The Bibliometric Landscape of Gene Editing Innovation and Regulation in the Worldwide
Abstract
:1. Introduction
2. Materials and Method
3. Results
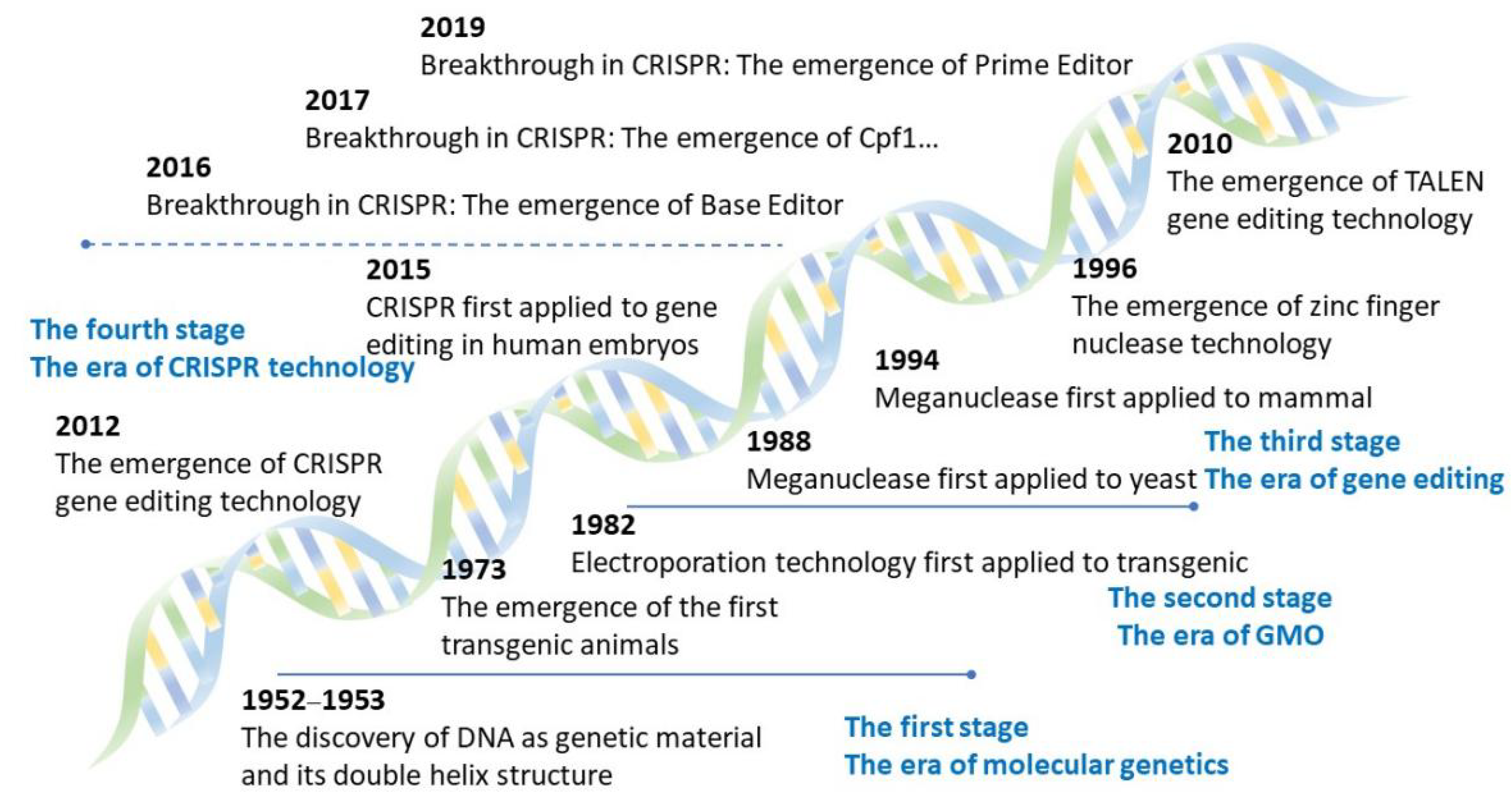
3.1. Origin and Eras of Gene Editing Technology
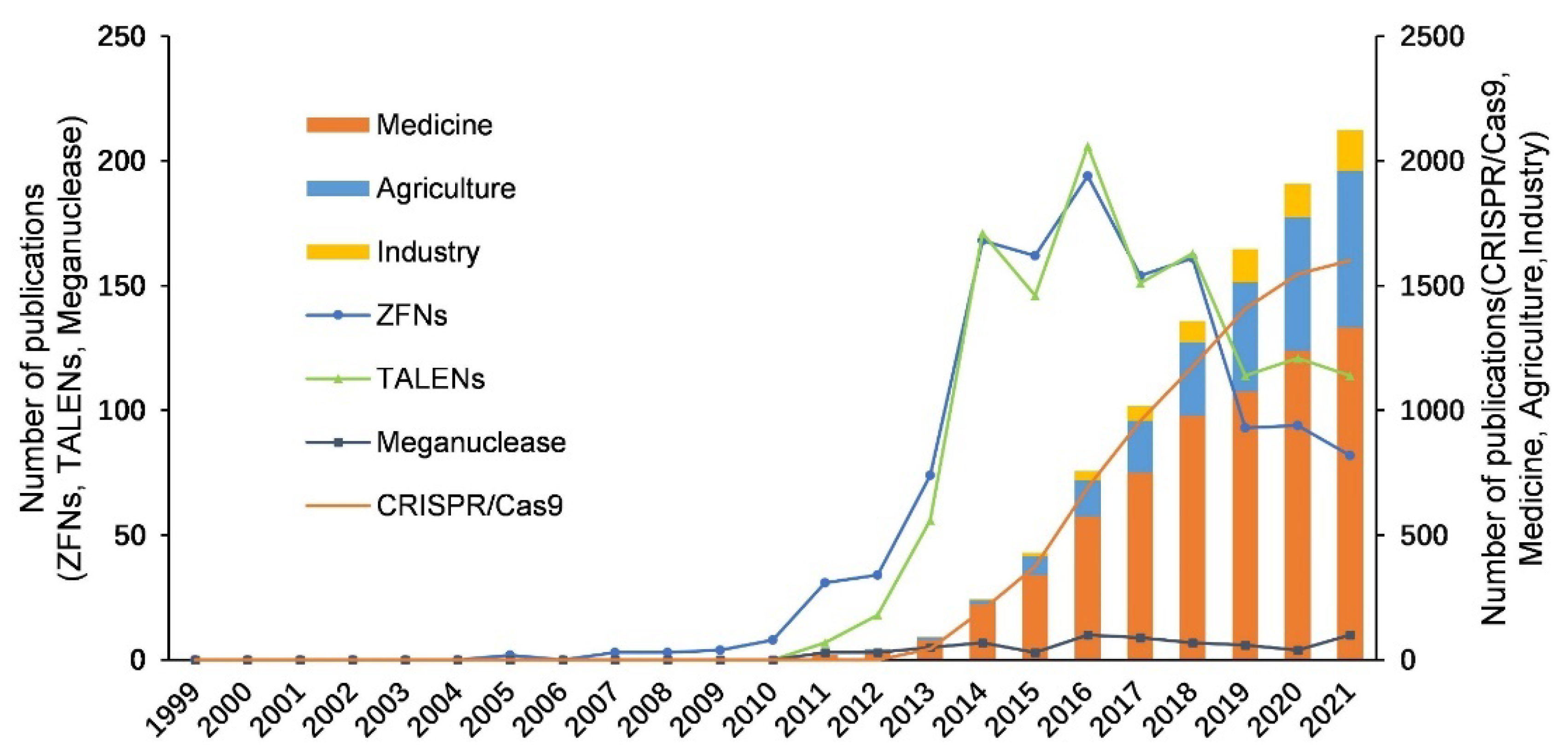
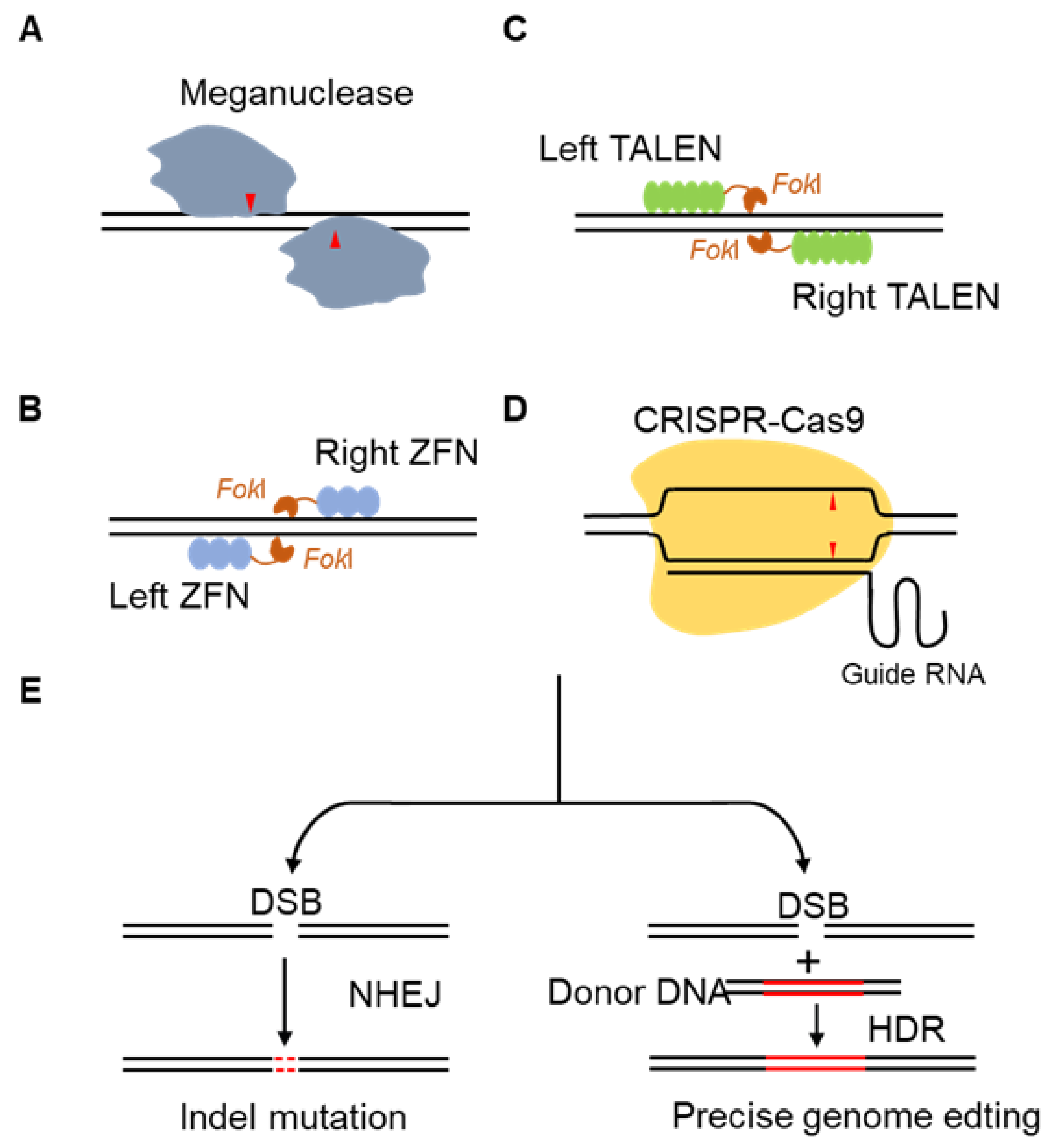
3.2. Development of Four Types of Gene Editing Technologies
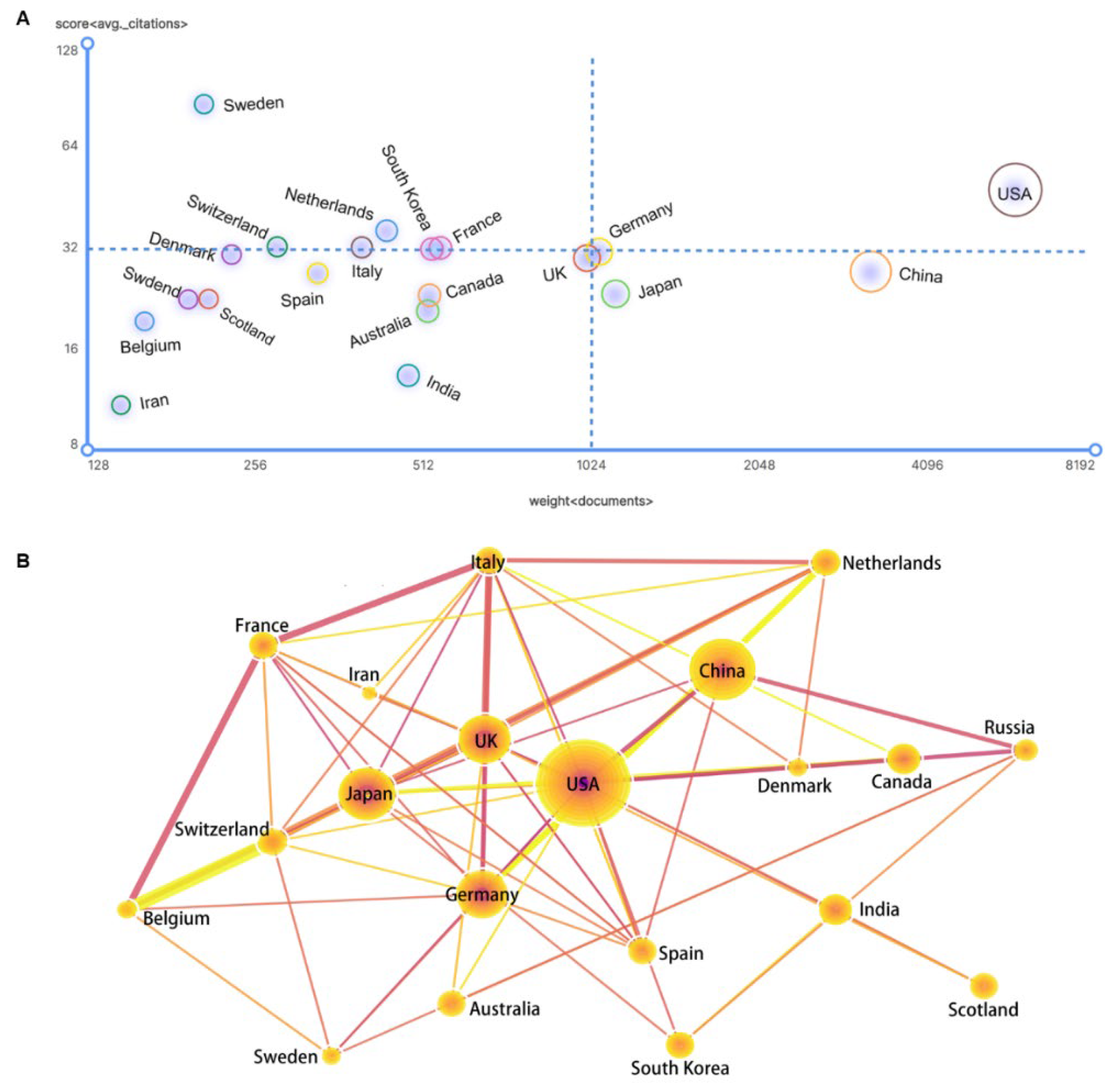
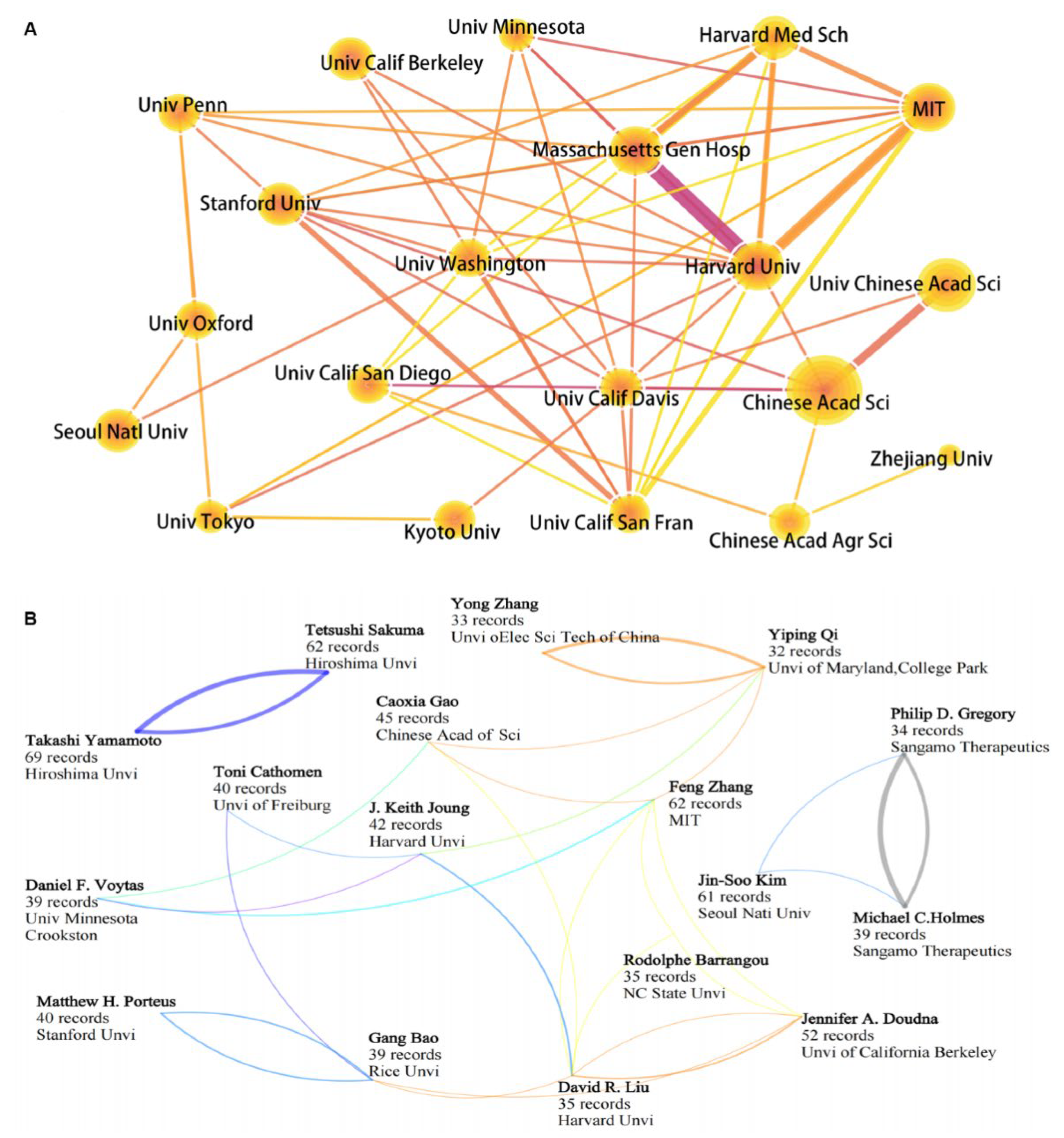
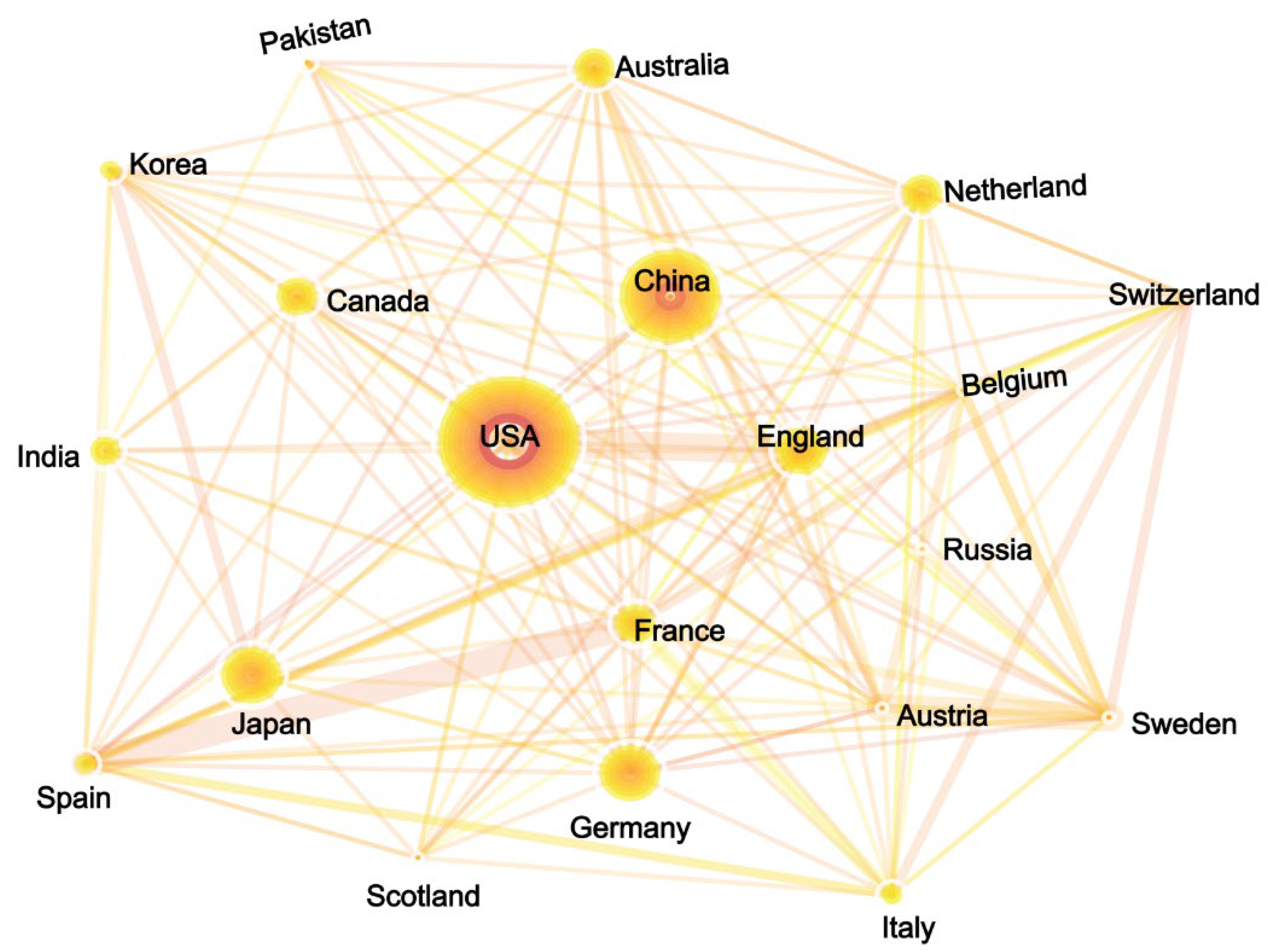
3.3. Competition and Cooperation of Key Actors in Gene Editing Technology
3.3.1. National Performance
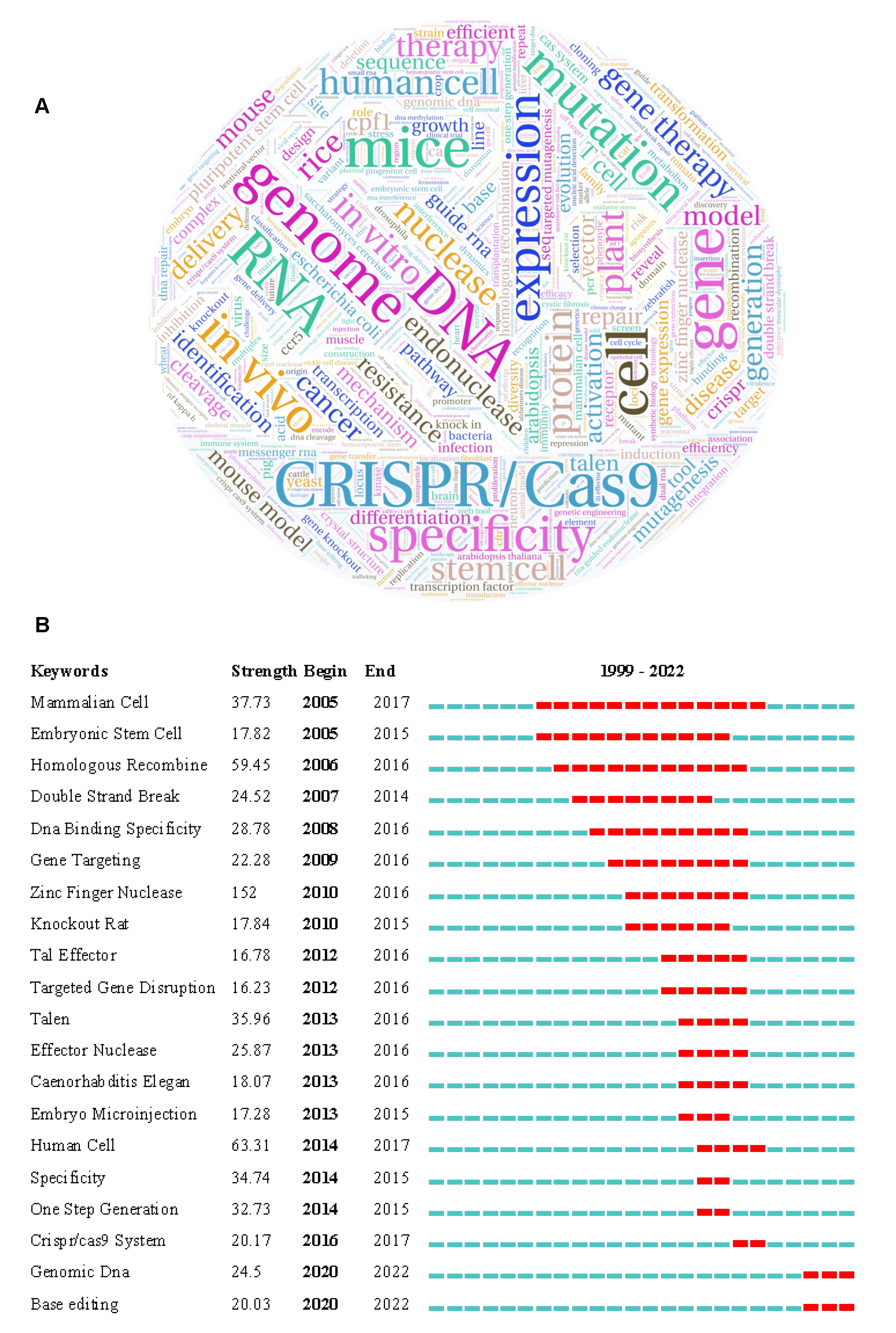
3.3.2. Key Institutions and Scientists Engaged in Gene Editing Technology
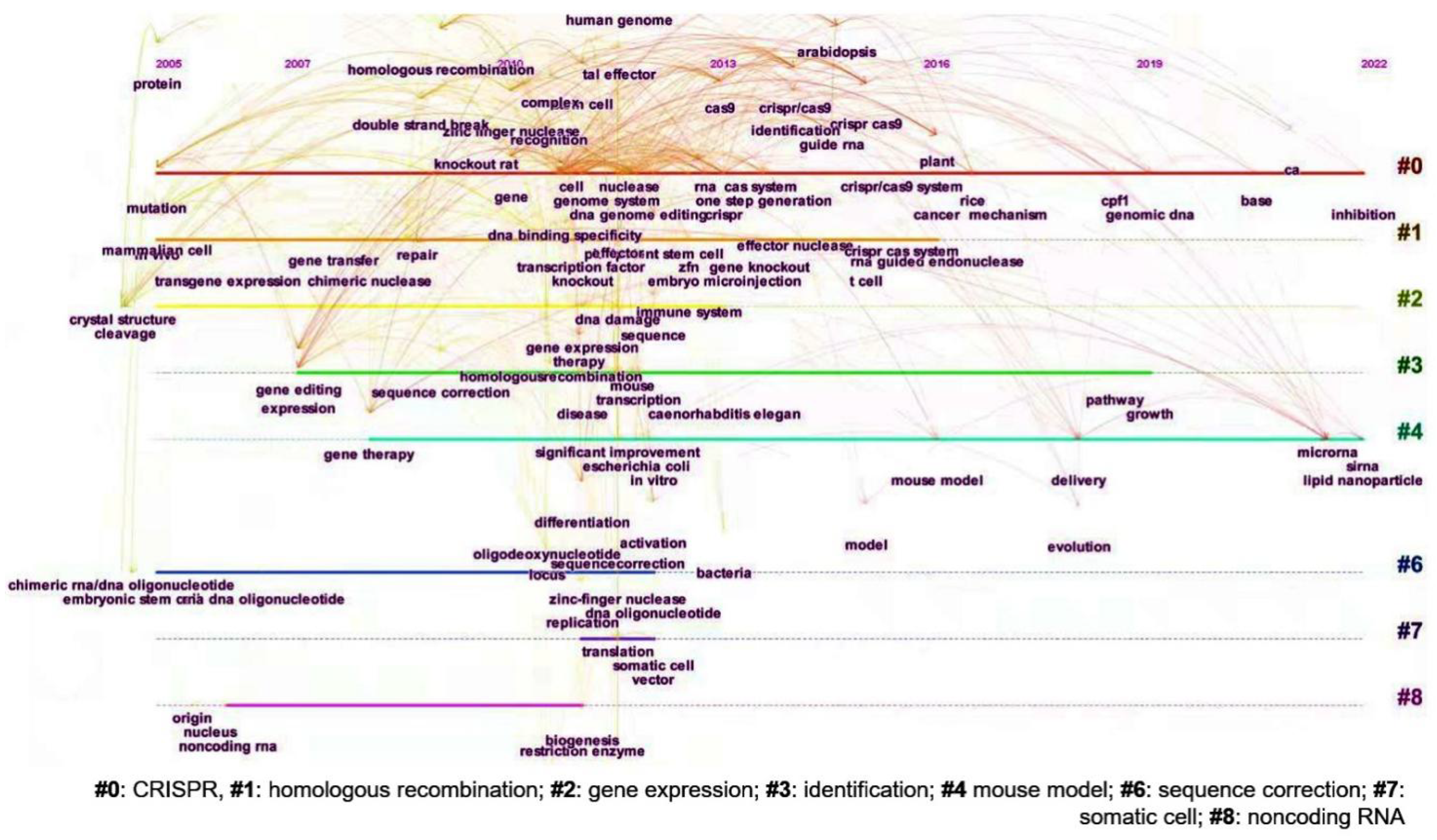
3.4. Research Hotspots and Evolution of Gene Editing Technology
4. Discussion and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. J. Sci. 2014, 6213, 1077–1087. [Google Scholar] [CrossRef]
- Fesnak, A.D.; June, C.H.; Levine, B.L. Engineered T cells: The promise and challenges of cancer immunotherapy. Nat. Rev. Cancer. 2016, 16, 566–581. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, K.; An, X. CRISPR/Cas System: Applications and Prospects for Maize Improvement. ACS Agric. Sci. Technol. 2021, 2, 174–183. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Martin-Laffon, J.; Kuntz, M.; Ricroch, A.E. Worldwide CRISPR patent landscape shows strong geographical biases. Nat. Biotechnol. 2019, 37, 613–620. [Google Scholar] [CrossRef]
- Huang, Y.; Porter, A.; Zhang, Y.; Barrangou, R. Collaborative networks in gene editing. Nat. Biotechnol. 2019, 37, 1107–1109. [Google Scholar] [CrossRef]
- Porter, A.L.; Kongthon, A.; Lu, J.C. Research profiling: Improving the literature review. Scientometrics 2002, 53, 351–370. [Google Scholar] [CrossRef]
- Bellis, N.D. Bibliometrics and Citation Analysis; The Scarecrow Press: Lanham, MD, USA, 2009. [Google Scholar]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Am. J. Psychiatry 2003, 160, 623–624. [Google Scholar] [CrossRef]
- Blum, B.; Bakalara, N.; Simpson, L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 1990, 60, 189–198. [Google Scholar] [CrossRef]
- Colleaux, L.; d’Auriol, L.; Betermier, M.; Cottarel, G.; Jacquier, A.; Galibert, F.; Dujon, B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell 1986, 44, 521–533. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Redondo, P.; Prieto, J.; Muñoz, I.G.; Alibés, A.; Stricher, F.; Serrano, L.; Cabaniols, J.P.; Daboussi, F.; Arnould, S.; Perez, C.; et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature 2008, 456, 107–111. [Google Scholar] [CrossRef]
- D’Halluin, K.; Vanderstraeten, C.; Van Hulle, J.; Rosolowska, J.; Van Den Brande, I.; Pennewaert, A.; D’Hont, K.; Bossut, M.; Jantz, D.; Ruiter, R.; et al. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol. J. 2013, 11, 933–941. [Google Scholar] [CrossRef]
- Daboussi, F.; Leduc, S.; Maréchal, A.; Dubois, G.; Guyot, V.; Perez-Michaut, C.; Amato, A.; Falciatore, A.; Juillerat, A.; Beurdeley, M.; et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 2014, 5, 3831. [Google Scholar] [CrossRef]
- Urnov, F.D.; Miller, J.C.; Lee, Y.L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef]
- Tesson, L.; Usal, C.; Ménoret, S.; Leung, E.; Niles, B.J.; Remy, S.; Santiago, Y.; Vincent, A.I.; Meng, X.; Zhang, L.; et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 2011, 29, 695–696. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Li, T.; Huang, S.; Zhao, X.; Wright, D.A.; Carpenter, S.; Spalding, M.H.; Weeks, D.P.; Yang, B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011, 39, 6315–6325. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef] [Green Version]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2014, 33, 41–52. [Google Scholar] [CrossRef]
- Tanihara, F.; Hirata, M.; Otoi, T. Current status of the application of gene editing in pigs. J. Reprod. Dev. 2021, 67, 177–187. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef]
