Hydrocephalus in Nfix−/− Mice Is Underpinned by Changes in Ependymal Cell Physiology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Statement of Animal Ethics
2.3. Nfix KO
2.4. NfixiFOXJ1-GFP
2.5. Tamoxifen Treatment
2.6. Perfusion and Immunofluorescence (IF) Histochemistry
2.7. Cell Culture
2.8. Lentiviral Treatment
2.9. Cell Culture Immunofluorescence (IF)
2.10. Immunofluorescence (IF) Imaging
2.10.1. Line Scanning Analysis
2.10.2. Fluorescence Intensity Analysis
2.11. Transmission Electron Microscopy (TEM)
2.12. RT2 qPCR Arrays
2.13. Hematoxylin
2.14. Light Microscopy
2.15. Statistical Analysis
3. Results
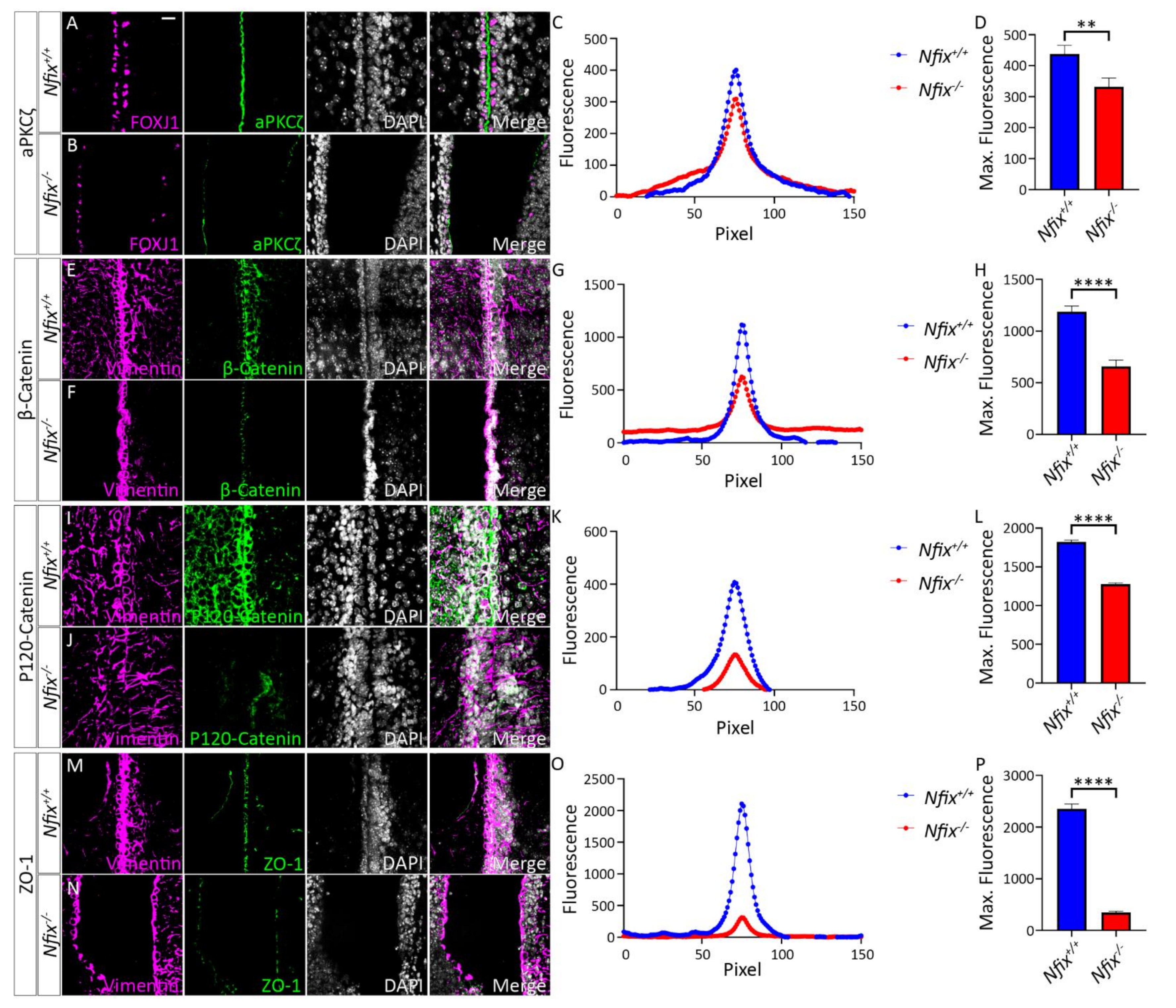
3.1. Nfix−/− Mice Demonstrate Disrupted Junctional Localisation