Drosophila as a Model for Human Viral Neuroinfections
Abstract
1. Introduction
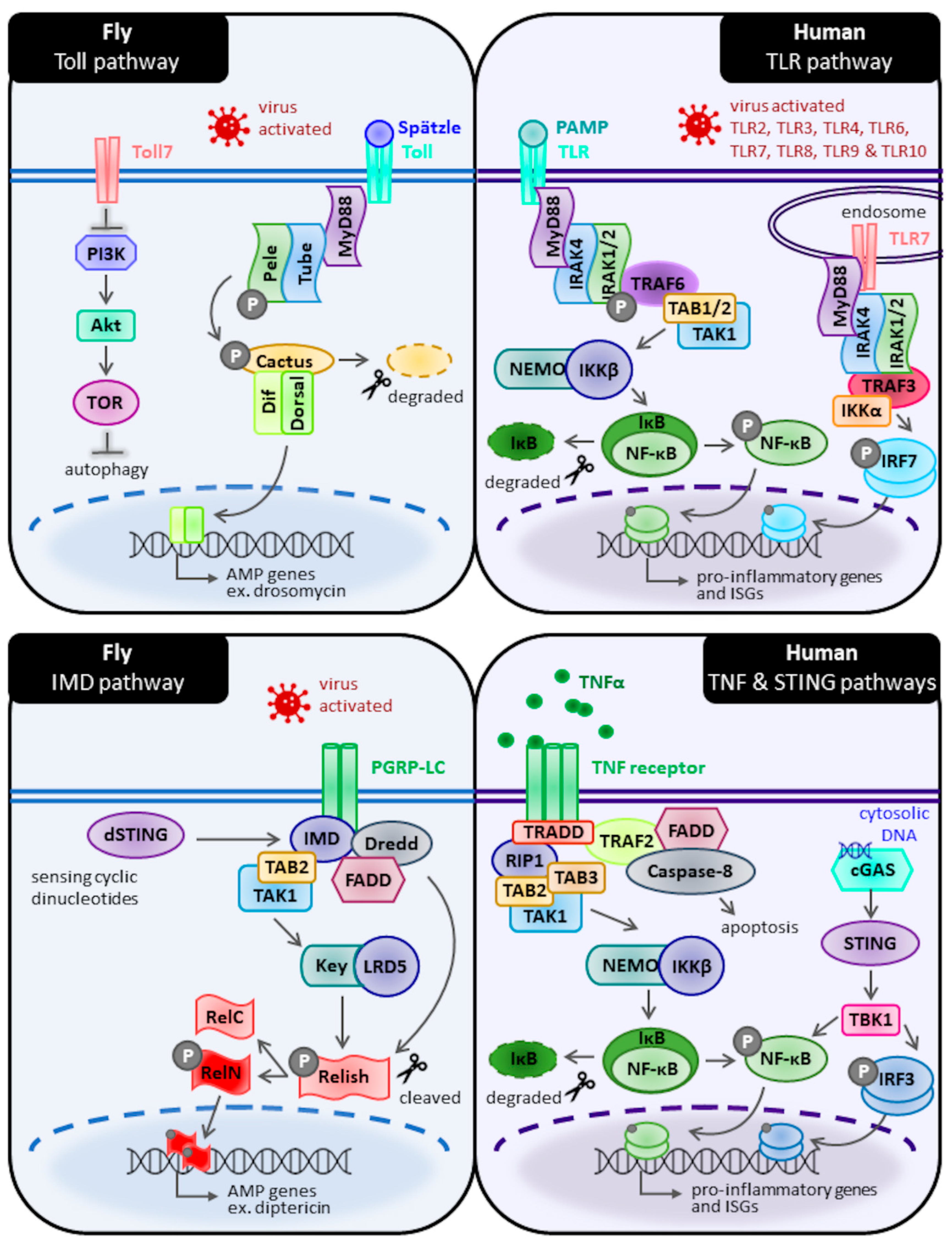
2. Antiviral Immunity in Drosophila—What to Expect and How It Differs from Humans
2.1. Toll Pathways
2.2. Immune Deficiency (IMD) Pathway
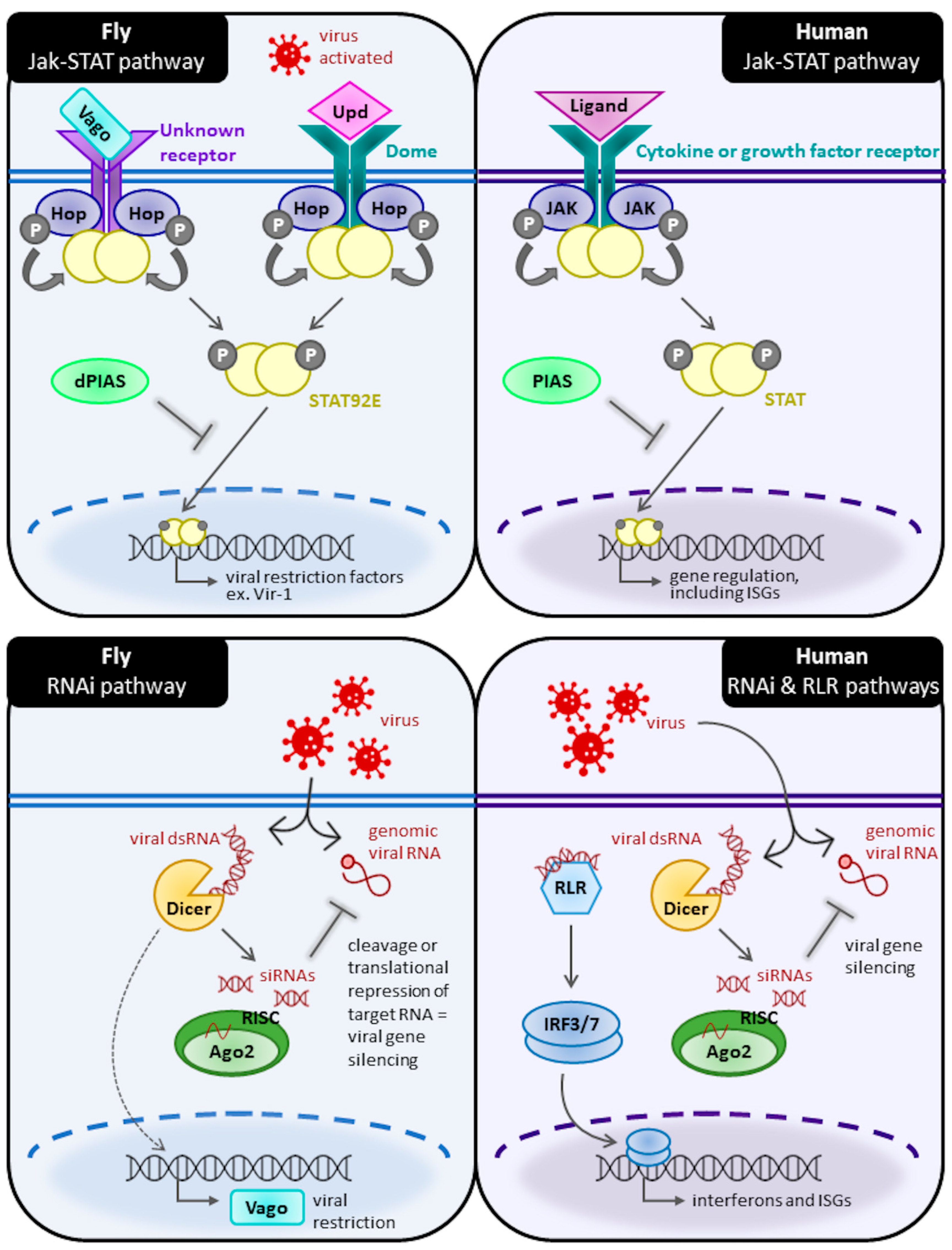
2.3. Jak-STAT Pathway
2.4. RNA Interference
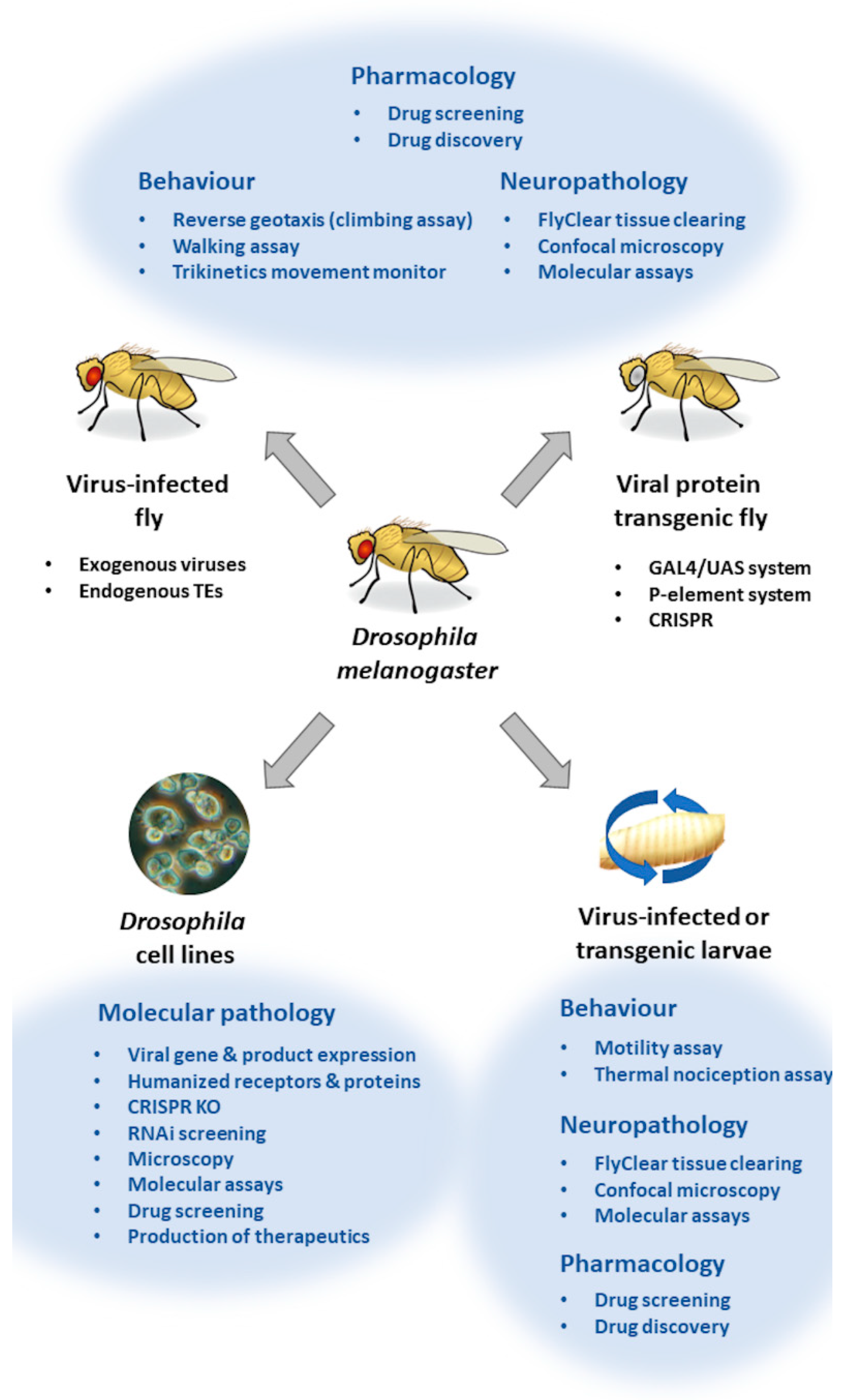
3. Neuroinfection and Neuroimmunity Models in Drosophila
3.1. In Vitro Drosophila Viral Neuroinfection Models
3.1.1. Studying Viral Entry
3.1.2. Assessing Viral Replication
3.1.3. RNAi Screens to Identify Antiviral Pathways
3.1.4. Examining Antiviral Immune Responses In Vitro
3.1.5. Production of Viral Proteins for Therapeutics Using Drosophila Cells
3.2. In Vivo Drosophila Viral Neuroinfection Models
3.2.1. Examining Antiviral Immune Responses In Vivo
3.2.2. Investigating Viral Clearance In Vivo
3.2.3. Investigating Viral Latency In Vivo
3.2.4. Infectious Neurodevelopmental Models
3.2.5. Infectious Neurodegenerative Models
3.3. Viral Transgenic Drosophila
3.4. Antiviral Drug Screening in Drosophila
4. Endogenous Retroviruses and Transposons in Drosophila
4.1. TEs through the Ages—From Development to Aging
4.2. ERVs, TDP-43 Pathology and Motor Neuron Disease
4.3. TEs and Tauopathy
4.4. TEs and Huntington Protein Expansion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jennings, B.H. Drosophila—A versatile model in biology & medicine. Mater. Today 2011, 14, 190–195. [Google Scholar] [CrossRef]
- Wangler, M.F.; Yamamoto, S.; Bellen, H.J. Fruit Flies in Biomedical Research. Genetics 2015, 199, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Van Sluijs, L.; Pijlman, G.P.; Kammenga, J.E. Why do Individuals Differ in Viral Susceptibility? A Story Told by Model Organisms. Viruses 2017, 9, 284. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nichols, C.D. Human Disease Models in Drosophila melanogaster and the Role of the Fly in Therapeutic Drug Discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef] [PubMed]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. Dis. Models Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A Systematic Analysis of Human Disease-Associated Gene Sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef]
- Harnish, J.; Link, N.; Yamamoto, S. Drosophila as a Model for Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 2724. [Google Scholar] [CrossRef]
- Younes, S.; Al-Sulaiti, A.; Nasser, E.A.A.; Najjar, H.; Kamareddine, L. Drosophila as a Model Organism in Host–Pathogen Interaction Studies. Front. Cell. Infect. Microbiol. 2020, 10, 214. [Google Scholar] [CrossRef]
- Myllymäki, H.; Valanne, S.; Rämet, M. The Drosophila Imd Signaling Pathway. J. Immunol. 2014, 192, 3455–3462. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.-H.; Rämet, M. The Drosophila Toll Signaling Pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Stokes, B.A.; Yadav, S.; Shokal, U.; Smith, L.C.; Eleftherianos, I. Bacterial and fungal pattern recognition receptors in homologous innate signaling pathways of insects and mammals. Front. Microbiol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Jaiswal, M.; Charng, W.-L.; Gambin, T.; Karaca, E.; Mirzaa, G.; Wiszniewski, W.; Sandoval, H.; Haelterman, N.A.; Xiong, B.; et al. A Drosophila Genetic Resource of Mutants to Study Mechanisms Underlying Human Genetic Diseases. Cell 2014, 159, 200–214. [Google Scholar] [CrossRef]
- Mussabekova, A.; Daeffler, L.; Imler, J.-L. Innate and intrinsic antiviral immunity in Drosophila. Experientia 2017, 74, 2039–2054. [Google Scholar] [CrossRef]
- Sabin, L.R.; Hanna, S.L.; Cherry, S. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 2010, 22, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cherry, S. Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 2014, 42, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Kidd, S. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell 1992, 71, 623–635. [Google Scholar] [CrossRef]
- Wu, L.P.; Anderson, K.V. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature 1998, 392, 93–97. [Google Scholar] [CrossRef]
- Ip, Y.T.; Reach, M.; Engstrom, Y.; Kadalayil, L.; Cai, H.; Gonzalez-Crespo, S.; Tatei, K.; Levine, M. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell 1993, 75, 753–763. [Google Scholar] [CrossRef]
- Ghosh, S.; Gifford, A.M.; Riviere, L.R.; Tempst, P.; Nolan, G.P.; Baltimore, D. Cloning of the p50 DNA binding subunit of NF-κB: Homology to rel and dorsal. Cell 1990, 62, 1019–1029. [Google Scholar] [CrossRef]
- Towb, P.; Sun, H.; Wasserman, S.A. Tube Is an IRAK-4 Homolog in a Toll Pathway Adapted for Development and Immunity. J. Innate Immun. 2009, 1, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Dunne, A.; Ejdebäck, M.; Ludidi, P.L.; O’Neill, L.A.J.; Gay, N.J. Structural Complementarity of Toll/Interleukin-1 Receptor Domains in Toll-like Receptors and the Adaptors Mal and MyD88. J. Biol. Chem. 2003, 278, 41443–41451. [Google Scholar] [CrossRef] [PubMed]
- Großhans, J.; Schnorrer, F.; Nüsslein-Volhard, C. Oligomerisation of Tube and Pelle leads to nuclear localisation of Dorsal. Mech. Dev. 1999, 81, 127–138. [Google Scholar] [CrossRef]
- Resch, K.; Jockusch, H.; Schmitt-John, T. Assignment1 of homologous genes, Peli1/PELI1 and Peli2/PELI2, for the Pelle adaptor protein Pellino to mouse chromosomes 11 and 14 and human chromosomes 2p13.3 and 14q21, respectively, by physical and radiation hybrid mapping. Cytogenet. Genome Res. 2001, 92, 172–174. [Google Scholar] [CrossRef]
- Jensen, L.E.; Whitehead, A.S. Pellino3, a Novel Member of the Pellino Protein Family, Promotes Activation of c-Jun and Elk-1 and May Act as a Scaffolding Protein. J. Immunol. 2003, 171, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Silverman, N.; Hong, M.; Liao, D.S.; Chung, Y.; Chen, Z.; Maniatis, T. The Role of Ubiquitination in Drosophila Innate Immunity. J. Biol. Chem. 2005, 280, 34048–34055. [Google Scholar] [CrossRef]
- Georgel, P.; Naitza, S.; Kappler, C.; Ferrandon, D.; Zachary, D.; Swimmer, C.; Kopczynski, C.; Duyk, G.; Reichhart, J.-M.; Hoffmann, J.A. Drosophila Immune Deficiency (IMD) Is a Death Domain Protein that Activates Antibacterial Defense and Can Promote Apoptosis. Dev. Cell 2001, 1, 503–514. [Google Scholar] [CrossRef]
- Stöven, S.; Silverman, N.; Junell, A.; Hedengren-Olcott, M.; Erturk, D.; Engström, Y.; Maniatis, T.; Hultmark, D. Caspase-mediated processing of the Drosophila NF-B factor Relish. Proc. Natl. Acad. Sci. USA 2003, 100, 5991–5996. [Google Scholar] [CrossRef]
- Meinander, A.; Runchel, C.; Tenev, T.; Chen, L.; Kim, C.-H.; Ribeiro, P.S.; Broemer, M.; Leulier, F.; Zvelebil, M.; Silverman, N.; et al. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012, 31, 2770–2783. [Google Scholar] [CrossRef]
- Leulier, F.; Vidal, S.; Saigo, K.; Ueda, R.; Lemaitre, B. Inducible Expression of Double-Stranded RNA Reveals a Role for dFADD in the Regulation of the Antibacterial Response in Drosophila Adults. Curr. Biol. 2002, 12, 996–1000. [Google Scholar] [CrossRef]
- Kleino, A.; Valanne, S.; Ulvila, J.; Kallio, J.; Myllymäki, H.; Enwald, H.; Stöven, S.; Poidevin, M.; Ueda, R.; Hultmark, D.; et al. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005, 24, 3423–3434. [Google Scholar] [CrossRef] [PubMed]
- Choe, K.-M.; Lee, H.; Anderson, K.V. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. USA 2005, 102, 1122–1126. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Kromer-Metzger, E.; Michaut, L.; Nicolas, E.; Meister, M.; Georgel, P.; Reichhart, J.M.; Hoffmann, J.A. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. USA 1995, 92, 9465–9469. [Google Scholar] [CrossRef] [PubMed]