- Wu, Y.; Liang, D.; Wang, Y.; Bai, M.; Tang, W.; Bao, S.; Yan, Z.; Li, D.; Li, J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013, 13, 659–662. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Gao, Y.; Lu, G.; Habben, J.E.; Mao, G.; Chen, G.; Wang, J.; Yang, F.; Zhao, X.; et al. A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol. Biol. 2020, 102, 373–388. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.S.; Domm, J.; Eustace, B.K.; Corbacioglu, S. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Lebwohl, D. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef]
- Inoue, K.; Oliveira, L.; Abeliovich, A. CRISPR Transcriptional Activation Analysis Unmasks an Occult γ-Secretase Processivity Defect in Familial Alzheimer’s Disease Skin Fibroblasts. Cell Rep. 2017, 21, 1727–1736. [Google Scholar] [CrossRef]
- Shi, J.; Wang, E.; Milazzo, J.P.; Wang, Z.; Kinney, J.B.; Vakoc, C.R. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 2015, 33, 661–667. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Gonçalves, E.; Beaver, C.M.; Migliardi, G.; Garnett, M.J. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Moon, J.; Kwon, H.J.; Yong, D.; Lee, I.C.; Kim, H.; Kang, H.; Lim, E.K.; Lee, K.S.; Jung, J.; Park, H.G.; et al. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9. ACS Sens. 2020, 5, 4017–4026. [Google Scholar] [CrossRef]
- Ma, L.; Boucher, J.I.; Paulsen, J.; Matuszewski, S.; Eide, C.A.; Ou, J.; Eickelberg, G.; Press, R.D.; Zhu, L.J.; Druker, B.J.; et al. CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc. Natl. Acad. Sci. USA 2017, 114, 11751–11756. [Google Scholar] [CrossRef]
- Živojević, K.; Mladenović, M.; Djisalov, M.; Mundzic, M.; Ruiz-Hernandez, E.; Gadjanski, I.; Knežević, N.Ž. Advanced mesoporous silica nanocarriers in cancer theranostics and gene editing applications. J. Control. Release Off. J. Control. Release Soc. 2021, 337, 193–211. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Morineau, C.; Bellec, Y.; Tellier, F.; Gissot, L.; Kelemen, Z.; Nogué, F.; Faure, J.D. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 2017, 15, 729–739. [Google Scholar] [CrossRef]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2020, 18, 1153–1168. [Google Scholar] [CrossRef]
- Karunarathna, N.L.; Wang, H.; Harloff, H.J.; Jiang, L.; Jung, C. Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 2020, 18, 2251–2266. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Ma, X.; Wang, B.; Zheng, Z.; Zhang, Y.; Xie, X.; Yang, B.; Zhao, Z.; Zhu, Q.; et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5’UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Li, R.; Li, R.; Li, X.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 415–427. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.; Zhang, C.; Chu, R.; Gu, Z.; Tan, H.; Zhao, D.; Fan, X.; Liu, Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 2164–2166. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, G.; Li, R.; Yan, S.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. The RNA Editing Factor SlORRM4 Is Required for Normal Fruit Ripening in Tomato. Plant Physiol. 2017, 175, 1690–1702. [Google Scholar] [CrossRef]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. Cell Mol. Biol. 2018, 94, 513–524. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. Cell Mol. Biol. 2017, 91, 714–724. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. New Strateg. Plant Improv. 2019, 39, 47. [Google Scholar] [CrossRef]
- Kuang, Y.; Li, S.; Ren, B.; Yan, F.; Spetz, C.; Li, X.; Zhou, X.; Zhou, H. Base-Editing-Mediated Artificial Evolution of OsALS1 In Planta to Develop Novel Herbicide-Tolerant Rice Germplasms. Mol. Plant 2020, 13, 565–572. [Google Scholar] [CrossRef]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Mansoor, S.; Pareek, A. Engineering abiotic stress tolerance via CRISPR/ Cas-mediated genome editing. J. Exp. Bot. 2020, 71, 470–479. [Google Scholar] [CrossRef]
