska, H.; Walczak, W.; Szweykowska-Kulinska, Z.; Jarmolowski, A. Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol. 2016, 172, 297–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amor, B.B.; Wirth, S.; Merchan, F.; Laporte, P.; D’Aubenton-Carafa, Y.; Hirsch, J.; Maizel, A.; Mallory, A.; Lucas, A.; Deragon, J.M.; et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009, 19, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Dun, H.; Lian, C.; Zhang, X.; Yin, W.; Xia, X. The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol. Biochem. 2017, 115, 418–438. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Duan, H.; Li, J.; Deng, X.W.; Yin, W.; Xia, X. Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol. Biol. 2013, 81, 525–539. [Google Scholar] [CrossRef]
- Shan, T.; Fu, R.; Xie, Y.; Chen, Q.; Wang, Y.; Li, Z.; Song, X.; Li, P.; Wang, B. Regulatory Mechanism of Maize (Zea mays L.) miR164 in Salt Stress Response. Russ. J. Genet. 2020, 56, 835–842. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.F.; Song, N.; Wei, J.P.; Wang, X.J.; Feng, H.; Yin, Z.Y.; Kang, Z.S. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol. Biochem. 2014, 80, 90–96. [Google Scholar] [CrossRef]
- Li, T.; Li, H.; Zhang, Y.X.; Liu, J.Y. Identification and analysis of seven H2O2-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 2011, 39, 2821–2833. [Google Scholar] [CrossRef]
- Kong, Y.; Elling, A.A.; Chen, B.; Deng, X. Differential Expression of microRNAs in Maize Inbred and Hybrid Lines during Salt and Drought Stress. Am. J. Plant Sci. 2010, 1, 69–76. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef]
- Carnavale Bottino, M.; Rosario, S.; Grativol, C.; Thiebaut, F.; Rojas, C.A.; Farrineli, L.; Hemerly, A.S.; Ferreira, P.C.G. High-Throughput Sequencing of Small RNA Transcriptome Reveals Salt Stress Regulated MicroRNAs in Sugarcane. PLoS ONE 2013, 8, e59423. [Google Scholar] [CrossRef]
- Paul, S.; Kundu, A.; Pal, A. Identification and validation of conserved microRNAs along with their differential expression in roots of Vigna unguiculata grown under salt stress. Plant Cell. Tissue Organ Cult. 2011, 105, 233–242. [Google Scholar] [CrossRef]
- Li, H.; Dong, Y.; Yin, H.; Wang, N.; Yang, J.; Liu, X.; Wang, Y.; Wu, J.; Li, X. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 2011, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Sahito, Z.A.; Wang, L.; Sun, Z.; Yan, Q.; Zhang, X.; Jiang, Q.; Ullah, I.; Tong, Y.; Li, X. The miR172c-NNC1 module modulates root plastic development in response to salt in soybean. BMC Plant Biol. 2017, 17, 229. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Yu, H.; Liu, M.; Lu, Y.; Ouyang, B. Identification of salt-stress responsive microRNAs from Solanum lycopersicum and Solanum pimpinellifolium. Plant Growth Regul. 2017, 83, 129–140. [Google Scholar] [CrossRef]
- Lelandais-Brière, C.; Naya, L.; Sallet, E.; Calenge, F.; Frugier, F.; Hartmann, C.; Gouzy, J.; Crespi, M. Genome-wide medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 2009, 21, 2780–2796. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kwak, K.J.; Jung, H.J.; Lee, H.J.; Kang, H. MicroRNA402 affects seed germination of arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol. 2010, 51, 1079–1083. [Google Scholar] [CrossRef]
- He, F.; Xu, C.; Fu, X.; Shen, Y.; Guo, L.; Leng, M.; Luo, K. The microRNA390/TRANS-ACTING SHORT INTERFERING RNA3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol. 2018, 177, 775–791. [Google Scholar] [CrossRef]
- Wen, F.L.; Yue, Y.; He, T.F.; Gao, X.M.; Zhou, Z.S.; Long, X.H. Identification of miR390-TAS3-ARF pathway in response to salt stress in Helianthus tuberosus L. Gene 2020, 738, 144460. [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, J.; Li, Z.; Hu, Q.; Yuan, N.; Zhou, M.; Xia, X.; Noorai, R.; Saski, C.; Li, S.; et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef]
- Pegler, J.L.; Nguyen, D.Q.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Molecular manipulation of the mir396/grf expression module alters the salt stress response of arabidopsis thaliana. Agronomy 2021, 11, 1751. [Google Scholar] [CrossRef]
