USP29 Deubiquitinates SETD8 and Regulates DNA Damage-Induced H4K20 Monomethylation and 53BP1 Focus Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. siRNA Oligos, Plasmids, and Transfection
| GFP | CAAGCUGACCCUGAAGUUCdTdT |
| Luc | CGUACGCGGAAUACUUCGAdTdT |
| SETD8 | GUACGGAGCGCCAUGAAGUdTdT |
| USP29#1 | CCCAUCAAGUUUAGAGGAUdTdT |
| USP29#2 | GGAAUAUGCUGAAGGAAAUdTdT |
| TRABID | AGAGGCUUCUUCAAUAAUAdTdT |
| USP18 | CCAGGGAGUUAUCAAGCAAdTdT |
| USP2 | CAGAUUGUGGUUACUGUUCUAdTdT |
| USP30 | CCAGAGUCCUGUUCGAUUUdTdT |
| USP33 | UCUCGACAGUGGCUUAAUUAAdTdT |
| USP51 | CCAUUUAGCUGUAGACCUUdTdT |
2.3. Antibodies
2.4. Immunoprecipitations, Pulldowns, and Western Blot
2.5. Immunofluorescence
2.6. Flow Cytometry
2.7. Clonogenic Survival
2.8. Statistical Analysis and Reproducibility
3. Results
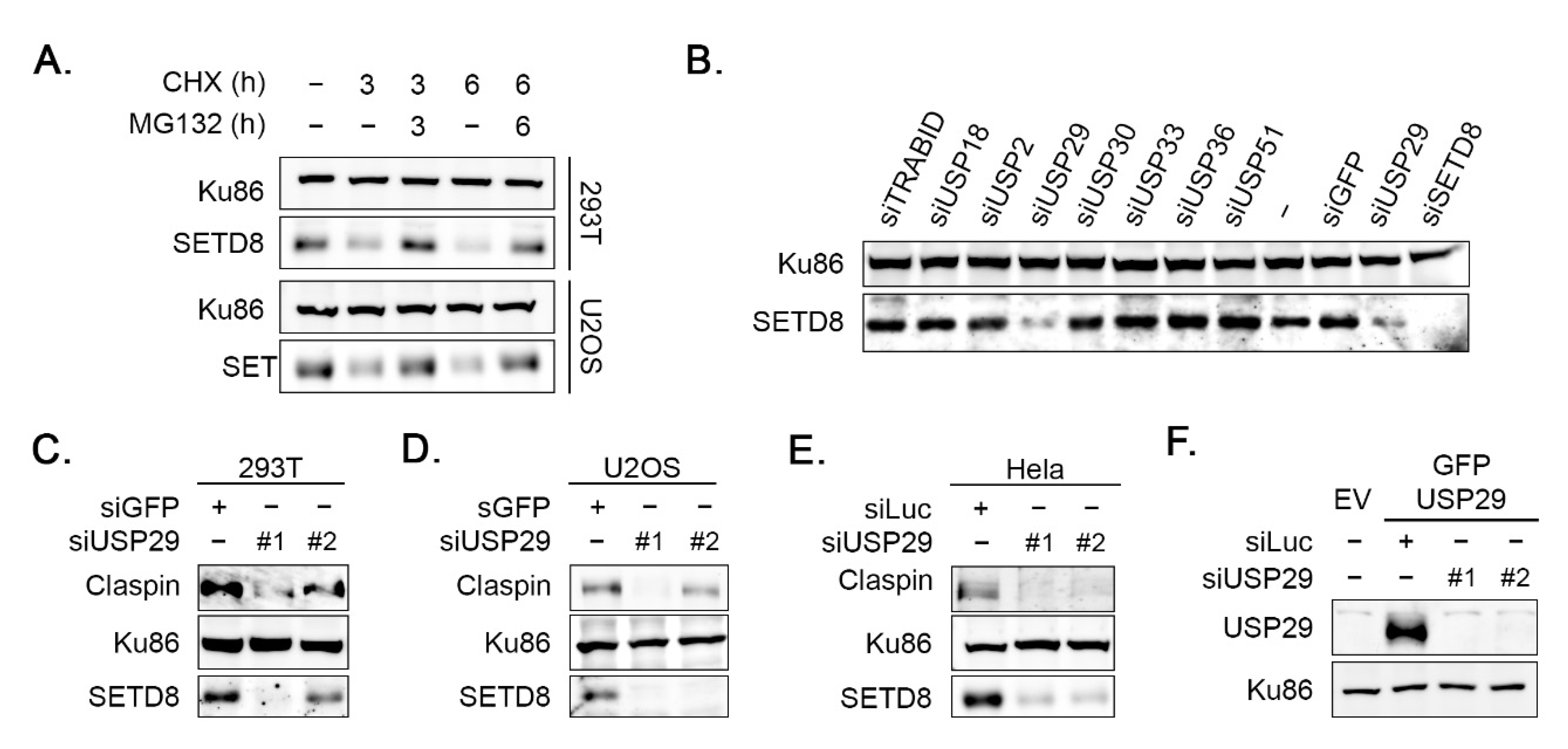
3.1. USP29 Controls SETD8 Protein Levels
3.2. USP29 Regulates SETD8 Levels in a Cell-Cycle-Independent Manner
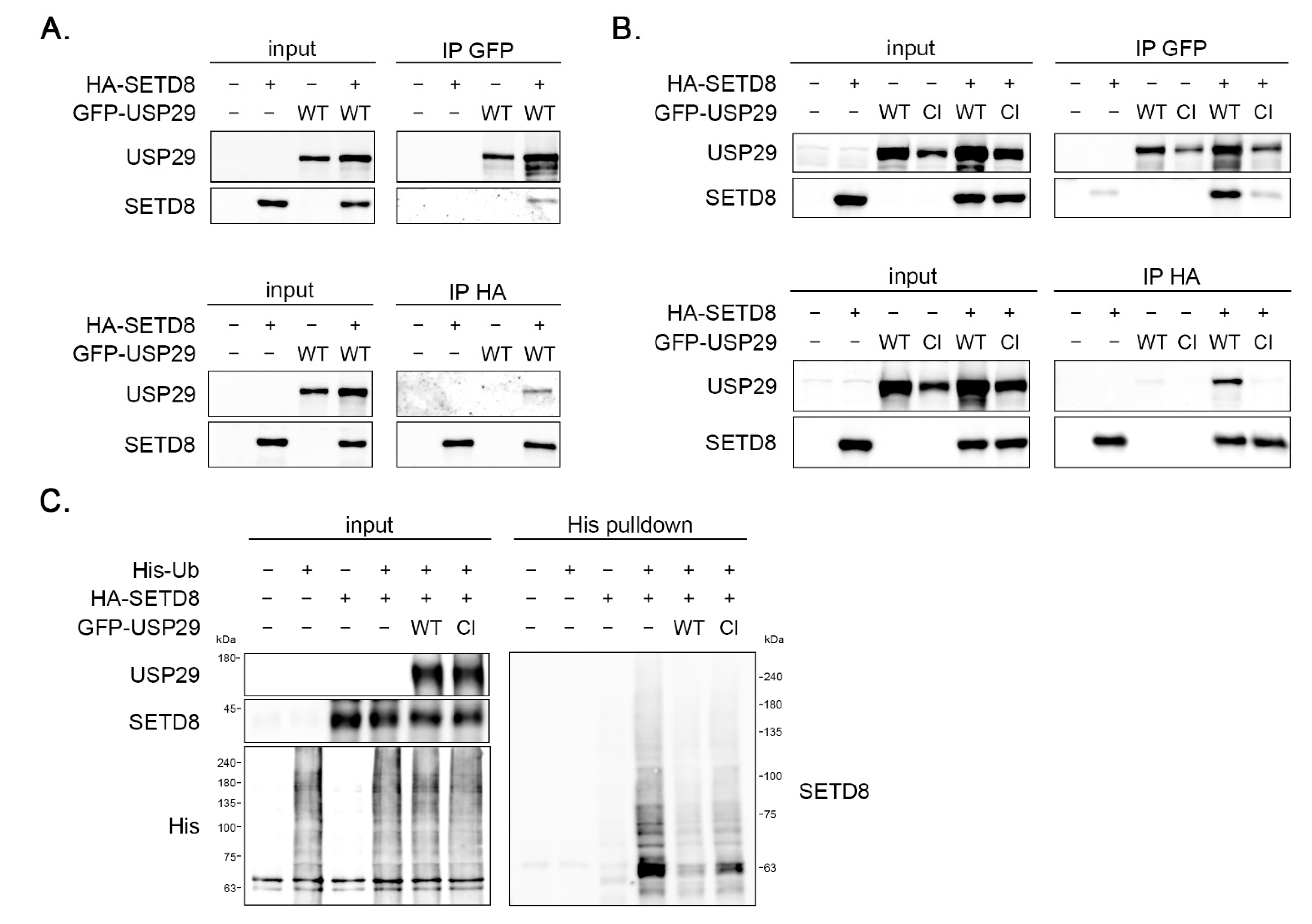
3.3. SETD8 and USP29 Interact In Vivo
3.4. USP29 Deubiquitinates SETD8 In Vivo
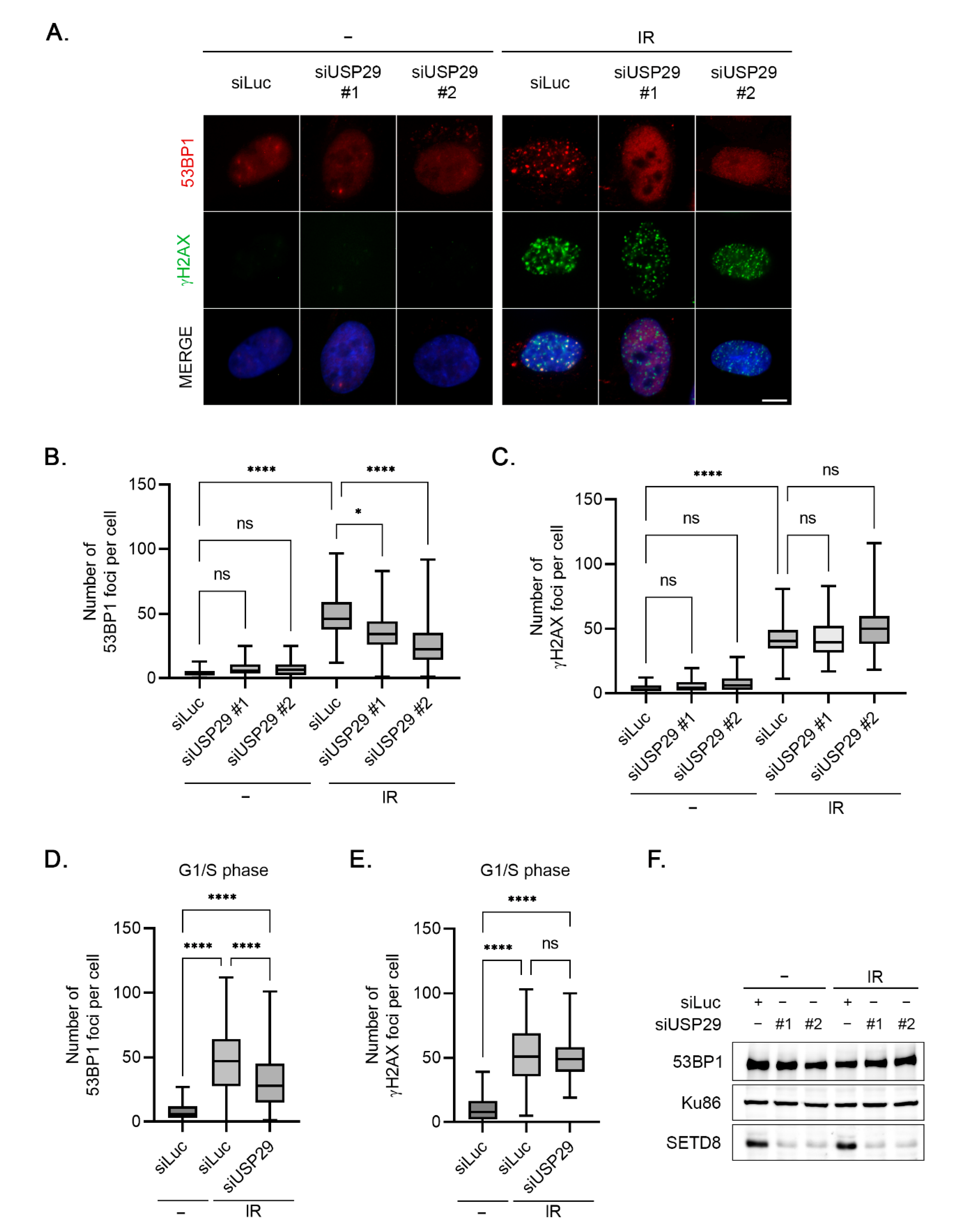
3.5. Accumulation of 53BP1 into Nuclear Foci upon IR Is Regulated by USP29
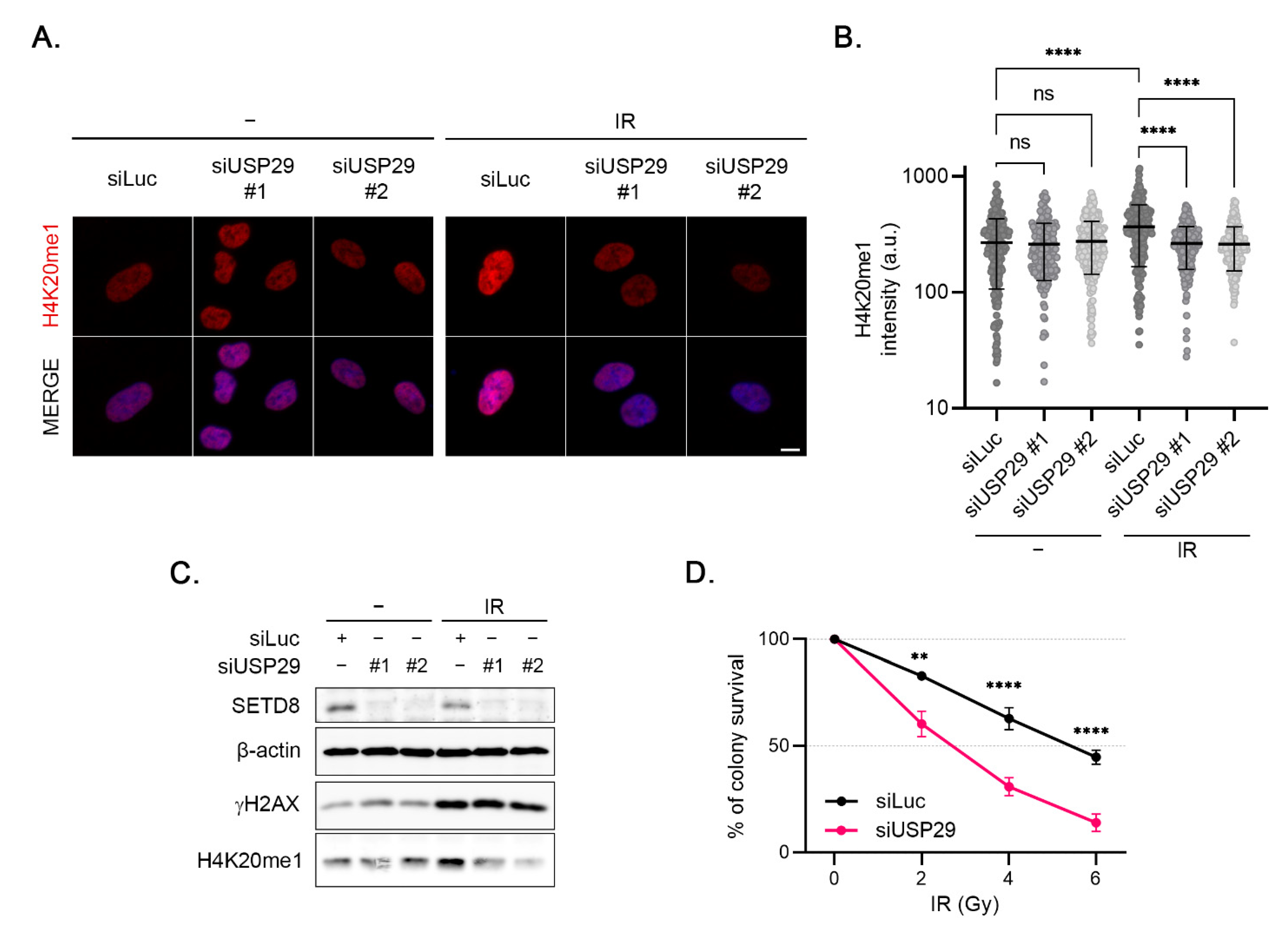
3.6. USP29 Controls IR-Induced H4K20 Monomethylation and Cell Survival