- Silverman, N.; Zhou, R.; Stöven, S.; Pandey, N.; Hultmark, D.; Maniatis, T. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000, 14, 2461–2471. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, L.P.; Anderson, K.V. The antibacterial arm of the Drosophila innate immune response requires an IκB kinase. Genes Dev. 2001, 15, 104–110. [Google Scholar] [CrossRef]
- Ertürk-Hasdemir, D.; Broemer, M.; Leulier, F.; Lane, W.S.; Paquette, N.; Hwang, D.; Kim, C.-H.; Stöven, S.; Meier, P.; Silverman, N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl. Acad. Sci. USA 2009, 106, 9779–9784. [Google Scholar] [CrossRef]
- Fukuyama, H.; Verdier, Y.; Guan, Y.; Makino-Okamura, C.; Shilova, V.; Liu, X.; Maksoud, E.; Matsubayashi, J.; Haddad, I.; Spirohn, K.; et al. Landscape of protein–protein interactions in Drosophila immune deficiency signaling during bacterial challenge. Proc. Natl. Acad. Sci. USA 2013, 110, 10717–10722. [Google Scholar] [CrossRef]
- Rutschmann, S.; Jung, A.C.; Zhou, R.; Silverman, N.; Hoffmann, J.A.; Ferrandon, D. Role of Drosophila IKKγ in a Toll-independent antibacterial immune response. Nat. Immunol. 2000, 1, 342–347. [Google Scholar] [CrossRef]
- Dushay, M.S.; Asling, B.; Hultmark, D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. USA 1996, 93, 10343–10347. [Google Scholar] [CrossRef]
- Hedengren, M.; Bengtåsling, B.; Dushay, M.S.; Ando, I.; Ekengren, S.; Wihlborg, M.; Hultmark, D. Relish, a Central Factor in the Control of Humoral but Not Cellular Immunity in Drosophila. Mol. Cell 1999, 4, 827–837. [Google Scholar] [CrossRef]
- Vidal, S.; Khush, R.S.; Leulier, F.; Tzou, P.; Nakamura, M.; Lemaitre, B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-κB-dependent innate immune responses. Genes Dev. 2001, 15, 1900–1912. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Hiroyasu, A.; Guzman, R.M.; Roberts, S.A.; Goodman, A.G. Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep. 2018, 23, 3537–3550.e6. [Google Scholar] [CrossRef] [PubMed]
- Hombrıía, J.C.-G.; Brown, S. The Fertile Field of Drosophila JAK/STAT Signalling. Curr. Biol. 2002, 12, R569–R575. [Google Scholar] [CrossRef]
- Makki, R.; Meister, M.; Pennetier, D.; Ubeda, J.-M.; Braun, A.; Daburon, V.; Krzemień, J.; Bourbon, H.-M.; Zhou, R.; Vincent, A.; et al. A Short Receptor Downregulates JAK/STAT Signalling to Control the Drosophila Cellular Immune Response. PLoS Biol. 2010, 8, e1000441. [Google Scholar] [CrossRef]
- Brown, S.; Hu, N.; Hombrıía, J.C.-G. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 2001, 11, 1700–1705. [Google Scholar] [CrossRef]
- Binari, R.; Perrimon, N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994, 8, 300–312. [Google Scholar] [CrossRef]
- Hou, X.S.; Melnick, M.B.; Perrimon, N. marelle Acts Downstream of the Drosophila HOP/JAK Kinase and Encodes a Protein Similar to the Mammalian STATs. Cell 1996, 84, 411–419. [Google Scholar] [CrossRef]
- Callus, B.A.; Mathey-Prevot, B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene 2002, 21, 4812–4821. [Google Scholar] [CrossRef]
- Karsten, P.; Häder, S.; Zeidler, M.P. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 2002, 117, 343–346. [Google Scholar] [CrossRef]
- Yan, R.; Small, S.; Desplan, C.; Dearolf, C.R.; Darnell, J.E. Identification of a Stat Gene That Functions in Drosophila Development. Cell 1996, 84, 421–430. [Google Scholar] [CrossRef]
- Hari, K.L.; Cook, K.R.; Karpen, G.H. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001, 15, 1334–1348. [Google Scholar] [CrossRef] [PubMed]
- Betz, A.; Lampen, N.; Martinek, S.; Young, M.W.; Darnell, J.E., Jr. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. USA 2001, 98, 9563–9568. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 2003, 82, 323–330. [Google Scholar] [CrossRef]
- Kataoka, Y.; Takeichi, M.; Uemura, T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells 2001, 6, 313–325. [Google Scholar] [CrossRef]
- Vourekas, A.; Zheng, K.; Fu, Q.; Maragkakis, M.; Alexiou, P.; Ma, J.; Pillai, R.S.; Mourelatos, Z.; Wang, P.J. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015, 29, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Ichigotani, Y.; Okuda, T.; Irimura, T.; Nakatsugawa, S.; Hamaguchi, M. Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2000, 1490, 163–169. [Google Scholar] [CrossRef]
- Wan, L.; Dockendorff, T.C.; Jongens, T.A.; Dreyfuss, G. Characterization of dFMR1, a Drosophila melanogaster Homolog of the Fragile X Mental Retardation Protein. Mol. Cell. Biol. 2000, 20, 8536–8547. [Google Scholar] [CrossRef]
- Ishizuka, A.; Siomi, M.C.; Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002, 16, 2497–2508. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.-E.; Smith, D.P.; Wang, X. R2D2, a Bridge Between the Initiation and Effector Steps of the Drosophila RNAi Pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef]
- Caudy, A.; Ketting, R.; Hammond, S.M.; Denli, A.M.; Bathoorn, A.M.P.; Tops, B.; Silva, J.M.; Myers, M.M.; Hannon, G.J.; Plasterk, R.H.A. A micrococcal nuclease homologue in RNAi effector complexes. Nature 2003, 425, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gupta, P.; Fressigne, L.; Bossé, G.D.; Wang, X.; Simard, M.J.; Hansen, D. TEG-1 CD2BP2 controls miRNA levels by regulating miRISC stability in C. elegans and human cells. Nucleic Acids Res. 2017, 45, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.C.; McLane, L.M.; Maqbool, T.; Panda, D.; Gordesky-Gold, B.; Cherry, S. A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev. 2013, 27, 1511–1525. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lozano, J.R.; Gonzalez-Escribano, M.F.; Wichmann, I.; Nuñez-Roldan, A. Cytoplasmic detection of a novel protein containing a nuclear localization sequence by human autoantibodies. Clin. Exp. Immunol. 1997, 107, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Gold, B.; Tartell, M.A.; Rausch, K.; Casas-Tinto, S.; Cherry, S. The Transcription Factor FoxK Participates with Nup98 To Regulate Antiviral Gene Expression. mBio 2015, 6, e02509-14. [Google Scholar] [CrossRef]
- Goto, A.; Okado, K.; Martins, N.; Cai, H.; Barbier, V.; Lamiable, O.; Troxler, L.; Santiago, E.; Kuhn, L.; Paik, D.; et al. The Kinase IKKβ Regulates a STING- and NF-κB-Dependent Antiviral Response Pathway in Drosophila. Immunity 2018, 49, 225–234.e4. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Panda, D.; Pascual-Garcia, P.; Dunagin, M.; Tudor, M.; Hopkins, K.C.; Xu, J.; Gold, B.; Raj, A.; Capelson, M.; Cherry, S. Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E3890–E3899. [Google Scholar] [CrossRef]
- Di Nunzio, F.; Fricke, T.; Miccio, A.; Valle-Casuso, J.C.; Perez, P.; Souque, P.; Rizzi, E.; Severgnini, M.; Mavilio, F.; Charneau, P.; et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 2013, 440, 8–18. [Google Scholar] [CrossRef]
- Contamine, D.; Petitjean, A.M.; Ashburner, M. Genetic resistance to viral infection: The molecular cloning of a Drosophila gene that restricts infection by the rhabdovirus sigma. Genetics 1989, 123, 525–533. [Google Scholar] [CrossRef]
- Hurley, E.P.; Staveley, B.E. Inhibition of Ref(2)P, the Drosophila homologue of the p62/SQSTM1 gene, increases lifespan and leads to a decline in motor function. BMC Res. Notes 2021, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.K.; Luo, Y.; Moon, S.; Ninova, M.; Marinov, G.K.; Chung, Y.D.; Aravin, A.A. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev. 2016, 30, 840–855. [Google Scholar] [CrossRef] [PubMed]
- Stetson, D.B.; Ko, J.S.; Heidmann, T.; Medzhitov, R. Trex1 Prevents Cell-Intrinsic Initiation of Autoimmunity. Cell 2008, 134, 587–598. [Google Scholar] [CrossRef]