- Macovei, A.; Tuteja, N. MicroRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Liu, D.; Chen, D.; Cheng, Y.; Zhang, X.; Song, L.; Hu, M.; Dong, J.; Shen, F. MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 2019, 16, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Jiang, J.; Qiao, G.; Lin, S.; Li, H.; Xie, L.; Zhuo, R. Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol. Biol. Rep. 2012, 39, 8645–8654. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-responsive microRNAs in Populus. Plant J. 2008, 55, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De La Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009, 70, 385–401. [Google Scholar] [CrossRef]
- Han, J.; Xie, H.; Kong, M.L.; Sun, Q.P.; Li, R.Z.; Pan, J.B. Computational identification of miRNAs and their targets in Phaseolus vulgaris. Genet. Mol. Res. 2014, 13, 310–322. [Google Scholar] [CrossRef]
- Gao, S.; Yang, L.; Zeng, H.Q.; Zhou, Z.S.; Yang, Z.M.; Li, H.; Sun, D.; Xie, F.; Zhang, B. A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, H.J.; Jung, H.J.; Maruyama, K.; Suzuki, N.; Kang, H. Overexpression of microRNA395c or 395e affects differently the seed germination of arabidopsis thaliana under stress conditions. Planta 2010, 232, 1447–1454. [Google Scholar] [CrossRef]
- Dong, Z.; Shi, L.; Wang, Y.; Chen, L.; Cai, Z.; Wang, Y.; Jin, J.; Li, X. Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. Int. J. Mol. Sci. 2013, 14, 2717–2738. [Google Scholar] [CrossRef]
- De Paola, D.; Cattonaro, F.; Finetti Sialer, M.M.; Catalano, D.; Fracchiolla, A.; Morgese, A.; Pignone, D.; Sonnante, G. MicroRNAs in the globe artichoke: Detection and analysis. Acta Hortic. 2013, 983, 187–192. [Google Scholar] [CrossRef]
- Pokoo, R.; Ren, S.; Wang, Q.; Motes, C.M.; Hernandez, T.D.; Ahmadi, S.; Monteros, M.J.; Zheng, Y.; Sunkar, R. Genotype- and tissue-specific miRNA profiles and their targets in three alfalfa (Medicago sativa L.) genotypes. BMC Genom. 2018, 19, 115–131. [Google Scholar] [CrossRef]
- Bi, F.; Meng, X.; Ma, C.; Yi, G. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genom. 2015, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, J.; Cai, Y.; Gong, X.; Xiong, X.; Qi, W.; Pang, Q.; Wang, X.; Wang, Y. Genome-wide identification and characterization of Eutrema salsugineum microRNAs for salt tolerance. Physiol. Plant. 2016, 157, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Nie, X.; Cui, L.; Deng, P.; Wang, M.; Song, W. Genome-wide identification and characterization of salinity stress-responsive miRNAS in wild emmer wheat (Triticum turgidum ssp. dicoccoides). Genes 2017, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.K.; Panda, A.K.; Rawal, H.C.; Sharma, T.R. Discovery of microRNA-target modules of African rice (Oryza glaberrima) under salinity stress. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ai, B.; Chen, Y.; Zhao, M.; Ding, G.; Xie, J.; Zhang, F. Overexpression of miR1861h increases tolerance to salt stress in rice (Oryza sativa L.). Genet. Resour. Crop Evol. 2021, 68, 87–92. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, X.; Zhou, Y.; Xie, J. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.). Biotechnol. Lett. 2016, 38, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, S.M.; Alaraidh, I.A.; Khan, M.A.; Migdadi, H.M.; Alghamdi, S.S.; Alsahli, A.A. Identification and characterization of salt-responsive micrornas in vicia faba by high-throughput sequencing. Genes 2019, 10, 303. [Google Scholar] [CrossRef]