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, K.; Rice, J.C.; Sarma, K.; Erdjument-Bromage, H.; Werner, J.; Wang, Y.; Chuikov, S.; Valenzuela, P.; Tempst, P.; Steward, R.; et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 2002, 9, 1201–1213. [Google Scholar] [CrossRef]
- Beck, D.B.; Oda, H.; Shen, S.S.; Reinberg, D. PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012, 26, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Milite, C.; Feoli, A.; Viviano, M.; Rescigno, D.; Cianciulli, A.; Balzano, A.L.; Mai, A.; Castellano, S.; Sbardella, G. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin. Epigenet. 2016, 8, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, L.; Sun, J.; Hu, X.; Kalvakolanu, D.V.; Ren, H.; Guo, B. Roles for the methyltransferase SETD8 in DNA damage repair. Clin. Epigenet. 2022, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.G.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Zhao, Y.; Brickner, J.R.; Majid, M.C.; Mosammaparast, N. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends Cell Biol. 2014, 24, 426–434. [Google Scholar] [CrossRef]
- Oda, H.; Hübner, M.R.; Beck, D.B.; Vermeulen, M.; Hurwitz, J.; Spector, D.L.; Reinberg, D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 2010, 40, 364–376. [Google Scholar] [CrossRef]
- Pei, H.; Zhang, L.; Luo, K.; Qin, Y.; Chesi, M.; Fei, F.; Bergsagel, P.L.; Wang, L.; You, Z.; Lou, Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011, 469, 124–128. [Google Scholar] [CrossRef]
- Dulev, S.; Tkach, J.; Lin, S.; Batada, N.N. SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Rep. 2014, 15, 1163–1174. [Google Scholar] [CrossRef]
- Tuzon, C.T.; Spektor, T.; Kong, X.; Congdon, L.M.; Wu, S.; Schotta, G.; Yokomori, K.; Rice, J.C. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep. 2014, 8, 430–438. [Google Scholar] [CrossRef]
- Huen, M.S.Y.; Chen, J. The DNA damage response pathways: At the crossroad of protein modifications. Cell Res. 2008, 18, 8–16. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, X.; Zhong, J.; Inuzuka, H.; Wan, L.; Li, X.; Wang, L.; Ye, X.; Sun, L.; Gao, D.; et al. SCF(β-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat. Commun. 2015, 6, 10185. [Google Scholar] [CrossRef]
- Yin, Y.; Yu, V.C.; Zhu, G.; Chang, D.C. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 2008, 7, 1423–1432. [Google Scholar] [CrossRef]
- Wu, S.; Wang, W.; Kong, X.; Congdon, L.M.; Yokomori, K.; Kirschner, M.W.; Rice, J.C. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010, 24, 2531–2542. [Google Scholar] [CrossRef]
- Abbas, T.; Shibata, E.; Park, J.; Jha, S.; Karnani, N.; Dutta, A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 2010, 40, 9–21. [Google Scholar] [CrossRef]