- Lewis, M.; Arnot, C.J.; Beeston, H.; McCoy, A.; Ashcroft, A.E.; Gay, N.J.; Gangloff, M. Cytokine Spätzle binds to the Drosophila immunoreceptor Toll with a neurotrophin-like specificity and couples receptor activation. Proc. Natl. Acad. Sci. USA 2013, 110, 20461–20466. [Google Scholar] [CrossRef]
- Lye, S.H.; Chtarbanova, S. Drosophila as a Model to Study Brain Innate Immunity in Health and Disease. Int. J. Mol. Sci. 2018, 19, 3922. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Forero, M.G.; Wentzell, J.S.; Durmus, I.; Wolf, R.; Anthoney, N.C.; Parker, M.; Jiang, R.; Hasenauer, J.; Strausfeld, N.J.; et al. A Toll-receptor map underlies structural brain plasticity. eLife 2020, 9, e52743. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Nishida, Y.; Ip, Y.T. Functional analysis of Toll-related genes in Drosophila. Dev. Growth Differ. 2010, 52, 771–783. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef]
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.N.; Wu, L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef]
- Lamiable, O.; Meignin, C.; Imler, J.-L. WntD and Diedel: Two immunomodulatory cytokines in Drosophila immunity. Fly 2016, 10, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Fei, S.; Xia, J.; Labropoulou, V.; Swevers, L.; Sun, J. Antimicrobial Peptides as Potential Antiviral Factors in Insect Antiviral Immune Response. Front. Immunol. 2020, 11, 2030. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, U.; Girard, J.R.; Goins, L.M.; Spratford, C.M. Drosophila as a Genetic Model for Hematopoiesis. Genetics 2019, 211, 367–417. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, Á.G.; Naylor, H.; Esteves, S.S.; Pais, I.S.; Martins, N.E.; Teixeira, L. The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog. 2014, 10, e1004507. [Google Scholar] [CrossRef]
- Moy, R.H.; Gold, B.; Molleston, J.M.; Schad, V.; Yanger, K.; Salzano, M.-V.; Yagi, Y.; Fitzgerald, K.A.; Stanger, B.Z.; Soldan, S.S.; et al. Antiviral Autophagy Restricts Rift Valley Fever Virus Infection and Is Conserved from Flies to Mammals. Immunity 2014, 40, 51–65. [Google Scholar] [CrossRef]
- Nakamoto, M.; Moy, R.H.; Xu, J.; Bambina, S.; Yasunaga, A.; Shelly, S.S.; Gold, B.; Cherry, S. Virus Recognition by Toll-7 Activates Antiviral Autophagy in Drosophila. Immunity 2012, 36, 658–667. [Google Scholar] [CrossRef]
- Iwanaszko, M.; Kimmel, M. NF-κB and IRF pathways: Cross-regulation on target genes promoter level. BMC Genom. 2015, 16, 307. [Google Scholar] [CrossRef]
- Platanitis, E.; Decker, T. Regulatory Networks Involving STATs, IRFs, and NFκB in Inflammation. Front. Immunol. 2018, 9, 2542. [Google Scholar] [CrossRef]
- Kounatidis, I.; Chtarbanova, S.; Cao, Y.; Hayne, M.; Jayanth, D.; Ganetzky, B.; Ligoxygakis, P. NF-κB Immunity in the Brain Determines Fly Lifespan in Healthy Aging and Age-Related Neurodegeneration. Cell Rep. 2017, 19, 836–848. [Google Scholar] [CrossRef]
- Huang, B.; Qi, Z.T.; Xu, Z.; Nie, P. Global characterization of interferon regulatory factor (IRF) genes in vertebrates: Glimpse of the diversification in evolution. BMC Immunol. 2010, 11, 22. [Google Scholar] [CrossRef]
- Nehyba, J.; Hrdličková, R.; Bose, H.R. Dynamic Evolution of Immune System Regulators: The History of the Interferon Regulatory Factor Family. Mol. Biol. Evol. 2009, 26, 2539–2550. [Google Scholar] [CrossRef] [PubMed]
- Yanai, H.; Chiba, S.; Hangai, S.; Kometani, K.; Inoue, A.; Kimura, Y.; Abe, T.; Kiyonari, H.; Nishio, J.; Taguchi-Atarashi, N.; et al. Revisiting the role of IRF3 in inflammation and immunity by conditional and specifically targeted gene ablation in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 5253–5258. [Google Scholar] [CrossRef] [PubMed]
- AL Hamrashdi, M.; Brady, G. Regulation of IRF3 activation in human antiviral signaling pathways. Biochem. Pharmacol. 2022, 200, 115026. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Y.; Sanchez, D.J.; Aliyari, R.; Lu, S.; Cheng, G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. USA 2012, 109, 4239–4244. [Google Scholar] [CrossRef]
- Margolis, S.R.; Wilson, S.C.; Vance, R.E. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef]
- Kaneko, T.; Yano, T.; Aggarwal, K.; Lim, J.-H.; Ueda, K.; Oshima, Y.; Peach, C.; Erturk-Hasdemir, D.; Goldman, W.; Oh, B.-H.; et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006, 7, 715–723. [Google Scholar] [CrossRef]
- Costa, A.; Jan, E.; Sarnow, P.; Schneider, D. The Imd Pathway Is Involved in Antiviral Immune Responses in Drosophila. PLoS ONE 2009, 4, e7436. [Google Scholar] [CrossRef]
- West, C.; Silverman, N. cGAS-like receptors put a sting into the evolution of immune defence. Nature 2021, 597, 34–35. [Google Scholar] [CrossRef]
- Holleufer, A.; Winther, K.G.; Gad, H.H.; Ai, X.; Chen, Y.; Li, L.; Wei, Z.; Deng, H.; Liu, J.; Frederiksen, N.A.; et al. Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature 2021, 597, 114–118. [Google Scholar] [CrossRef]
- Cai, H.; Holleufer, A.; Simonsen, B.; Schneider, J.; Lemoine, A.; Gad, H.H.; Huang, J.; Huang, J.; Chen, D.; Peng, T.; et al. 2′3′-cGAMP triggers a STING- and NF-κB–dependent broad antiviral response in Drosophila. Sci. Signal. 2020, 13, eabc4537. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cherry, S. Zika virus infection activates sting-dependent antiviral autophagy in the Drosophila brain. Autophagy 2019, 15, 174–175. [Google Scholar] [CrossRef] [PubMed]
- Winkler, B.; Funke, D.; Benmimoun, B.; Spéder, P.; Rey, S.; Logan, M.A.; Klämbt, C. Brain inflammation triggers macrophage invasion across the blood-brain barrier in Drosophila during pupal stages. Sci. Adv. 2021, 7, eabh0050. [Google Scholar] [CrossRef] [PubMed]
- Myllymäki, H.; Rämet, M. JAK/STAT Pathway in Drosophila Immunity. Scand. J. Immunol. 2014, 79, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA Interference–Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. J. Immunol. 2013, 190, 650–658. [Google Scholar] [CrossRef]
- Schneider, J.; Imler, J.-L. Sensing and signalling viral infection in drosophila. Dev. Comp. Immunol. 2021, 117, 103985. [Google Scholar] [CrossRef]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.-L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Morin-Poulard, I.; Vincent, A.; Crozatier, M. The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAK-STAT 2013, 2, e25700. [Google Scholar] [CrossRef]
- Swevers, L.; Liu, J.; Smagghe, G. Defense Mechanisms against Viral Infection in Drosophila: RNAi and Non-RNAi. Viruses 2018, 10, 230. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA Silencing Pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Soares, Z.G.; Gonçalves, A.N.A.; de Oliveira, K.P.V.; Marques, J.T. Viral RNA recognition by the Drosophila small interfering RNA pathway. Microbes Infect. 2014, 16, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.; Imler, J.-L. Antiviral immunity in drosophila. Curr. Opin. Immunol. 2009, 21, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Wu, X.; Pan, Y.; Fang, Y.; Zhang, J.; Xie, M.; Yang, F.; Yu, T.; Ma, P.; Li, W.; Shu, Y. The Biogenesis and Functions of piRNAs in Human Diseases. Mol. Ther. Nucleic Acids 2020, 21, 108–120. [Google Scholar] [CrossRef]
- Yamashiro, H.; Siomi, M.C. PIWI-Interacting RNA in Drosophila: Biogenesis, Transposon Regulation, and Beyond. Chem. Rev. 2018, 118, 4404–4421. [Google Scholar] [CrossRef]
- Huang, X.; Wong, G. An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative diseases. Transl. Neurodegener. 2021, 10, 9. [Google Scholar] [CrossRef]