- Zhou, J.; Xin, X.; He, Y.; Chen, H.; Li, Q.; Tang, X.; Zhong, Z.; Deng, K.; Zheng, X.; Akher, S.A.; et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 2019, 38, 475–485. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR-Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef]
- Cai, Z.; Xian, P.; Cheng, Y.; Ma, Q.; Lian, T.; Nian, H.; Ge, L. CRISPR/Cas9-mediated gene editing of GmJAGGED1 increased yield in the low-latitude soybean variety Huachun 6. Plant Biotechnol. J. 2021, 19, 1898–1900. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef]
- Wang, W.; Pan, Q.; Tian, B.; He, F.; Chen, Y.; Bai, G.; Akhunova, A.; Trick, H.N.; Akhunov, E. Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. Cell Mol. Biol. 2019, 100, 251–264. [Google Scholar] [CrossRef]
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965. [Google Scholar] [CrossRef]
- Tong, Y.; Charusanti, P.; Zhang, L.; Weber, T.; Lee, S.Y. CRISPR-Cas9 Based Engineering of Actinomycetal Genomes. ACS Synth. Biol. 2015, 4, 1020–1029. [Google Scholar] [CrossRef]
- Cai, P.; Duan, X.; Wu, X.; Gao, L.; Ye, M.; Zhou, Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021, 49, 7791–7805. [Google Scholar] [CrossRef]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Chiu, C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.T.; Ryu, J.; Kang, B.C.; Kim, J.S.; Kim, S.G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017, 8, 14406. [Google Scholar] [CrossRef]
- Jiang, Y.; Qian, F.; Yang, J.; Liu, Y.; Dong, F.; Xu, C.; Sun, B.; Chen, B.; Xu, X.; Li, Y.; et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017, 8, 15179. [Google Scholar] [CrossRef]
- Zheng, K.; Wang, Y.; Li, N.; Jiang, F.F.; Wu, C.X.; Liu, F.; Chen, H.C.; Liu, Z.F. Highly efficient base editing in bacteria using a Cas9-cytidine deaminase fusion. Commun. Biol. 2018, 1, 32. [Google Scholar] [CrossRef]
- Kim, K.; Ryu, S.M.; Kim, S.T.; Baek, G.; Kim, D.; Lim, K.; Chung, E.; Kim, S.; Kim, J.S. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 2017, 35, 435–437. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Ryu, S.M.; Koo, T.; Kim, K.; Lim, K.; Baek, G.; Kim, S.T.; Kim, H.S.; Kim, D.E.; Lee, H.; Chung, E.; et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018, 36, 536–539. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Cho, S.I.; Lee, S.; Mok, Y.G.; Lim, K.; Lee, J.; Lee, J.M.; Chung, E.; Kim, J.S. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell 2022, 185, 1764–1776.e12. [Google Scholar] [CrossRef]
- Banno, S.; Nishida, K.; Arazoe, T.; Mitsunobu, H.; Kondo, A. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat. Microbiol. 2018, 3, 423–429. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, J.; Yin, W.; Zhang, Z.; Song, Y.; Chang, X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 2016, 13, 1029–1035. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Pavan, E.; Ormazabal, M.; Peruzzo, P.; Vaena, E.; Rozenfeld, P.; Dardis, A. CRISPR/Cas9 Editing for Gaucher Disease Modelling. Int. J. Mol. Sci. 2020, 21, 3268. [Google Scholar] [CrossRef]
- Fareh, M.; Zhao, W.; Hu, W.; Casan, J.; Kumar, A.; Symons, J.; Zerbato, J.M.; Fong, D.; Voskoboinik, I.; Ekert, P.G.; et al. Reprogrammed CRISPR-Cas13b suppresses SARS-CoV-2 replication and circumvents its mutational escape through mismatch tolerance. Nat. Commun. 2021, 12, 4270. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef]
- Mahas, A.; Aman, R.; Mahfouz, M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2019, 20, 263. [Google Scholar] [CrossRef]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676.e14. [Google Scholar] [CrossRef]
- Pâques, F.; Duchateau, P. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr. Gene Ther. 2007, 7, 49–66. [Google Scholar] [CrossRef]
- Belfort, M.; Roberts, R.J. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997, 25, 3379–3388. [Google Scholar] [CrossRef]
- Khalil, A.M. The genome editing revolution: Review. J. Genet. Eng. Biotechnol. 2020, 18, 68. [Google Scholar] [CrossRef]