- Centore, R.C.; Havens, C.G.; Manning, A.L.; Li, J.-M.; Flynn, R.L.; Tse, A.; Jin, J.; Dyson, N.J.; Walter, J.C.; Zou, L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 2010, 40, 22–33. [Google Scholar] [CrossRef]
- Pérez-Castro, A.J.; Freire, R. Rad9B responds to nucleolar stress through ATR and JNK signalling, and delays the G1-S transition. J. Cell Sci. 2012, 125, 1152–1164. [Google Scholar] [CrossRef]
- Martín, Y.; Cabrera, E.; Amoedo, H.; Hernández-Pérez, S.; Domínguez-Kelly, R.; Freire, R. USP29 controls the stability of checkpoint adaptor Claspin by deubiquitination. Oncogene 2015, 34, 1058–1063. [Google Scholar] [CrossRef]
- Semple, J.I.; Smits, V.A.J.; Fernaud, J.-R.; Mamely, I.; Freire, R. Cleavage and degradation of Claspin during apoptosis by caspases and the proteasome. Cell Death Differ. 2007, 14, 1433–1442. [Google Scholar] [CrossRef][Green Version]
- Refolio, E.; Cavero, S.; Marcon, E.; Freire, R.; San-Segundo, P.A. The Ddc2/ATRIP checkpoint protein monitors meiotic recombination intermediates. J. Cell Sci. 2011, 124, 2488–2500. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Wu, S.; Rice, J.C. A new regulator of the cell cycle: The PR-Set7 histone methyltransferase. Cell Cycle 2011, 10, 68–72. [Google Scholar] [CrossRef]