- Naik, H.K.; Varadahalli, R.D. Genomic identification of salt induced microRNAs in niger (Guizotia abyssinica Cass.). Plant Gene 2020, 23, 100242. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Brown, C.W.; Pegler, J.L.; Eamens, A.L.; Grof, C.P.L. Molecular manipulation of microRNA397 abundance influences the development and salt stress response of arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 7879. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.; Li, J. Global identification and analysis of micrornas involved in salt stress responses in two alfalfa (Medicago sativa millennium) Lines. Can. J. Plant Sci. 2020, 100, 445–455. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, P.; Kang, H.; Liu, L.; Cao, Q.; Sun, J.; Dong, T.; Zhu, M.; Li, Z.; Xu, T. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genom. 2020, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pegler, J.L.; Nguyen, D.Q.; Grof, C.P.L.; Eamens, A.L. Profiling of the salt stress responsive MicroRNA landscape of C4 genetic model species Setaria viridis (L.) beauv. Agronomy 2020, 10, 837. [Google Scholar] [CrossRef]
- Zeeshan, M.; Qiu, C.W.; Naz, S.; Cao, F.; Wu, F. Genome-wide discovery of mirnas with differential expression patterns in responses to salinity in the two contrasting wheat cultivars. Int. J. Mol. Sci. 2021, 22, 2556. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; Beric, A.; Meyers, B.C. Despacito: The slow evolutionary changes in plant microRNAs. Curr. Opin. Plant Biol. 2018, 42, 16–22. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Khalid, N.; Islam, W.; Sanaullah, T.; Anwar, M.; Khan, S.; Ye, W.; Lou, Y. Zinc finger protein transcription factors: Integrated line of action for plant antimicrobial activity. Microb. Pathog. 2019, 132, 141–149. [Google Scholar] [CrossRef]
- Jagannadham, P.T.K.; Muthusamy, S.K.; Chidambaranathan, P. Micromics: A Novel Approach to Understand the Molecular Mechanisms in Plant Stress Tolerance. In Recent Approaches in Omics for Plant Resilience to Climate Change; Springer: Cham, Denmark, 2019; pp. 93–108. [Google Scholar]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Himabindu, Y.; Chakradhar, T.; Reddy, M.C.; Kanygin, A.; Redding, K.E.; Chandrasekhar, T. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 2016, 124, 39–63. [Google Scholar] [CrossRef]
- Liu, S.; Wu, L.; Qi, H.; Xu, M. LncRNA/circRNA–miRNA–mRNA networks regulate the development of root and shoot meristems of Populus. Ind. Crops Prod. 2019, 133, 333–347. [Google Scholar] [CrossRef]
- Mittal, D.; Sharma, N.; Sharma, V.; Sopory, S.K.; Sanan-Mishra, N. Role of microRNAs in rice plant under salt stress. Ann. Appl. Biol. 2016, 168, 2–18. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Molecular manipulation of the miR399/PHO2 expression module alters the salt stress response of arabidopsis thaliana. Plants 2021, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marlétaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef]
- Tang, Y.; Du, G.; Xiang, J.; Hu, C.; Li, X.; Wang, W.; Zhu, H.; Qiao, L.; Zhao, C.; Wang, J.; et al. Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.). Genomics 2022, 114, 171–184. [Google Scholar] [CrossRef]
- Kim, J.K.; Cao, J.; Wu, R. Regulation and interaction of multiple protein factors with the proximal promoter regions of a rice high pl α-amylase gene. MGG Mol. Gen. Genet. 1992, 232, 383–393. [Google Scholar] [CrossRef]
- Pastuglia, M.; Roby, D.; Dumas, C.; Cock, J.M. Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 1997, 9, 49–60. [Google Scholar] [CrossRef]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003, 33, 259–270. [Google Scholar] [CrossRef]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 2003, 15, 1591–1604. [Google Scholar] [CrossRef]
- Li, Z.X.; Li, S.G.; Zhang, L.F.; Han, S.Y.; Li, W.F.; Xu, H.Y.; Yang, W.H.; Liu, Y.L.; Fan, Y.R.; Qi, L.W. Over-expression of miR166a inhibits cotyledon formation in somatic embryos and promotes lateral root development in seedlings of Larix leptolepis. Plant Cell. Tissue Organ Cult. 2016, 127, 461–473. [Google Scholar] [CrossRef]
- Xi, D.; Chen, X.; Wang, Y.; Zhong, R.; He, J.; Shen, J.; Ming, F. Arabidopsis ANAC092 regulates auxin-mediated root development by binding to the ARF8 and PIN4 promoters. J. Integr. Plant Biol. 2019, 61, 1015–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauclair, L.; Yu, A.; Bouché, N. MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010, 62, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Muhammad Shazwan, S.; Muhammad Aliff, M.; Asral Wirda, A.A.; Hayati, A.R.; Maizatul Azma, M.; Nur Syahrina, A.R.; Nazefah, A.H.; Jameela, S.; Nur Fariha, M.M. Microrna expression in antiphospholipid syndrome: A systematic review and microrna target genes analysis. Malays. J. Pathol. 2016, 38, 273–283. [Google Scholar]