- Silva, G.; Poirot, L.; Galetto, R.; Smith, J.; Montoya, G.; Duchateau, P.; Pâques, F. Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr. Gene Ther. 2011, 11, 11–27. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Miller, J.C.; Holmes, M.C.; Wang, J.; Guschin, D.Y.; Lee, Y.L.; Rupniewski, I.; Beausejour, C.M.; Waite, A.J.; Wang, N.S.; Kim, K.A.; et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007, 25, 778–785. [Google Scholar] [CrossRef]
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161. [Google Scholar] [CrossRef]
- Durai, S.; Mani, M.; Kandavelou, K.; Wu, J.; Porteus, M.H.; Chandrasegaran, S. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005, 33, 5978–5990. [Google Scholar] [CrossRef]
- Gabriel, R.; Lombardo, A.; Arens, A.; Miller, J.C.; Genovese, P.; Kaeppel, C.; Nowrouzi, A.; Bartholomae, C.C.; Wang, J.; Friedman, G.; et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011, 29, 816–823. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, S.; Huang, X.; Orwig, K.E.; Sheng, Y. Rapid assembly of customized TALENs into multiple delivery systems. PLoS ONE 2013, 8, e80281. [Google Scholar] [CrossRef]
- Reyon, D.; Tsai, S.Q.; Khayter, C.; Foden, J.A.; Sander, J.D.; Joung, J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012, 30, 460–465. [Google Scholar] [CrossRef]
- Zhu, H.; Lau, C.H.; Goh, S.L.; Liang, Q.; Chen, C.; Du, S.; Phang, R.Z.; Tay, F.C.; Tan, W.K.; Li, Z.; et al. Baculoviral transduction facilitates TALEN-mediated targeted transgene integration and Cre/LoxP cassette exchange in human-induced pluripotent stem cells. Nucleic Acids Res. 2013, 41, e180. [Google Scholar] [CrossRef]
- Mussolino, C.; Alzubi, J.; Fine, E.J.; Morbitzer, R.; Cradick, T.J.; Lahaye, T.; Bao, G.; Cathomen, T. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014, 42, 6762–6773. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 2579–2586. [Google Scholar] [CrossRef]
- Samanta, M.K.; Dey, A.; Gayen, S. CRISPR/Cas9: An advanced tool for editing plant genomes. Transgenic Res. 2016, 25, 561–573. [Google Scholar] [CrossRef]
- Hilton, I.B.; Gersbach, C.A. Enabling functional genomics with genome engineering. Genome Res. 2015, 25, 1442–1455. [Google Scholar] [CrossRef]
- Koike-Yusa, H.; Li, Y.; Tan, E.P.; Velasco-Herrera, M.; Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014, 32, 267–273. [Google Scholar] [CrossRef]
- Ma, H.; Dang, Y.; Wu, Y.; Jia, G.; Anaya, E.; Zhang, J.; Abraham, S.; Choi, J.G.; Shi, G.; Qi, L.; et al. A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell Rep. 2014, 12, 673–683. [Google Scholar] [CrossRef]
- Korkmaz, G.; Lopes, R.; Ugalde, A.P.; Nevedomskaya, E.; Han, R.; Myacheva, K.; Zwart, W.; Elkon, R.; Agami, R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 192–198. [Google Scholar] [CrossRef]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef]
- Tabebordbar, M.; Zhu, K.; Cheng, J.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef]
- Yin, H.; Xue, W.; Chen, S.; Bogorad, R.L.; Benedetti, E.; Grompe, M.; Koteliansky, V.; Sharp, P.A.; Jacks, T.; Anderson, D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014, 32, 551–553. [Google Scholar] [CrossRef]
- Strecker, J.; Jones, S.; Koopal, B.; Schmid-Burgk, J.; Zetsche, B.; Gao, L.; Makarova, K.; Koonin, E.; Zhang, F. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 2019, 10, 212. [Google Scholar] [CrossRef]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e7. [Google Scholar] [CrossRef]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339.e5. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da, C.M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Mok, B.Y.; de Moraes, M.H.; Zeng, J.; Bosch, D.E.; Kotrys, A.V.; Raguram, A.; Hsu, F.; Radey, M.C.; Peterson, S.B.; Mootha, V.K.; et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 2020, 583, 631–637. [Google Scholar] [CrossRef]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef]