- Heath, C.G.; Viphakone, N.; Wilson, S.A. The role of TREX in gene expression and disease. Biochem. J. 2016, 473, 2911–2935. [Google Scholar] [CrossRef]
- Tao, S.-S.; Wu, G.-C.; Zhang, Q.; Zhang, T.-P.; Leng, R.-X.; Pan, H.-F.; Ye, D.-Q. TREX1 As a Potential Therapeutic Target for Autoimmune and Inflammatory Diseases. Curr. Pharm. Des. 2019, 25, 3239–3247. [Google Scholar] [CrossRef]
- Barckmann, B.; El-Barouk, M.; Pélisson, A.; Mugat, B.; Li, B.; Franckhauser, C.; Lavier, A.-S.F.; Mirouze, M.; Fablet, M.; Chambeyron, S. The somatic piRNA pathway controls germline transposition over generations. Nucleic Acids Res. 2018, 46, 9524–9536. [Google Scholar] [CrossRef]
- Keegan, R.M.; Talbot, L.R.; Chang, Y.-H.; Metzger, M.J.; Dubnau, J. Intercellular viral spread and intracellular transposition of Drosophila gypsy. PLoS Genet. 2021, 17, e1009535. [Google Scholar] [CrossRef] [PubMed]
- Tafesh-Edwards, G.; Eleftherianos, I. Drosophila immunity against natural and nonnatural viral pathogens. Virology 2020, 540, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Nayak, N.; Mishra, M. Drosophila melanogaster as a model to understand the mechanisms of infection mediated neuroinflammation in neurodegenerative diseases. J. Integr. Neurosci. 2022, 21, 66. [Google Scholar] [CrossRef] [PubMed]
- Luhur, A.; Klueg, K.M.; Zelhof, A.C. Generating and working with Drosophila cell cultures: Current challenges and opportunities. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e339. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.-H.; Kim, J.; Kim, H.-W.; Cho, J.W. Herpes simplex viruses (1 and 2) and varicella-zoster virus infections in an adult population with aseptic meningitis or encephalitis: A nine-year retrospective clinical study. Medicine 2021, 100, e27856. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Bohannon, K.P.; Longnecker, R. Drosophila Schneider 2 (S2) cells: A novel tool for studying HSV-induced membrane fusion. Virology 2013, 437, 100–109. [Google Scholar] [CrossRef]
- Liu, W.; Vigdorovich, V.; Zhan, C.; Patskovsky, Y.; Bonanno, J.B.; Nathenson, S.G.; Almo, S.C. Increased Heterologous Protein Expression in Drosophila S2 Cells for Massive Production of Immune Ligands/Receptors and Structural Analysis of Human HVEM. Mol. Biotechnol. 2015, 57, 914–922. [Google Scholar] [CrossRef]
- Mao, H.; Zhao, X.; Zhu, H.; Guo, J.; Ma, Z. Expression and immunogenicity of recombinant glycoprotein D of herpes simplex virus 1 in Drosophila S2 cells. Prep. Biochem. Biotechnol. 2016, 46, 384–391. [Google Scholar] [CrossRef]
- Klein, R.S. Encephalitic Arboviruses of Africa: Emergence, Clinical Presentation and Neuropathogenesis. Front. Immunol. 2021, 12, 769942. [Google Scholar] [CrossRef]
- Yeh, J.X.; Schultz, K.L.W.; Calvert, V.; Petricoin, E.F.; Griffin, D.E. The NF-κB/leukemia inhibitory factor/STAT3 signaling pathway in antibody-mediated suppression of Sindbis virus replication in neurons. Proc. Natl. Acad. Sci. USA 2020, 117, 29035–29045. [Google Scholar] [CrossRef]
- Rose, P.P.; Hanna, S.L.; Spiridigliozzi, A.; Wannissorn, N.; Beiting, D.P.; Ross, S.R.; Hardy, R.W.; Bambina, S.A.; Heise, M.T.; Cherry, S. Natural Resistance-Associated Macrophage Protein Is a Cellular Receptor for Sindbis Virus in Both Insect and Mammalian Hosts. Cell Host Microbe 2011, 10, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Moser, T.S.; Jones, R.G.; Thompson, C.B.; Coyne, C.B.; Cherry, S. A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics. PLoS Pathog. 2010, 6, e1000954. [Google Scholar] [CrossRef] [PubMed]
- Bengali, Z.; Satheshkumar, P.S.; Yang, Z.; Weisberg, A.S.; Paran, N.; Moss, B. Drosophila S2 Cells Are Non-Permissive for Vaccinia Virus DNA Replication Following Entry via Low pH-Dependent Endocytosis and Early Transcription. PLoS ONE 2011, 6, e17248. [Google Scholar] [CrossRef] [PubMed]
- Stübgen, J.-P. Neuromuscular Disorders Associated With Hepatitis B Virus Infection. J. Clin. Neuromuscul. Dis. 2011, 13, 26–37. [Google Scholar] [CrossRef]
- Zhang, P.; Raney, A.K.; McLachlan, A. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J. Virol. 1993, 67, 1472–1481. [Google Scholar] [CrossRef]
- Li, J.; Ou, J.-H. Differential Regulation of Hepatitis B Virus Gene Expression by the Sp1 Transcription Factor. J. Virol. 2001, 75, 8400–8406. [Google Scholar] [CrossRef]
- Zhang, P.; McLachlan, A. Differentiation-Specific Transcriptional Regulation of the Hepatitis B Virus Nucleocapsid Gene in Human Hepatoma Cell Lines. Virology 1994, 202, 430–440. [Google Scholar] [CrossRef]
- Wang, H.D.; Yuh, C.H.; Dang, C.V.; Johnson, D.L. The hepatitis B virus X protein increases the cellular level of TATA-binding protein, which mediates transactivation of RNA polymerase III genes. Mol. Cell. Biol. 1995, 15, 6720–6728. [Google Scholar] [CrossRef]
- Wang, H.D.; Trivedi, A.; Johnson, D.L. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 1997, 17, 6838–6846. [Google Scholar] [CrossRef]
- Wang, H.-D.; Trivedi, A.; Johnson, D.L. Regulation of RNA Polymerase I-Dependent Promoters by the Hepatitis B Virus X Protein via Activated Ras and TATA-Binding Protein. Mol. Cell. Biol. 1998, 18, 7086–7094. [Google Scholar] [CrossRef]
- Anywaine, Z.; Lule, S.A.; Hansen, C.; Warimwe, G.; Elliott, A. Clinical manifestations of Rift Valley fever in humans: Systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2022, 16, e0010233. [Google Scholar] [CrossRef]
- Wijchers, P.J.; Hoekman, M.F.; Burbach, J.P.H.; Smidt, M.P. Identification of forkhead transcription factors in cortical and dopaminergic areas of the adult murine brain. Brain Res. 2006, 1068, 23–33. [Google Scholar] [CrossRef]
- Hao, L.; Sakurai, A.; Watanabe, T.; Sorensen, E.; Nidom, C.A.; Newton, M.A.; Ahlquist, P.; Kawaoka, Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 2008, 454, 890–893. [Google Scholar] [CrossRef]
- Martins, N.; Imler, J.-L.; Meignin, C. Discovery of novel targets for antivirals: Learning from flies. Curr. Opin. Virol. 2016, 20, 64–70. [Google Scholar] [CrossRef]
- Dietrich, I.; Jansen, S.; Fall, G.; Lorenzen, S.; Rudolf, M.; Huber, K.; Heitmann, A.; Schicht, S.; Ndiaye, E.H.; Watson, M.; et al. RNA Interference Restricts Rift Valley Fever Virus in Multiple Insect Systems. mSphere 2017, 2, e00090-17. [Google Scholar] [CrossRef]
- Ulvila, J.; Hultmark, D.; Rämet, M. RNA Silencing in the Antiviral Innate Immune Defence—Role of DEAD-box RNA Helicases. Scand. J. Immunol. 2010, 71, 146–158. [Google Scholar] [CrossRef]
- Johnson, J.E.; Nasar, F.; Coleman, J.W.; Price, R.E.; Javadian, A.; Draper, K.; Lee, M.; Reilly, P.A.; Clarke, D.K.; Hendry, R.M.; et al. Neurovirulence properties of recombinant vesicular stomatitis virus vectors in non-human primates. Virology 2007, 360, 36–49. [Google Scholar] [CrossRef]
- Mueller, S.; Gausson, V.; Vodovar, N.; Deddouche, S.; Troxler, L.; Perot, J.; Pfeffer, S.; Hoffmann, J.A.; Saleh, M.-C.; Imler, J.-L. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 19390–19395. [Google Scholar] [CrossRef]
- Fisher, C.; Streicker, D.G.; Schnell, M.J. The spread and evolution of rabies virus: Conquering new frontiers. Nat. Rev. Genet. 2018, 16, 241–255. [Google Scholar] [CrossRef]
- Robinson-Papp, J.; Schütz, S.G. HIV-related neuropathy: Current perspectives. HIV/AIDS Res. Palliat. Care 2013, 5, 243–251. [Google Scholar] [CrossRef][Green Version]
- Pilatti, L.; Astray, R.M.; Rocca, M.P.; Barbosa, F.F.; Jorge, S.A.C.; Butler, M.; Augusto, E.D.F.P. Purification of rabies virus glycoprotein produced in Drosophila melanogaster S2 cells: An efficient immunoaffinity method. Biotechnol. Prog. 2020, 36, e3046. [Google Scholar] [CrossRef]