- Liebsch, D.; Palatnik, J.F. MicroRNA miR396, GRF transcription factors and GIF co-regulators: A conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef]
- Dalmadi, Á.; Miloro, F.; Bálint, J.; Várallyay, É.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef] [PubMed]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. Micrornas as potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Sabbione, A.; Daurelio, L.; Vegetti, A.; Talón, M.; Tadeo, F.; Dotto, M. Genome-wide analysis of AGO, DCL and RDR gene families reveals RNA-directed DNA methylation is involved in fruit abscission in Citrus sinensis. BMC Plant Biol. 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Dutta, T.; Neelapu, N.R.R.; Wani, S.H.; Surekha, C. Salt stress tolerance and small RNA. In Plant Small RNA; Elsevier: Amsterdam, The Netherlands, 2020; pp. 191–207. [Google Scholar]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Windels, D.; Bielewicz, D.; Ebneter, M.; Jarmolowski, A.; Szweykowska-Kulinska, Z.; Vazquez, F. miR393 is required for production of proper Auxin signalling outputs. PLoS ONE 2014, 9, e95972. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, Z.; Song, G.; Yao, H.; Han, L. Antioxidant enzyme activity and microRNA are associated with growth of Poa pratensis callus under salt stress. Plant Biotechnol. Rep. 2020, 14, 429–438. [Google Scholar] [CrossRef]
- Makkar, H.; Arora, S.; Khuman, A.K.; Chaudhary, B. Target-Mimicry-Based miR167 Diminution Confers Salt-Stress Tolerance During In Vitro Organogenesis of Tobacco (Nicotiana tabacum L.). J. Plant Growth Regul. 2022, 41, 1462–1480. [Google Scholar] [CrossRef]
- Hobecker, K.V.; Reynoso, M.A.; Bustos-Sanmamed, P.; Wen, J.; Mysore, K.S.; Crespi, M.; Blanco, F.A.; Zanetti, M.E. The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 2017, 174, 2469–2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, D.; Chun, H.J.; Kang, S.; Shin, G.; Park, S.J.; Hong, H.; Kim, C.; Kim, D.H.; Lee, S.Y.; Kim, M.C.; et al. A role for arabidopsis miR399f in salt, drought, and ABA signaling. Mol. Cells 2016, 39, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Li, W.; Wang, T.; Zhang, Y.; Li, Y. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 175–194. [Google Scholar] [CrossRef]
- Long, R.; Li, M.; Li, X.; Gao, Y.; Zhang, T.; Sun, Y.; Kang, J.; Wang, T.; Cong, L.; Yang, Q. A Novel miRNA Sponge Form Efficiently Inhibits the Activity of miR393 and Enhances the Salt Tolerance and ABA Insensitivity in Arabidopsis thaliana. Plant Mol. Biol. Report. 2017, 35, 409–415. [Google Scholar] [CrossRef]
- Song, J.B.; Gao, S.; Sun, D.; Li, H.; Shu, X.X.; Yang, Z.M. MiR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 2013, 13, 210. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Yan, J.; Wang, K.; Luo, H.; Zhang, W. MiR319 mediated salt tolerance by ethylene. Plant Biotechnol. J. 2019, 17, 2370–2383. [Google Scholar] [CrossRef]
- Guo, J.; Wang, J.; Xi, L.; Huang, W.D.; Liang, J.; Chen, J.G. RACK1 is a negative regulator of ABA responses in arabidopsis. J. Exp. Bot. 2009, 60, 3819–3833. [Google Scholar] [CrossRef]
- Denver, J.B.; Ullah, H. miR393s regulate salt stress response pathway in Arabidopsis thaliana through scaffold protein RACK1A mediated ABA signaling pathways. Plant Signal. Behav. 2019, 14, 1600394. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, Y.F.; Randlett, M.D.; Zhao, X.C.; Findell, J.L.; Kieber, J.J.; Schaller, G.E. Localization of the Raf-like Kinase CTR1 to the Endoplasmic Reticulum of Arabidopsis through Participation in Ethylene Receptor Signaling Complexes. J. Biol. Chem. 2003, 278, 34725–34732. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice 2019, 12, 1–11. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).