- Sakuma, T.; Nakade, S.; Sakane, Y.; Suzuki, K.T.; Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016, 11, 118–133. [Google Scholar] [CrossRef]
- Cox, D.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Kuiken, T.; Kuzma, J. Genome Editing in Latin America: Regional Regulatory Overview. 2021. Available online: https://publications.iadb.org/publications/english/document/Genome-Editing-in-Latin-America-Regional-Regulatory-Overview.pdf(accessed on 5 June 2022). [CrossRef]
- Gatica-Arias, A. The regulatory current status of plant breeding technologies in some Latin American and the Caribbean countries. Plant Cell Tissue Organ Cult. 2020, 141, 229–242. [Google Scholar] [CrossRef]
- Waltz, E. CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 2016, 34, 582. [Google Scholar] [CrossRef]
- Mallapaty, S. China’s approval of gene-edited crops energizes researchers. Nature 2022, 602, 559–560. [Google Scholar] [CrossRef]
- Small, H. A SCI-map case study: Building a map of AIDS research. Scientometrics 1994, 30, 229–241. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy. Angew. Chem. 2018, 57, 1491–1496. [Google Scholar] [CrossRef]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef]
- Akcakaya, P.; Bobbin, M.L.; Guo, J.A.; Malagon-Lopez, J.; Clement, K.; Garcia, S.P.; Fellows, M.D.; Porritt, M.J.; Firth, M.A.; Carreras, A.; et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 2018, 561, 416–419. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- Mullard, A. CRISPR technologies are going to need a bigger toolbox. Nat. Rev. Drug Discov. 2021, 20, 808–809. [Google Scholar] [CrossRef]
- Altae-Tran, H.; Kannan, S.; Demircioglu, F.E.; Oshiro, R.; Nety, S.P.; McKay, L.J.; Dlakić, M.; Inskeep, W.P.; Makarova, K.S.; Macrae, R.K.; et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 2021, 374, 57–65. [Google Scholar] [CrossRef]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef]
- Teng, F.; Cui, T.; Feng, G.; Guo, L.; Xu, K.; Gao, Q.; Li, T.; Li, J.; Zhou, Q.; Li, W. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018, 4, 63. [Google Scholar] [CrossRef] [Green Version]

| GE Technologies | Research Status | Typical Application Cases | ||||||
|---|---|---|---|---|---|---|---|---|
| Creators | Institutions and Countries | Years | Refs | Application Fields | Research Objects | Target Genes | Refs | |
| Meganuclease | Colleaux. L | CNRS Laboratoire and Université Pierre et Marie Curie (France) | 1986 | [17] | Medicine and Health | Human xeroderma pigmentosum | XPC | [21] |
| Agriculture | Cotton insect resistant | hppd and epsps | [22] | |||||
| Industry | Phaeodactylum tricornutum’s productivity of triglycerides | UDP-glucose pyrophosphorylase gene | [23] | |||||
| ZFNs | Srinivasan Chandrasegaran | Johns Hopkins University (JHU, USA) | 1996 | [18] | Medicine and Health | K562, CD4+ T cells (X-linked severe combined immune deficiency (SCID)) | IL2RG | [24] |
| Agriculture | Maize herbicide tolerance | Ipk1 and Zp15 | [25] | |||||
| TALENs | Daniel F. Voytas | University of Minnesota (USA) | 2010 | [19] | Medicine and Health | Rat immunoglobulinM (rat model) | IgM | [26] |
| Agriculture | Rice disease-resistant (Xanthomonas oryzae) | Os11N3 | [27] | |||||
| Industry | Saccharomyces cerevisiae | URA3, LYS2 and ADE2 | [28] | |||||
| CRISPR/Cas9 | Jennifer A. Doudna and Emmanuelle Charpentier | Howard Hughes Medical Institute (HHMI, USA) and The Laboratory for Molecular Infection Medicine Sweden (MIMS, Sweden) | 2012 | [20] | Medicine and Health | Human 293FT and mouse cells (first mammalian model) | SpCas9, SpRNase III, EMX1, PVALB and Th | [29] |
| Cynomolgus monkey (mammalian model) | Nr0b1, Ppar-γ and Rag1 | [30] | ||||||
| Duchenne muscular dystrophy(mouse model) | DMD | [31,32,33] | ||||||
| Human hereditary tyrosinemia (mouse model) | Fah | [34] | ||||||
| Human hemophilia (mouse model) | Serpinc1 | [35] | ||||||
| Human intestinal neoplasia (mouse model) | APC, P53 (TP53), KRAS and SMAD4 | [36,37] | ||||||