- Batista, F.R.; Moraes, M.; Büntemeyer, H.; Noll, T. Influence of culture conditions on recombinant Drosophila melanogaster S2 cells producing rabies virus glycoprotein cultivated in serum-free medium. Biologicals 2009, 37, 108–118. [Google Scholar] [CrossRef]
- Yokomizo, A.Y.; Jorge, S.A.C.; Astray, R.M.; Fernandes, I.; Ribeiro, O.G.; Horton, D.S.P.Q.; Tonso, A.; Tordo, N.; Pereira, C.A. Rabies virus glycoprotein expression in Drosophila S2 cells. I. Functional recombinant protein in stable co-transfected cell line. Biotechnol. J. 2007, 2, 102–109. [Google Scholar] [CrossRef]
- Deml, L.; Wolf, H.; Wagner, R. High level expression of hepatitis B virus surface antigen in stably transfected Drosophila Schneider-2 cells. J. Virol. Methods 1999, 79, 191–203. [Google Scholar] [CrossRef]
- Deml, L.; Schirmbeck, R.; Reimann, J.; Wolf, H.; Wagner, R. Purification and characterization of hepatitis B virus surface antigen particles produced in Drosophila Schneider-2 cells. J. Virol. Methods 1999, 79, 205–217. [Google Scholar] [CrossRef]
- Horng, J.-H.; Lin, W.-H.; Wu, C.-R.; Lin, Y.-Y.; Wu, L.-L.; Chen, D.-S.; Chen, P.-J. HBV X protein-based therapeutic vaccine accelerates viral antigen clearance by mobilizing monocyte infiltration into the liver in HBV carrier mice. J. Biomed. Sci. 2020, 27, 70. [Google Scholar] [CrossRef]
- Culp, J.S.; Johansen, H.; Hellmig, B.; Beck, J.; Matthews, T.J.; Delers, A.; Rosenberg, M. Regulated Expression Allows High Level Production and Secretion of HIV-1 gp120 Envelope Glycoprotein in Drosophila Schneider Cells. Nat. Biotechnol. 1991, 9, 173–177. [Google Scholar] [CrossRef]
- Yang, L.; Song, Y.; Li, X.; Huang, X.; Liu, J.; Ding, H.; Zhu, P.; Zhou, P. HIV-1 Virus-Like Particles Produced by Stably Transfected Drosophila S2 Cells: A Desirable Vaccine Component. J. Virol. 2012, 86, 7662–7676. [Google Scholar] [CrossRef]
- Chilian, M.; Parra, K.V.; Sandoval, A.; Ramirez, J.; Yoon, W.H. CRISPR/Cas9-mediated tissue-specific knockout and cDNA rescue using sgRNAs that target exon-intron junctions in Drosophila melanogaster. STAR Protoc. 2022, 3, 101465. [Google Scholar] [CrossRef]
- Cichewicz, K.; Hirsh, J. ShinyR-DAM: A program analyzing Drosophila activity, sleep and circadian rhythms. Commun. Biol. 2018, 1, 25. [Google Scholar] [CrossRef]
- Pende, M.; Becker, K.; Wanis, M.; Saghafi, S.; Kaur, R.; Hahn, C.; Pende, N.; Foroughipour, M.; Hummel, T.; Dodt, H.-U. High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun. 2018, 9, 4731. [Google Scholar] [CrossRef]
- Jr, H.H.B.; Dunmire, S.K.; A Hogquist, K. Infectious mononucleosis. Clin. Transl. Immunol. 2015, 4, e33. [Google Scholar] [CrossRef]
- Mundo, L.; Del Porro, L.; Granai, M.; Siciliano, M.C.; Mancini, V.; Santi, R.; Marcar, L.; Vrzalikova, K.; Vergoni, F.; Di Stefano, G.; et al. Frequent traces of EBV infection in Hodgkin and non-Hodgkin lymphomas classified as EBV-negative by routine methods: Expanding the landscape of EBV-related lymphomas. Mod. Pathol. 2020, 33, 2407–2421. [Google Scholar] [CrossRef]
- Arbach, H.; Viglasky, V.; Lefeu, F.; Guinebretieère, J.-M.; Ramirez, V.; Bride, N.; Boualaga, N.; Bauchet, T.; Peyrat, J.-P.; Mathieu, M.-C.; et al. Epstein-Barr Virus (EBV) Genome and Expression in Breast Cancer Tissue: Effect of EBV Infection of Breast Cancer Cells on Resistance to Paclitaxel (Taxol). J. Virol. 2006, 80, 845–853. [Google Scholar] [CrossRef]
- Mackrides, N.; Campuzano-Zuluaga, G.; Maque-Acosta, Y.; Moul, A.; Hijazi, N.; Ikpatt, F.O.; Levy, R.; Verdun, R.E.; Kunkalla, K.; Natkunam, Y.; et al. Epstein-Barr virus-positive follicular lymphoma. Mod. Pathol. 2017, 30, 519–529. [Google Scholar] [CrossRef]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef]
- Zivadinov, R.; Guan, Y.; Jakimovski, D.; Ramanathan, M.; Weinstock-Guttman, B. The role of Epstein-Barr virus in multiple sclerosis: From molecular pathophysiology to in vivo imaging. Neural Regen. Res. 2019, 14, 373–386. [Google Scholar] [CrossRef]
- Sherri, N.; Salloum, N.; Mouawad, C.; Haidar-Ahmad, N.; Shirinian, M.; Rahal, E.A. Epstein-Barr Virus DNA Enhances Diptericin Expression and Increases Hemocyte Numbers in Drosophila melanogaster via the Immune Deficiency Pathway. Front. Microbiol. 2018, 9, 1268. [Google Scholar] [CrossRef]
- Madi, J.R.; Al Outa, A.; Ghannam, M.; Hussein, H.M.; Shehab, M.; Hasan, Z.A.K.H.; Fayad, A.A.; Shirinian, M.; Rahal, E.A. Drosophila melanogaster as a Model System to Assess the Effect of Epstein-Barr Virus DNA on Inflammatory Gut Diseases. Front. Immunol. 2021, 12, 586930. [Google Scholar] [CrossRef]
- Alosaimi, M.F.; Hoenig, M.; Jaber, F.; Platt, C.D.; Jones, J.; Wallace, J.; Debatin, K.-M.; Schulz, A.; Jacobsen, E.; Möller, P.; et al. Immunodeficiency and EBV-induced lymphoproliferation caused by 4-1BB deficiency. J. Allergy Clin. Immunol. 2019, 144, 574–583.e5. [Google Scholar] [CrossRef]
- van Montfrans, J.M.; Hoepelman, A.I.; Otto, S.; van Gijn, M.; van de Corput, L.; de Weger, R.A.; Monaco-Shawver, L.; Banerjee, P.P.; Sanders, E.A.; der Zijde, C.M.J.; et al. CD27 deficiency is associated with combined immunodeficiency and persistent symptomatic EBV viremia. J. Allergy Clin. Immunol. 2012, 129, 787–793.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gordesky-Gold, B.; Leney-Greene, M.; Weinbren, N.L.; Tudor, M.; Cherry, S. Inflammation-Induced, STING-Dependent Autophagy Restricts Zika Virus Infection in the Drosophila Brain. Cell Host Microbe 2018, 24, 57–68.e3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, S.; Gou, H.; Zhang, J.; Li, C. Crosstalk Between Autophagy and the cGAS–STING Signaling Pathway in Type I Interferon Production. Front. Cell Dev. Biol. 2021, 9, 748485. [Google Scholar] [CrossRef]
- Levine, B.; Deretic, V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007, 7, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Lennemann, N.J.; Coyne, C.B. Catch Me If You Can: The Link between Autophagy and Viruses. PLoS Pathog. 2015, 11, e1004685. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Kang, J.-H.; Lee, S. Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int. J. Mol. Sci. 2020, 21, 3369. [Google Scholar] [CrossRef]
- Contamine, D. Role of the Drosophila genome in Sigma virus multiplication I. Role of the ret(2)P gene; Selection of host-adapted mutants at the nonpermissive allele Pp. Virology 1981, 114, 474–488. [Google Scholar] [CrossRef]
- Willard, K.A.; Elling, C.L.; Stice, S.L.; Brindley, M.A. The Oxysterol 7-Ketocholesterol Reduces Zika Virus Titers in Vero Cells and Human Neurons. Viruses 2018, 11, 20. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Pattnaik, A.; Annamalai, A.S.; Franco, R.; Pattnaik, A.K. Mechanistic Target of Rapamycin Signaling Activation Antagonizes Autophagy To Facilitate Zika Virus Replication. J. Virol. 2020, 94, e01575-20. [Google Scholar] [CrossRef]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy Is an Essential Component of Drosophila Immunity against Vesicular Stomatitis Virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef]
- Chen, Q.; Lin, L.; Smith, S.; Huang, J.; Berger, S.; Zhou, J. CTCF-Dependent Chromatin Boundary Element between the Latency-Associated Transcript and ICP0 Promoters in the Herpes Simplex Virus Type 1 Genome. J. Virol. 2007, 81, 5192–5201. [Google Scholar] [CrossRef] [PubMed]
- Elgueta, D.; Murgas, P.; Riquelme, E.; Yang, G.; Cancino, G.I. Consequences of Viral Infection and Cytokine Production During Pregnancy on Brain Development in Offspring. Front. Immunol. 2022, 13, 816619. [Google Scholar] [CrossRef] [PubMed]