- Sanders, S.L.; Portoso, M.; Mata, J.; Bähler, J.; Allshire, R.C.; Kouzarides, T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004, 119, 603–614. [Google Scholar] [CrossRef]
- Botuyan, M.V.; Lee, J.; Ward, I.M.; Kim, J.-E.; Thompson, J.R.; Chen, J.; Mer, G. Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair. Cell 2006, 127, 1361–1373. [Google Scholar] [CrossRef]
- Jørgensen, S.; Schotta, G.; Sørensen, C.S. Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013, 41, 2797–2806. [Google Scholar] [CrossRef]
- Liu, J.; Chung, H.-J.; Vogt, M.; Jin, Y.; Malide, D.; He, L.; Dundr, M.; Levens, D. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011, 30, 846–858. [Google Scholar] [CrossRef]
- Mosbech, A.; Lukas, C.; Bekker-Jensen, S.; Mailand, N. The Deubiquitylating Enzyme USP44 Counteracts the DNA Double-strand Break Response Mediated by the RNF8 and RNF168 Ubiquitin Ligases. J. Biol. Chem. 2013, 288, 16579–16587. [Google Scholar] [CrossRef]
- Vissers, J.H.; Nicassio, F.; van Lohuizen, M.; Di Fiore, P.; Citterio, E. The many faces of ubiquitinated histone H2A: Insights from the DUBs. Cell Div. 2008, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Fukuura, K.; Inoue, Y.; Miyajima, C.; Watanabe, S.; Tokugawa, M.; Morishita, D.; Ohoka, N.; Komada, M.; Hayashi, H. The ubiquitin-specific protease USP17 prevents cellular senescence by stabilizing the methyltransferase SET8 and transcriptionally repressing p21. J. Biol. Chem. 2019, 294, 16429–16439. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Z.; Xia, C.; Yan, K.; Fang, Z.; Fan, Y. SETD8 stabilized by USP17 epigenetically activates SREBP1 pathway to drive lipogenesis and oncogenesis of ccRCC. Cancer Lett. 2022, 527, 150–163. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Reyes, Y.; Paz-Cabrera, M.C.; Freire, R.; Smits, V.A.J. USP29 Deubiquitinates SETD8 and Regulates DNA Damage-Induced H4K20 Monomethylation and 53BP1 Focus Formation. Cells 2022, 11, 2492. https://doi.org/10.3390/cells11162492
Hernández-Reyes Y, Paz-Cabrera MC, Freire R, Smits VAJ. USP29 Deubiquitinates SETD8 and Regulates DNA Damage-Induced H4K20 Monomethylation and 53BP1 Focus Formation. Cells. 2022; 11(16):2492. https://doi.org/10.3390/cells11162492
Chicago/Turabian StyleHernández-Reyes, Yeray, María Cristina Paz-Cabrera, Raimundo Freire, and Veronique A. J. Smits. 2022. "USP29 Deubiquitinates SETD8 and Regulates DNA Damage-Induced H4K20 Monomethylation and 53BP1 Focus Formation" Cells 11, no. 16: 2492. https://doi.org/10.3390/cells11162492
APA StyleHernández-Reyes, Y., Paz-Cabrera, M. C., Freire, R., & Smits, V. A. J. (2022). USP29 Deubiquitinates SETD8 and Regulates DNA Damage-Induced H4K20 Monomethylation and 53BP1 Focus Formation. Cells, 11(16), 2492. https://doi.org/10.3390/cells11162492