| Human lung adenocarcinoma (mouse model) | KRAS, p53 and LKB1 | [38] | ||||||
| Cataracts (mouse model) | Crygc | [39] | ||||||
| Human obesity (mouse model) | FTO | [30] | ||||||
| Resistance to Clostridium septicum alpha-toxin or 6-thioguanine (mouse ES cells) | Whole genome | [40] | ||||||
| Functional genomics in human cells | Anthrax and diphtheria toxin host genes | [41] | ||||||
| Human autologous CD34+ cells | BCL11A | [42] | ||||||
| Human hepatocytes | TTR | [43] | ||||||
| Human fibroblast | APP | [44] | ||||||
| Mouse acute myeloid leukemia cell | RPA3,Brd4, Smarca4, Eed, Suz12 and Rnf20 | [45] | ||||||
| 204 human cancer cell lines | 18,009 genes | [46] | ||||||
| SARS-CoV-2 | SARS-CoV-2 N1/N2/N3, pH1N1 H1 and pH1N1/H275Y N1 | [47] | ||||||
| Human leukemic | BCR-ABL | [48] | ||||||
| Human HeLa cells | Telomerase gene | [49] | ||||||
| Agriculture | Rice and Wheat (first applied to plants) | OsPDS, OsBADH2, OsMPK2 and TaMLO | [34] | |||||
| Rice herbicide tolerance | OsALS and OsFTIP1e | [41] | ||||||
| Tomato fruit size, quantity and nutritional value | SELF-PRUNING, OVATE, FASCIATED, FRUIT WEIGHT 2.2, MULTIFLORA and LYCOPENE BETA CYCLASE | [50] | ||||||
| Tomato storability | SP5G | [51] | ||||||
| Wheat grain weight enhancement | TaGASR7 | [52] | ||||||
| Sativa oleic acid content | FAD2 | [53] | ||||||
| Brassica napus L. (yellow seed traits, and increase protein content and fatty acid content) | BnTT8 | [54] | ||||||
| Oil content of rapeseed | BnSFAR4 and BnSFAR5 | [55] | ||||||
| Rice grain quality | Waxy | [56] | ||||||
| Tomato fruit nutrition | GABA-T/SSADH | [57] | ||||||
| Rice amylose | Wx | [58] | ||||||
| Tomato high quality seedless fruit | SlAGL6 | [59] | ||||||
| Mitochondrial function and fruit ripening in tomato | SlORRM4 | [60] | ||||||
| Tomato ripening regulation | lncRNA1459 | [61] | ||||||
| Grape resistance to Botrytis cinerea | VvWRKY52 | [62] | ||||||
| Citrus (improvement of citrus canker resistance) | CsLOB1 | [63] | ||||||
| Wheat resistance to mildew | TaEDR1 | [64] | ||||||
| Rice resistance to salinity tolerance | OsRR22 | [65] | ||||||
| Rice herbicide-tolerant | OsALS1 | [66] | ||||||
| Potato resistance to the herbicide | ALS1 | [67] | ||||||
| Rice disease resistance | OsSWEET13 and ten EBEPthXo2 | [68] | ||||||
| Rice broad-spectrum disease resistance | Bsr-k1 | [69] | ||||||
| Duncan grapefruit (disease resistant citrus varieties) | CsLOB1 | [70] | ||||||
| Rice resistance to plant hoppers and stem borers | OsCYP71A1 | [8] | ||||||
| Rice tolerance to abiotic stresses | OsSRFP1 | [71] | ||||||
| Rice seed setting rate, the total number of grains, number of full grains per panicle and 1000-grain weight | OsLOGL5 | [40] | ||||||
| Rice grain size, width and weight | OsGS3, OsGW2 and OsGn1a | [72] | ||||||
| Rice ear length, grain size, cold tolerance | OsPIN5b, GS3 and OsMYB30 | [73] | ||||||
| Rice grain number, ear structure, particle size | Gn1a, DEPT, GS3 and IPA1 | [74] | ||||||
| Soybean NSPP and leaflet shape | GmJAG1 | [75] | ||||||
| Maize grain yield | ARGOS8-v1, ARGOS8-v2 and Zmcle7 | [76,77] | ||||||
| Wheat grain shape and weight | TaGW7 | [78] | ||||||
| Industry | Yeast biosynthesizing monoterpenes | LIS | [79] | |||||
| Actinomycetes | actIORF1 and actVB | [80] | ||||||
| Pichia pastoris | RAD52 | [81] | ||||||
| CRISPR /Cas12a(Cpf1) | Feng Zhang | Broad Institute of MIT and Harvard (USA) | 2015 | [76] | Medicine and Health | First mammalian model (HEK 293T cell) | DNMT1, EMX1, VEGFA and GRIN2 | [82] |
| SARS-CoV-2 | SARS-CoV-2 N gene, E gene | [83] | ||||||
| Agriculture | Soybean fatty acid desaturases | FAD2-1A and FAD2-1B | [84] | |||||
| Industry | Corynebacterium glutamicum genome | crtYf and recT | [85] | |||||
| Base Editor (BE) | David R. Liu | Harvard University (USA) | 2016 | [86] | Medicine and Health | Albinism (mouse model) | Dmd and Tyr | [87] |
| Agriculture | Rice, Wheat and Maize genome | OsCDC48, OsNRT1.1B, OsSPL14, TaLOX2 and ZmCENH3 | [88] | |||||