- Schuler-Faccini, L.; del Campo, M.; García-Alix, A.; Ventura, L.O.; Boquett, J.A.; van der Linden, V.; Pessoa, A.; Júnior, H.V.D.L.; Ventura, C.V.; Leal, M.C.; et al. Neurodevelopment in Children Exposed to Zika in utero: Clinical and Molecular Aspects. Front. Genet. 2022, 13, 758715. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.S.; Link, N.; Jang, G.M.; Sharp, P.P.; Zhu, T.; Swaney, D.L.; Johnson, J.R.; Von Dollen, J.; Ramage, H.R.; Satkamp, L.; et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 2018, 175, 1931–1945.e18. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Maddirevula, S.; Ewida, N.; Alsahli, S.; Abdel-Salam, G.M.H.; Zaki, M.S.; Al Tala, S.; Alhashem, A.; Softah, A.; Al-Owain, M.; et al. Genomic and phenotypic delineation of congenital microcephaly. Genet. Med. 2019, 21, 545–552. [Google Scholar] [CrossRef]
- Link, N.; Chung, H.; Jolly, A.; Withers, M.; Tepe, B.; Arenkiel, B.R.; Shah, P.S.; Krogan, N.J.; Aydin, H.; Geckinli, B.B.; et al. Mutations in ANKLE2, a ZIKA Virus Target, Disrupt an Asymmetric Cell Division Pathway in Drosophila Neuroblasts to Cause Microcephaly. Dev. Cell 2019, 51, 713–729.e6. [Google Scholar] [CrossRef]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.-S.; Lee, S.-A.; Ge, J.; Wang, S.; Goldman, S.; Zlokovic, B.V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef]
- Saylor, D.; Dickens, A.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-associated neurocognitive disorder—Pathogenesis and prospects for treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef]
- Arendt, T.; Mosch, B.; Morawski, M. Neuronal Aneuploidy in Health and Disease:A Cytomic Approach to Understand the Molecular Individuality of Neurons. Int. J. Mol. Sci. 2009, 10, 1609–1627. [Google Scholar] [CrossRef]
- Shepherd, C.; Yang, Y.; Halliday, G. Region- and Cell-specific Aneuploidy in Brain Aging and Neurodegeneration. Neuroscience 2018, 374, 326–334. [Google Scholar] [CrossRef]
- Battaglia, P.A.; Ponti, D.; Naim, V.; Venanzi, S.; Psaila, R.; Gigliani, F. The HIV-Tat protein induces chromosome number aberrations by affecting mitosis. Cell Motil. Cytoskelet. 2005, 61, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, Y.-S. Effect of HIV-1 Tat on the formation of the mitotic spindle by interaction with ribosomal protein S3. Sci. Rep. 2018, 8, 8680. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.; Kapadia, R.; Soule, T.; Luo, H.; Schellenberg, K.L.; Douville, R.N.; Pfeffer, G. Neuromuscular Complications of SARS-CoV-2 and Other Viral Infections. Front. Neurol. 2022, 13, 914411. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.; Towery, L.; McCown, A.; Carlson, K.A. Impaired Geotaxis as a Novel Phenotype of Nora Virus Infection of Drosophila melanogaster. Scientifica 2020, 2020, 1804510. [Google Scholar] [CrossRef] [PubMed]
- Macke, A.; Lopez, W.; Carlson, D.J.; Carlson, K.A. Nora Virus VP4b and ORF1 Circulate in Hemolymph of Infected D. melanogaster with Coordinate Expression of Vago and Vir-1. Vaccines 2020, 8, 491. [Google Scholar] [CrossRef]
- Lopez, W.; Page, A.M.; Carlson, D.J.; Ericson, B.L.; Cserhati, M.F.; Guda, C.; Carlson, K.A. Analysis of immune-related genes during Nora virus infection of Drosophila melanogaster using next generation sequencing. AIMS Microbiol. 2018, 4, 123–139. [Google Scholar] [CrossRef]
- Hughes, T.T.; Allen, A.L.; Bardin, J.E.; Christian, M.N.; Daimon, K.; Dozier, K.D.; Hansen, C.L.; Holcomb, L.M.; Ahlander, J. Drosophila as a genetic model for studying pathogenic human viruses. Virology 2012, 423, 1–5. [Google Scholar] [CrossRef]
- Niu, D.; Wei, H.-J.; Lin, L.; George, H.; Wang, T.; Lee, I.-H.; Zhao, H.-Y.; Wang, Y.; Kan, Y.N.; Shrock, E.; et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 2017, 357, 1303–1307. [Google Scholar] [CrossRef]
- Adamson, A.L.; Wright, N.; LaJeunesse, D.R. Modeling Early Epstein-Barr Virus Infection in Drosophila melanogaster: The BZLF1 Protein. Genetics 2005, 171, 1125–1135. [Google Scholar] [CrossRef]
- Adamson, A.; LaJeunesse, D. A Study of Epstein-Barr Virus BRLF1 Activity in aDrosophilaModel System. Sci. World J. 2012, 2012, 347597. [Google Scholar] [CrossRef]
- Lee, S.B.; Park, J.; Jung, J.U.; Chung, J. Nef induces apoptosis by activating JNK signaling pathway and inhibits NF-κB-dependent immune responses in Drosophila. J. Cell Sci. 2005, 118, 1851–1859. [Google Scholar] [CrossRef]
- Varin, A.; Manna, S.K.; Quivy, V.; Decrion, A.-Z.; Van Lint, C.; Herbein, G.; Aggarwal, B.B. Exogenous Nef Protein Activates NF-κB, AP-1, and c-Jun N-Terminal Kinase and Stimulates HIV Transcription in Promonocytic Cells. Role in AIDS pathogenesis. J. Biol. Chem. 2003, 278, 2219–2227. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kumar, A. Differential signaling mechanism for HIV-1 Nef-mediated production of IL-6 and IL-8 in human astrocytes. Sci. Rep. 2015, 5, 9867. [Google Scholar] [CrossRef] [PubMed]
- Mangino, G.; Famiglietti, M.; Capone, C.; Veroni, C.; Percario, Z.A.; Leone, S.; Fiorucci, G.; Lülf, S.; Romeo, G.; Agresti, C.; et al. HIV-1 Myristoylated Nef Treatment of Murine Microglial Cells Activates Inducible Nitric Oxide Synthase, NO2 Production and Neurotoxic Activity. PLoS ONE 2015, 10, e0130189. [Google Scholar] [CrossRef]
- Leulier, F.; Marchal, C.; Miletich, I.; Limbourg-Bouchon, B.; Benarous, R.; Lemaitre, B. Directed expression of the HIV-1 accessory protein Vpu in Drosophila fat-body cells inhibits Toll-dependent immune responses. EMBO Rep. 2003, 4, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Bour, S.; Perrin, C.; Akari, H.; Strebel, K. The Human Immunodeficiency Virus Type 1 Vpu Protein Inhibits NF-κB Activation by Interfering with βTrCP-mediated Degradation of IκB. J. Biol. Chem. 2001, 276, 15920–15928. [Google Scholar] [CrossRef] [PubMed]
- Marchal, C.; Vinatier, G.; Sanial, M.; Plessis, A.; Pret, A.-M.; Limbourg-Bouchon, B.; Théodore, L.; Netter, S. The HIV-1 Vpu Protein Induces Apoptosis in Drosophila via Activation of JNK Signaling. PLoS ONE 2012, 7, e34310. [Google Scholar] [CrossRef]
- Sauter, D.; Hotter, D.; Van Driessche, B.; Stürzel, C.M.; Kluge, S.F.; Wildum, S.; Yu, H.; Baumann, B.; Wirth, T.; Plantier, J.-C.; et al. Differential Regulation of NF-κB-Mediated Proviral and Antiviral Host Gene Expression by Primate Lentiviral Nef and Vpu Proteins. Cell Rep. 2015, 10, 586–599. [Google Scholar] [CrossRef]
- Langer, S.; Hammer, C.; Hopfensperger, K.; Klein, L.; Hotter, D.; De Jesus, P.D.; Herbert, K.M.; Pache, L.; Smith, N.; Van Der Merwe, J.A.; et al. HIV-1 Vpu is a potent transcriptional suppressor of NF-κB-elicited antiviral immune responses. eLife 2019, 8, e41930. [Google Scholar] [CrossRef]
- Bimler, L.; Ronzulli, S.L.; Song, A.Y.; Johnson, S.K.; Jones, C.A.; Kim, T.; Le, D.T.; Tompkins, S.M.; Paust, S. Matrix Protein 2 Extracellular Domain-Specific Monoclonal Antibodies Are an Effective and Potentially Universal Treatment for Influenza A. J. Virol. 2020, 95, e01027-20. [Google Scholar] [CrossRef]
- Adamson, A.L.; Chohan, K.; Swenson, J.; LaJeunesse, D. A Drosophila Model for Genetic Analysis of Influenza Viral/Host Interactions. Genetics 2011, 189, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Lam, V.K.; Tokusumi, T.; Cerabona, D.; Schulz, R.A. Specific cell ablation in Drosophila using the toxic viral protein M2(H37A). Fly 2010, 4, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Soler, Y.; Perry, M.; Reynolds, J.L.; El-Hage, N. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in the Nervous System: Implications of COVID-19 in Neurodegeneration. Front. Neurol. 2020, 11, 583459. [Google Scholar] [CrossRef] [PubMed]