| Industry | Escherichia coli and Brucella melitensis genome | rppH and lacZ | [86] | |||||
| Adenine Base Editors (ABEs) | David R. Liu | Harvard University (USA) | 2017 | [89] | Medicine and Health | Duchenne muscular dystrophy (mouse model) | Tyr | [89] |
| Prime Editors (PEs) | David R. Liu | Harvard University (USA) | 2019 | [90] | Agriculture | Rice and Wheat genome | OsALS, OsCDC48, OsDEP1, OsEPSPS, OsGAPDH, OsLDMAR, TaGW2, TaUbi10, TaLOX2, TaMLO, TaDME and TaGASR7 | [90] |
| DddA-derived cytosine Base Editors (DdCBEs) | David R. Liu | Harvard University (USA) | 2020 | [91] | Medicine and Health | Human mitochondrial (HEK 293T cell) | ND1, ND4, ND5.2, COX3.1 and RNR2 | [91] |
| Target-AID | Akihiko Kondo | Kobe University (Japan) | 2016 | [92] | Agriculture | Tomato hormone | DELLA and ETR1 | [41] |
| Industry | Escherichia coli genome | galK and rpoB | [92] | |||||
| dCas9-AIDx | Yan Song and Xing Chang | Chinese Academy of Sciences and Shanghai Jiao Tong University School of Medicine (China) | 2016 | [93] | Medicine and Health | Chronic myeloid leukemia cells | GFP | [93] |
| CRISPR /Cas13a (C2c2) | Feng Zhang | Broad Institute of MIT and Harvard (USA) | 2016 | [77] | Medicine and Health | Dengue and Zika virus | the P. aeruginosa acyltransferase gene and the S. aureus thermonuclease gene | [94] |
| Human monocytic cell | GBA1 | [95] | ||||||
| CRISPR /Cas13b (C2c6) | Feng Zhang | Broad Institute of MIT and Harvard (USA) | 2017 | [78] | Medicine and Health | SARS-CoV-1 | RNA genome | [96] |
| CRISPR /Cas13d | Feng Zhang | Broad Institute of MIT and Harvard (USA) | 2018 | [79] | Medicine and Health | SARS-CoV-2 | RNA genome | [97] |
| Agriculture | Nicotiana benthamiana and Arabidopsis thaliana RNA virus | GFP | [98] | |||||
| Industry | Ruminococcus flavefaciens (frontotemporal dementia) | ANXA4 | [99] | |||||
| Rank | Organizations | Number of Records | Average Citations |
|---|---|---|---|
| 1 | Chinese Academy of Sciences | 599 | 37.53 |
| 2 | University of Chinese Academy of Sciences | 311 | 30.90 |
| 3 | Harvard Medical School | 291 | 40.30 |
| 4 | Stanford University | 261 | 46.75 |
| 5 | Harvard University | 251 | 280.71 |
| 6 | University of California, Berkeley | 220 | 118.13 |
| 7 | Chinese Academy of Agricultural Sciences | 192 | 22.41 |
| 8 | Massachusetts Institute of Technology | 173 | 298.36 |
| 9 | University of California, San Diego | 160 | 39.74 |
| 10 | University of California, San Francisco | 155 | 54.07 |
| 11 | Seoul National University | 153 | 70.65 |
| 12 | The University of Tokyo | 152 | 36.21 |
| 13 | University of Oxford | 151 | 22.07 |
| 14 | University of Minnesota | 148 | 46.14 |
| 15 | University of Pennsylvania | 147 | 56.56 |
| 16 | Zhejiang University | 146 | 20.82 |
| 17 | University of Washington | 144 | 40.56 |
| 18 | Kyoto University | 138 | 24.96 |
| 19 | Massachusetts Gen Hospital | 137 | 173.14 |
| 20 | University of California, Davis | 136 | 27.55 |
| Author | Number of Records | Total Citations | Average Citations | Year Range | Recent 3-Year Rate | Organizations | Top Research Fields (Number of Records) |
|---|---|---|---|---|---|---|---|
| Takashi Yamamoto | 69 | 1866 | 27.04 | 2012–2021 | 20% | Hiroshima University | Cell Biology (24); Multidisciplinary Sciences (21); Developmental Biology (13) |
| Tetsushi Sakuma | 62 | 1763 | 28.44 | 2012–2021 | 19% | Hiroshima University | Cell Biology (22); Multidisciplinary Sciences (19); Developmental Biology (11) |
| Feng Zhang | 62 | 36,524 | 589.1 | 2012–2021 | 25% | Massachusetts Institute of Technology | Biochemistry Molecular Biology (22); Cell Biology (21); Multidisciplinary Sciences (15) |
| Jin-Soo Kim | 61 | 9354 | 153.34 | 2009–2021 | 24% | Seoul National University | Biotechnology Applied Microbiology (24); Biochemistry Molecular Biology (15); Multidisciplinary Sciences (14) |
| Jennifer A. Doudna | 52 | 14,594 | 280.65 | 2012–2021 | 44% | University of California, Berkeley | Multidisciplinary Sciences (23); Biochemistry Molecular Biology (14); Cell Biology (7) |
| Caixia Gao | 45 | 3965 | 88.11 | 2014–2021 | 58% | Chinese Academy of Sciences | Biotechnology Applied Microbiology (15); Plant Sciences (11); Biochemistry Molecular Biology (9) |
| J. Keith Joung | 42 | 16,411 | 390.74 | 2010–2021 | 20% | Harvard University | Biotechnology Applied Microbiology (15); Biochemistry Molecular Biology (9); Multidisciplinary Sciences (8) |
| Matthew H. Porteus | 40 | 2762 | 69.05 | 2013–2021 | 44% | Stanford University | Medicine Research Experimental (11); Multidisciplinary Sciences (9); Biotechnology Applied Microbiology (8) |
| Toni Cathomen | 40 | 2033 | 50.83 | 2009–2021 | 28% | University of Freiburg | Medicine Research Experimental (15); Genetics Heredity (12); Biotechnology Applied Microbiology (11) |
| Gang Bao | 39 | 5242 | 134.41 | 2013–2021 | 39% | Rice University | Medicine Research Experimental (12); Biotechnology Applied Microbiology (8); Genetics Heredity (7) |
| Daniel F. Voytas | 39 | 4877 | 125.05 | 2011–2021 | 24% | University Minnesota Crookston | Plant Sciences (18); Biotechnology Applied Microbiology (10); Multidisciplinary Sciences (7) |
| Michael C. Holmes | 39 | 9602 | 246.21 | 2007–2021 | 18% | Sangamo Therapeutics | Biotechnology Applied Microbiology (14); Medicine Research Experimental (13); Genetics Heredity (9) |
| David R. Liu | 35 | 8268 | 236.23 | 2013–2021 | 38% | Harvard University | Multidisciplinary Sciences (17); Biochemistry Molecular Biology (8); Biotechnology Applied Microbiology (6); |
| Rodolphe Barrangou | 35 | 1996 | 57.03 | 2013–2021 | 49% | North Carolina State University | Microbiology (10); Biochemistry Molecular Biology (8); Biotechnology Applied Microbiology (7) |
| Philip D. Gregory | 34 | 10,762 | 316.53 | 2007–2018 | 31% | Sangamo Therapeutics | Biotechnology Applied Microbiology (9); Multidisciplinary Sciences (7); Biochemical Research Methods (5) |
| Yong Zhang | 33 | 1654 | 50.12 | 2015–2021 | 60% | University of Electronic Science Technology of China | Plant Sciences (14); Biochemistry Molecular Biology (11); Biotechnology Applied Microbiology (8) |
| Bing Yang | 33 | 2103 | 63.73 | 2011–2021 | 44% | Iowa State University | Plant Sciences (19); Biotechnology Applied Microbiology (12); Biochemistry Molecular Biology (10) |
| YiPing Qi | 32 | 1682 | 52.56 | 2015–2021 | 69% | University of Maryland, College Park | Plant Sciences (25); Biotechnology Applied Microbiology (6); Biochemistry Molecular Biology (5) |
| Xingxu Huang | 28 | 1447 | 51.68 | 2014–2021 | 32% | Shanghai Tech University | Biochemistry Molecular Biology (9); Cell Biology (9); Multidisciplinary Sciences (7) |
| Huimin Zhao | 28 | 1618 | 57.79 | 2012–2021 | 31% | University of Illinois, Urbana-Champaign | Biotechnology Applied Microbiology (14); Biochemical Research Methods (6); Biochemistry Molecular Biology (3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Pu, A.; Liu, Q.; Hou, Q.; Zhang, Y.; An, X.; Long, Y.; Jiang, Y.; Dong, Z.; Wu, S.; et al. The Bibliometric Landscape of Gene Editing Innovation and Regulation in the Worldwide. Cells 2022, 11, 2682. https://doi.org/10.3390/cells11172682
Wei X, Pu A, Liu Q, Hou Q, Zhang Y, An X, Long Y, Jiang Y, Dong Z, Wu S, et al. The Bibliometric Landscape of Gene Editing Innovation and Regulation in the Worldwide. Cells. 2022; 11(17):2682. https://doi.org/10.3390/cells11172682
Chicago/Turabian StyleWei, Xun, Aqing Pu, Qianqian Liu, Quancan Hou, Yong Zhang, Xueli An, Yan Long, Yilin Jiang, Zhenying Dong, Suowei Wu, and et al. 2022. "The Bibliometric Landscape of Gene Editing Innovation and Regulation in the Worldwide" Cells 11, no. 17: 2682. https://doi.org/10.3390/cells11172682
APA StyleWei, X., Pu, A., Liu, Q., Hou, Q., Zhang, Y., An, X., Long, Y., Jiang, Y., Dong, Z., Wu, S., & Wan, X. (2022). The Bibliometric Landscape of Gene Editing Innovation and Regulation in the Worldwide. Cells, 11(17), 2682. https://doi.org/10.3390/cells11172682