- van de Leemput, J.; Han, Z. Drosophila, a powerful model to study virus-host interactions and pathogenicity in the fight against SARS-CoV-2. Cell Biosci. 2021, 11, 110. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Lee, J.-G.; van de Leemput, J.; Lee, H.; Han, Z. Functional analysis of SARS-CoV-2 proteins in Drosophila identifies Orf6-induced pathogenic effects with Selinexor as an effective treatment. Cell Biosci. 2021, 11, 59. [Google Scholar] [CrossRef]
- Lee, J.-G.; Huang, W.; Lee, H.; van de Leemput, J.; Kane, M.A.; Han, Z. Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell Biosci. 2021, 11, 58. [Google Scholar] [CrossRef]
- Syed, Y.Y. Selinexor: First Global Approval. Drugs 2019, 79, 1485–1494. [Google Scholar] [CrossRef]
- Bush, K.M.; Barber, K.R.; Martinez, J.A.; Tang, S.-J.; Wairkar, Y.P. Drosophila model of anti-retroviral therapy induced peripheral neuropathy and nociceptive hypersensitivity. Biol. Open 2021, 10, bio054635. [Google Scholar] [CrossRef]
- Polydefkis, M.; Yiannoutsos, C.; Cohen, B.; Hollander, H.; Schifitto, G.; Clifford, D.; Simpson, D.; Katzenstein, D.; Shriver, S.; Hauer, P.; et al. Reduced intraepidermal nerve fiber density in HIV-associated sensory neuropathy. Neurology 2002, 58, 115–119. [Google Scholar] [CrossRef]
- Kruger, L.; Denton, T.T. A standardized method for incorporation of drugs into food for use with Drosophila melanogaster. Anal. Biochem. 2020, 599, 113740. [Google Scholar] [CrossRef]
- Coulom, H. Chronic Exposure to Rotenone Models Sporadic Parkinson’s Disease in Drosophila melanogaster. J. Neurosci. 2004, 24, 10993–10998. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Pallos, J.; Slepko, N.; Apostol, B.L.; Bodai, L.; Chang, L.-W.; Chiang, A.-S.; Thompson, L.M.; Marsh, J.L. Identification of combinatorial drug regimens for treatment of Huntington’s disease using Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 3777–3781. [Google Scholar] [CrossRef]
- Ayroles, J.F.; Carbone, M.A.; A Stone, E.; Jordan, K.W.; Lyman, R.F.; Magwire, M.M.; Rollmann, S.M.; Duncan, L.H.; Lawrence, F.; Anholt, R.R.H.; et al. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 2009, 41, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, L.F.; Schlosser, T.; Manning, S.A.; Parisot, J.P.; Street, I.P.; Richardson, H.E.; Humbert, P.O.; Brumby, A.M. An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Dis. Model. Mech. 2013, 6, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Cvetković, V.; Mitrović, T.; Stamenkovic, S.; Stojadinović, B.; Jovanović, B.; Đorđević, M.; Radulović, N. Toxicity of commonly used solvent dimethyl sulfoxide against Drosophila melanogaster larvae: Determination of LC50, LOEC and NOEC values. In Proceedings of the 11th Symposium on the Flora of Southeastern Serbia and Neighbouring Regions, Vlasina, Serbia, 13–16 June 2013. [Google Scholar]
- Bass, T.M.; Grandison, R.C.; Wong, R.; Martinez, P.; Partridge, L.; Piper, M.D.W. Optimization of Dietary Restriction Protocols in Drosophila. J. Gerontol. Ser. A 2007, 62, 1071–1081. [Google Scholar] [CrossRef]
- Piper, M.D.W.; Blanc, E.; Leitão-Gonçalves, R.; Yang, M.; He, X.; Linford, N.J.; Hoddinott, M.P.; Hopfen, C.; Soultoukis, G.A.; Niemeyer, C.; et al. A holidic medium for Drosophila melanogaster. Nat. Methods 2014, 11, 100–105. [Google Scholar] [CrossRef]
- Ormerod, K.G.; LePine, O.K.; Abbineni, P.S.; Bridgeman, J.M.; Coorssen, J.R.; Mercier, A.J.; Tattersall, G.J. Drosophila development, physiology, behavior, and lifespan are influenced by altered dietary composition. Fly 2017, 11, 153–170. [Google Scholar] [CrossRef]
- Sykes, C. Time- and Temperature-Controlled Transport: Supply Chain Challenges and Solutions. Pharm. Ther. 2018, 43, 154–170. [Google Scholar]
- Millet-Boureima, M.C.; Selber-Hnatiw, M.S.; Gamberi, C. Drug discovery and chemical probing in Drosophila. Genome 2021, 64, 147–159. [Google Scholar] [CrossRef]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef]
- Nair, A.B.; Jacob, S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Bergman, C.M.; Quesneville, H.; Anxolabéhère, D.; Ashburner, M. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006, 7, R112. [Google Scholar] [CrossRef] [PubMed]
- Krug, L.; Chatterjee, N.; Borges-Monroy, R.; Hearn, S.; Liao, W.-W.; Morrill, K.; Prazak, L.; Rozhkov, N.; Theodorou, D.; Hammell, M.; et al. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 2017, 13, e1006635. [Google Scholar] [CrossRef]
- Hemmer, L.W.; Dias, G.B.; Smith, B.; Van Vaerenberghe, K.; Howard, A.; Bergman, C.; Blumenstiel, J.P. Hybrid dysgenesis in Drosophila virilis results in clusters of mitotic recombination and loss-of-heterozygosity but leaves meiotic recombination unaltered. Mob. DNA 2020, 11, 10. [Google Scholar] [CrossRef]
- Roy, M.; Viginier, B.; Saint-Michel, É; Arnaud, F.; Ratinier, M.; Fablet, M. Viral infection impacts transposable element transcript amounts in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 12249–12257. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Viginier, B.; A Mayeux, C.; Ratinier, M.; Fablet, M. Infections by Transovarially Transmitted DMelSV in Drosophila Have No Impact on Ovarian Transposable Element Transcripts but Increase Their Amounts in the Soma. Genome Biol. Evol. 2021, 13, evab207. [Google Scholar] [CrossRef] [PubMed]
- Fablet, M.; Jacquet, A.; Rebollo, R.; Haudry, A.; Rey, C.; Salces-Ortiz, J.; Bajad, P.; Burlet, N.; Jantsch, M.F.; Guerreiro, M.P.G.; et al. Dynamic Interactions Between the Genome and an Endogenous Retrovirus: Tirant in Drosophila simulans Wild-Type Strains. G3 2019, 9, 855–865. [Google Scholar] [CrossRef]
- Kim, A.; Terzian, C.; Santamaria, P.; Pélisson, A.; Purd’Homme, N.; Bucheton, A. Retroviruses in invertebrates: The gypsyretrotransposon is apparently an infectious retrovirus of Drosophilamelanogaster. Proc. Natl. Acad. Sci. USA 1994, 91, 1285–1289. [Google Scholar] [CrossRef]
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011, 69, 141–151. [Google Scholar] [CrossRef]
- Li, W.; Lee, M.-H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef]
- Phan, K.; He, Y.; Fu, Y.; Dzamko, N.; Bhatia, S.; Gold, J.; Rowe, D.; Ke, Y.D.; Ittner, L.M.; Hodges, J.R.; et al. Pathological manifestation of human endogenous retrovirus K in frontotemporal dementia. Commun. Med. 2021, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Muotri, A.R.; Chu, V.T.; Marchetto, M.C.N.; Deng, W.; Moran, J.V.; Gage, F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005, 435, 903–910. [Google Scholar] [CrossRef]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Treiber, C.D.; Waddell, S. Transposon expression in the Drosophila brain is driven by neighboring genes and diversifies the neural transcriptome. Genome Res. 2020, 30, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- E Jönsson, M.; Garza, R.; Sharma, Y.; Petri, R.; Södersten, E.; Johansson, J.G.; A Johansson, P.; Atacho, D.A.; Pircs, K.; Madsen, S.; et al. Activation of endogenous retroviruses during brain development causes an inflammatory response. EMBO J. 2021, 40, e106423. [Google Scholar] [CrossRef]
- Brattås, P.L.; Jönsson, M.E.; Fasching, L.; Wahlestedt, J.N.; Shahsavani, M.; Falk, R.; Falk, A.; Jern, P.; Parmar, M.; Jakobsson, J. TRIM28 Controls a Gene Regulatory Network Based on Endogenous Retroviruses in Human Neural Progenitor Cells. Cell Rep. 2017, 18, 1–11. [Google Scholar] [CrossRef]
- Li, W.; Prazak, L.; Chatterjee, N.; Grüninger, S.; Krug, L.; Theodorou, D.; Dubnau, J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013, 16, 529–531. [Google Scholar] [CrossRef]
- Yang, N.; Srivastav, S.P.; Rahman, R.; Ma, Q.; Dayama, G.; Li, S.; Chinen, M.; Lei, E.P.; Rosbash, M.; Lau, N.C. Transposable element landscapes in aging Drosophila. PLoS Genet. 2022, 18, e1010024. [Google Scholar] [CrossRef]
- Fabian, D.K.; Dönertaş, H.M.; Fuentealba, M.; Partridge, L.; Thornton, J.M. Transposable Element Landscape in Drosophila Populations Selected for Longevity. Genome Biol. Evol. 2021, 13, evab031. [Google Scholar] [CrossRef]
- Liguori, F.; Amadio, S.; Volonté, C. Fly for ALS: Drosophila modeling on the route to amyotrophic lateral sclerosis modifiers. Experientia 2021, 78, 6143–6160. [Google Scholar] [CrossRef] [PubMed]
- Tam, O.H.; Rozhkov, N.V.; Shaw, R.; Kim, D.; Hubbard, I.; Fennessey, S.; Propp, N.; Fagegaltier, D.; Harris, B.T.; Ostrow, L.W.; et al. Postmortem Cortex Samples Identify Distinct Molecular Subtypes of ALS: Retrotransposon Activation, Oxidative Stress, and Activated Glia. Cell Rep. 2019, 29, 1164–1177.e5. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Klima, R.; Feiguin, F. TDP-43 prevents retrotransposon activation in the Drosophila motor system through regulation of Dicer-2 activity. BMC Biol. 2020, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-H.; Dubnau, J. The Gypsy Endogenous Retrovirus Drives Non-Cell-Autonomous Propagation in a Drosophila TDP-43 Model of Neurodegeneration. Curr. Biol. 2019, 29, 3135–3152.e4. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Keegan, R.M.; Prazak, L.; Dubnau, J. Cellular labeling of endogenous retrovirus replication (CLEVR) reveals de novo insertions of the gypsy retrotransposable element in cell culture and in both neurons and glial cells of aging fruit flies. PLoS Biol. 2019, 17, e3000278. [Google Scholar] [CrossRef] [PubMed]
- Azpurua, J.; El-Karim, E.G.; Tranquille, M.; Dubnau, J. A behavioral screen for mediators of age-dependent TDP-43 neurodegeneration identifies SF2/SRSF1 among a group of potent suppressors in both neurons and glia. PLoS Genet. 2021, 17, e1009882. [Google Scholar] [CrossRef]
- Paz, S.; Krainer, A.R.; Caputi, M. HIV-1 transcription is regulated by splicing factor SRSF1. Nucleic Acids Res. 2014, 42, 13812–13823. [Google Scholar] [CrossRef]
- Hautbergue, G.M.; Castelli, L.M.; Ferraiuolo, L.; Sanchez-Martinez, A.; Cooper-Knock, J.; Higginbottom, A.; Lin, Y.-H.; Bauer, C.S.; Dodd, J.E.; Myszczynska, M.; et al. SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat. Commun. 2017, 8, 16063. [Google Scholar] [CrossRef]
- Dias-Santagata, D.; Fulga, T.; Duttaroy, A.; Feany, M.B. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J. Clin. Investig. 2007, 117, 236–245. [Google Scholar] [CrossRef]
- Wittmann, C.W.; Wszolek, M.F.; Shulman, J.M.; Salvaterra, P.M.; Lewis, J.; Hutton, M.; Feany, M.B. Tauopathy in Drosophila: Neurodegeneration Without Neurofibrillary Tangles. Science 2001, 293, 711–714. [Google Scholar] [CrossRef]
- Khurana, V.; Lu, Y.; Steinhilb, M.L.; Oldham, S.; Shulman, J.M.; Feany, M. TOR-Mediated Cell-Cycle Activation Causes Neurodegeneration in a Drosophila Tauopathy Model. Curr. Biol. 2006, 16, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef]
- Guo, C.; Jeong, H.-H.; Hsieh, Y.-C.; Klein, H.-U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef] [PubMed]
- Casale, A.M.; Liguori, F.; Ansaloni, F.; Cappucci, U.; Finaurini, S.; Spirito, G.; Persichetti, F.; Sanges, R.; Gustincich, S.; Piacentini, L. Transposable element activation promotes neurodegeneration in a Drosophila model of Huntington’s disease. iScience 2022, 25, 103702. [Google Scholar] [CrossRef]
- Floreani, L.; Ansaloni, F.; Mangoni, D.; Agostoni, E.; Sanges, R.; Persichetti, F.; Gustincich, S. Analysis of LINE1 Retrotransposons in Huntington’s Disease. Front. Cell. Neurosci. 2021, 15, 743797. [Google Scholar] [CrossRef] [PubMed]
- Nitta, Y.; Sugie, A. Studies of neurodegenerative diseases using Drosophila and the development of novel approaches for their analysis. Fly 2022, 16, 275–298. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Lee, I.-S.; Jantrapirom, S.; Suda, K.; Yoshida, H. Drosophila models to study causative genes for human rare intractable neurological diseases. Exp. Cell Res. 2021, 403, 112584. [Google Scholar] [CrossRef]
- Duhart, J.C.; Mosca, T.J. Genetic regulation of central synapse formation and organization in Drosophila melanogaster. Genetics 2022, 221. [Google Scholar] [CrossRef]
| Fly Pathway | Drosophila Gene | Human Pathway | Human Gene; Protein Name | FlyBase a | Literature b | Reference |
|---|---|---|---|---|---|---|
| Toll | Cactus | TLR | NFKBIA; IκBα | O | H | [17,18] |
| Dif | NFKB1; NF-κB p50/RELA; NF-κB p65 | H | [12,18,19] | |||
| Dif | RELB | O | ||||
| Dorsal | NFKB1; NF-κB p50 | H | [20] | |||
| Dorsal | RELA; NF-κB p65 | O | ||||
| MyD88 | MYD88 | O | O | [21,22] | ||
| Pelle | IRAK1 | O | [21,23] | |||
| Pelle | IRAK4 | O | ||||
| Pellino | PELI1 & PELI2 | O | H | [24,25] | ||
| Tube | IRAK4 | O | [21,23] | |||
| Tube | MAL | H | [22] | |||
| IMD | Bendless | TNFR | UBE2N; UBC13 | O | H | [26] |
| Dredd | CASP8; Caspase-8 | H | [26,27,28,29] | |||
| Dredd | CASP10 | O | ||||
| Fadd | FADD | O | O | [26,30] | ||
| Iap2 | BIRC3; cIAP2 | H | [29,31] | |||
| Iap2 | BIRC2; cIAP1 | O | ||||
| Imd | RIPK1; RIP1 | H | [27,32,33] | |||
| Ird5 | IKBKB; IKKβ | O | H | [34,35,36,37] | ||
| Ird5 | CHUK; IKKα | O | ||||
| Kenny; IKKγ | IKBKG; NEMO/IKKγ | O | H | [34,38] | ||
| Kenny; IKKγ | OPTN | O | ||||
| Relish | NFKBIA; IκBα/ NFKB1; NF-κB p50/RELA; NF-κB p65 | O | H | [39,40] | ||
| Tab2 | TAB2 & TAB3 | O | H | [11,31] | ||
| Tak1 | MAP3K7; TAK1 | O | H | [31,41] | ||
| UEV1a | UBE2V1; UEV1a | H | [26] | |||
| UEV1a | UBE2V2; UEV2 | O | ||||
| dSTING | cGAS/STING | STING1; STING | O | O | [42] | |
| Jak-STAT | Cg14225/Latran | Jak-STAT | IL6ST; GP130 | H | [43,44] | |
| Domeless | LIFR & CNTFR | H | [43,45] | |||
| Domeless | PTPRQ | O | ||||
| Hopscotch | JAK1 & JAK2 | O | H | [46] | ||
| Marelle | STAT5A/STAT5B; STAT5 & STAT6 | O | H | [47] | ||
| Socs36E | SOCS4 & SOCS5 | O | H | [48,49] | ||
| STAT92E | STAT3 & STAT5A/STAT5B; STAT5 | O | H | [47,50] | ||
| Su(Var)2-10/dPIAS | PIAS1 | O | H | [51,52] | ||
| RNAi | Ago-1, Ago-2, & Ago-3 | RNAi | AGO1, AGO2, & AGO3 | H | [53,54] | |
| Ago-1 | AGO2 | O | ||||
| Ago-3 | PIWIL2 | O | ||||
| Armitage | MOV10L1 | O | H | [55] | ||
| Aubergine | PIWIL1; Hiwi | O | H | [53] | ||
| Dicer | DICER1 | O | H | [56] | ||
| Fmr1 | FMR1 | O | H | [57,58] | ||
| Piwi | PIWIL1; Hiwi | H | [59] | |||
| Piwi | PIWIL3; Hiwi3 | O | ||||
| R2d2 | TARBP2 | H | [60] | |||
| Rm62/Dmp68 | DDX5; P68 | H | [58] | |||
| Vasa intronic gene | SERBP1; PAI-RBP1 | O | H | [61,62] | ||
| Restriction factors | Dcp2 | Restriction factor | DCP2 | O | H | [63] |
| Ge-1 | EDC4; RCD-8 | O | H | [64] | ||
| FoxK | FOXK1 & FOXK2 | O | O | [65] | ||
| Nazo | C19orf12 | O | [66] | |||
| Nup98 | NUP98 | O | H | [65,67,68,69] | ||
| Ref(2)p | SQSTM1; P62 | O | H | [70,71] | ||
| TREX | TREX1 | O | H | [72,73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benoit, I.; Di Curzio, D.; Civetta, A.; Douville, R.N. Drosophila as a Model for Human Viral Neuroinfections. Cells 2022, 11, 2685. https://doi.org/10.3390/cells11172685
Benoit I, Di Curzio D, Civetta A, Douville RN. Drosophila as a Model for Human Viral Neuroinfections. Cells. 2022; 11(17):2685. https://doi.org/10.3390/cells11172685
Chicago/Turabian StyleBenoit, Ilena, Domenico Di Curzio, Alberto Civetta, and Renée N. Douville. 2022. "Drosophila as a Model for Human Viral Neuroinfections" Cells 11, no. 17: 2685. https://doi.org/10.3390/cells11172685
APA StyleBenoit, I., Di Curzio, D., Civetta, A., & Douville, R. N. (2022). Drosophila as a Model for Human Viral Neuroinfections. Cells, 11(17), 2685. https://doi.org/10.3390/cells11172685