of Ependymal Adhesion Proteins and Abnormal Ependymal Cell Morphology
3.1.1. Nfix−/− Mice Have Reduced Junctional Localisation of Key Adhesion Proteins
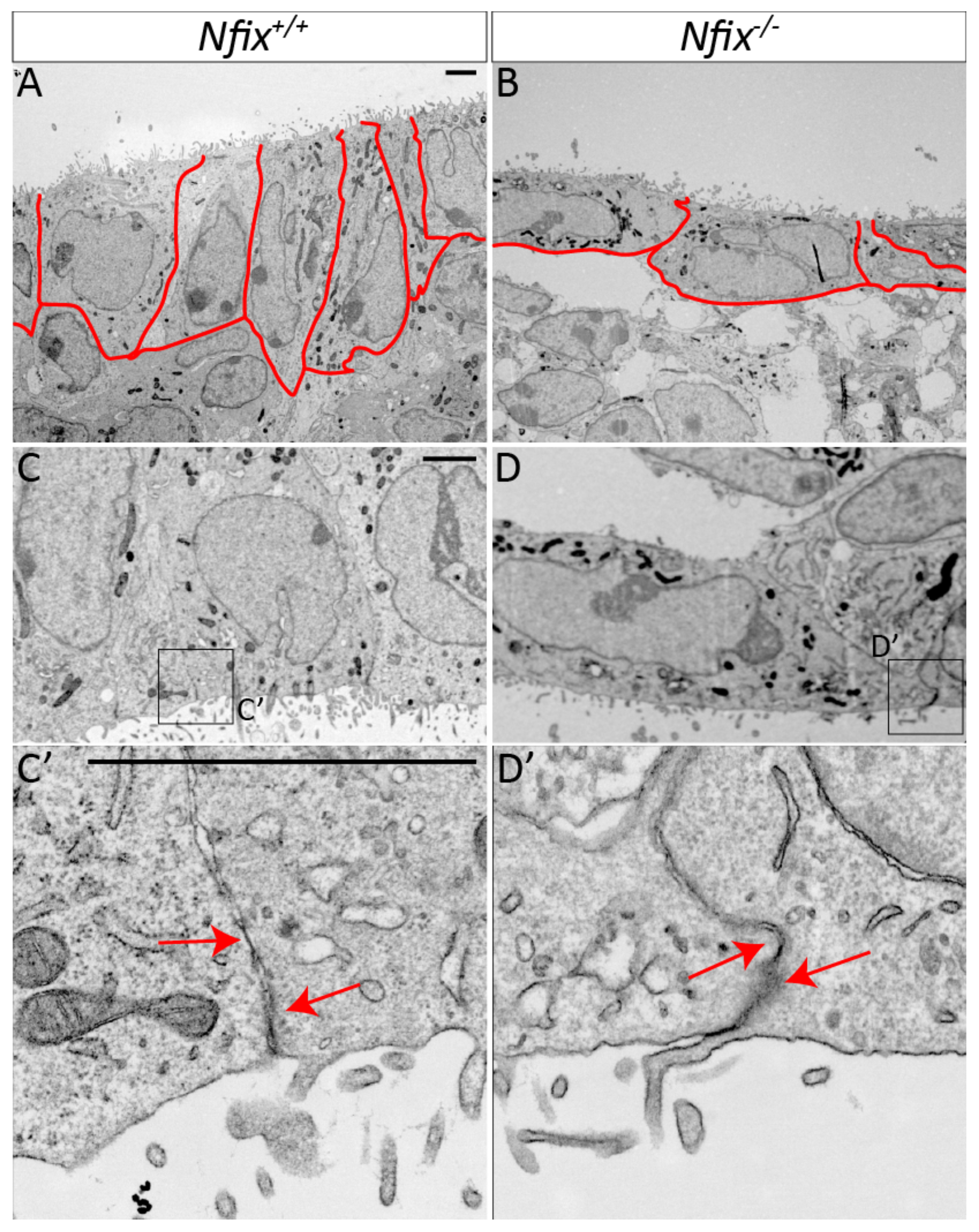
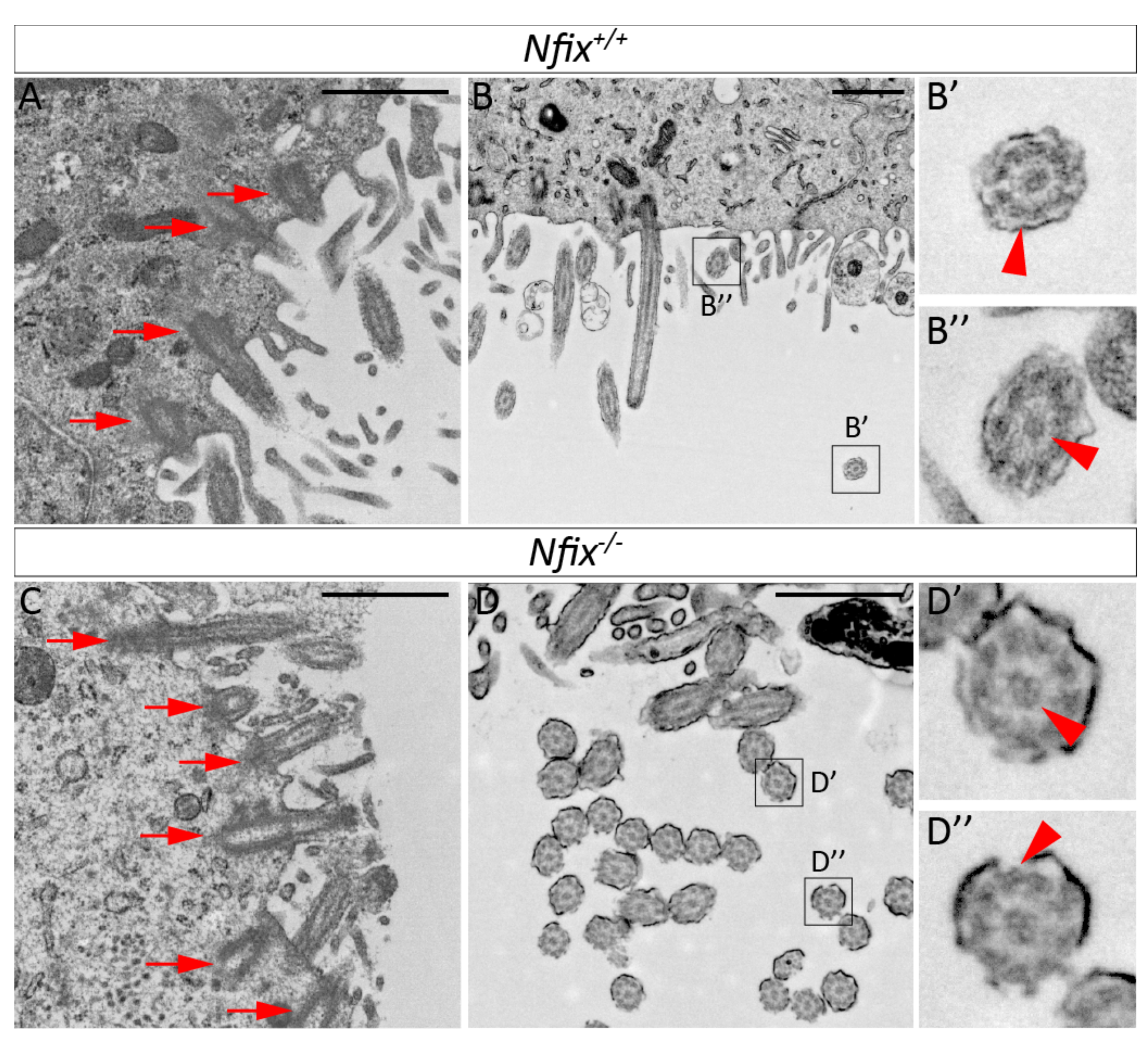
3.1.2. Nfix−/− Ependymal Cells Demonstrate Abnormal Morphology
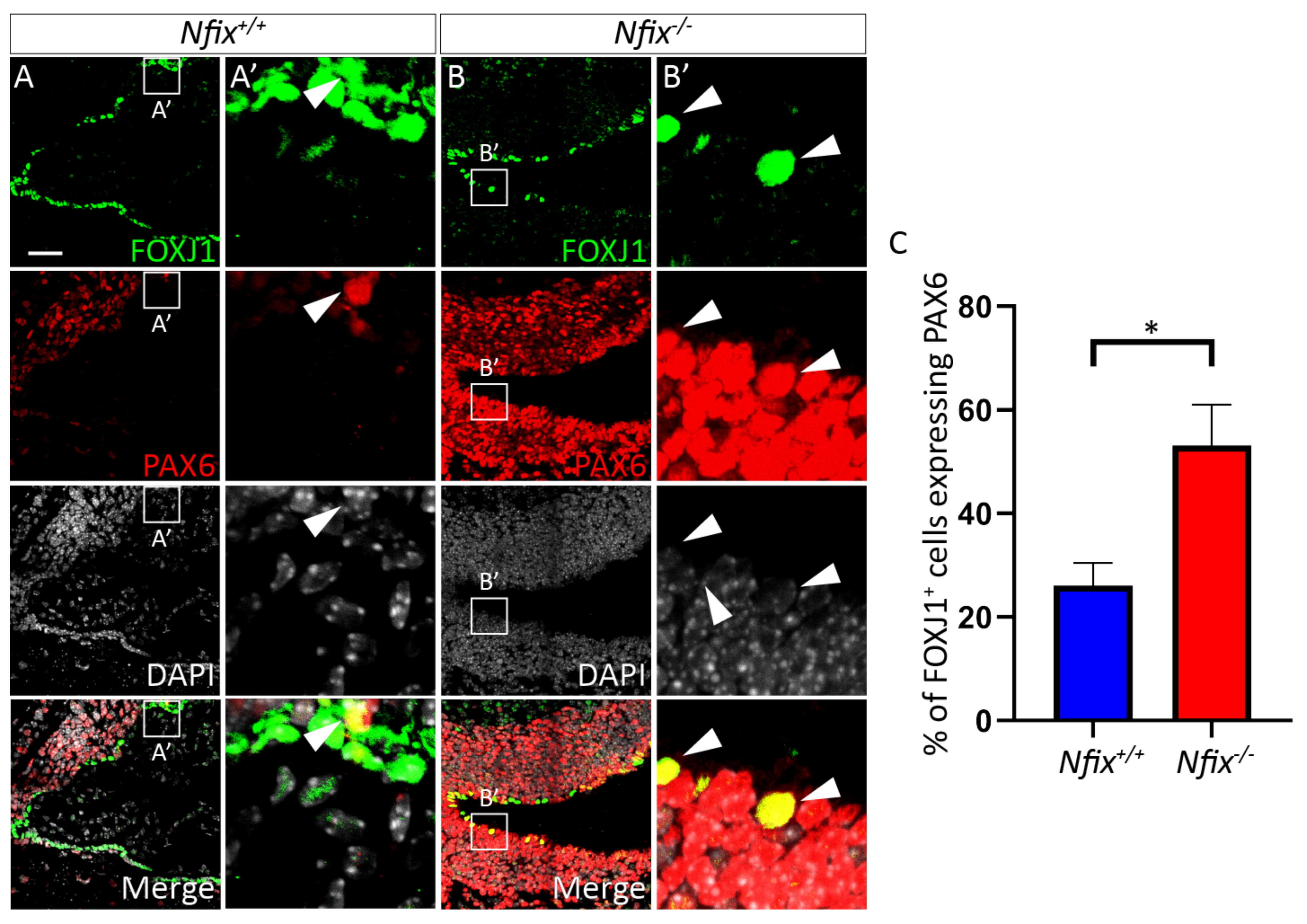
3.1.3. Nfix−/− Ependymal Cells Retain Expression of PAX6
3.2. Reduced Expression of NFIX in Epithelial Cells Is Associated with Abnormal Localisation of Key Adhesion Proteins
3.3. Profiling of MCF7 Cell mRNA Transcription Reveals Downregulation of Adhesion Genes in NFIX KD Cells
3.4. NFIX Is Essential for Maintenance of Terminal Ependymal Cell Identity and Function in Adult Mice
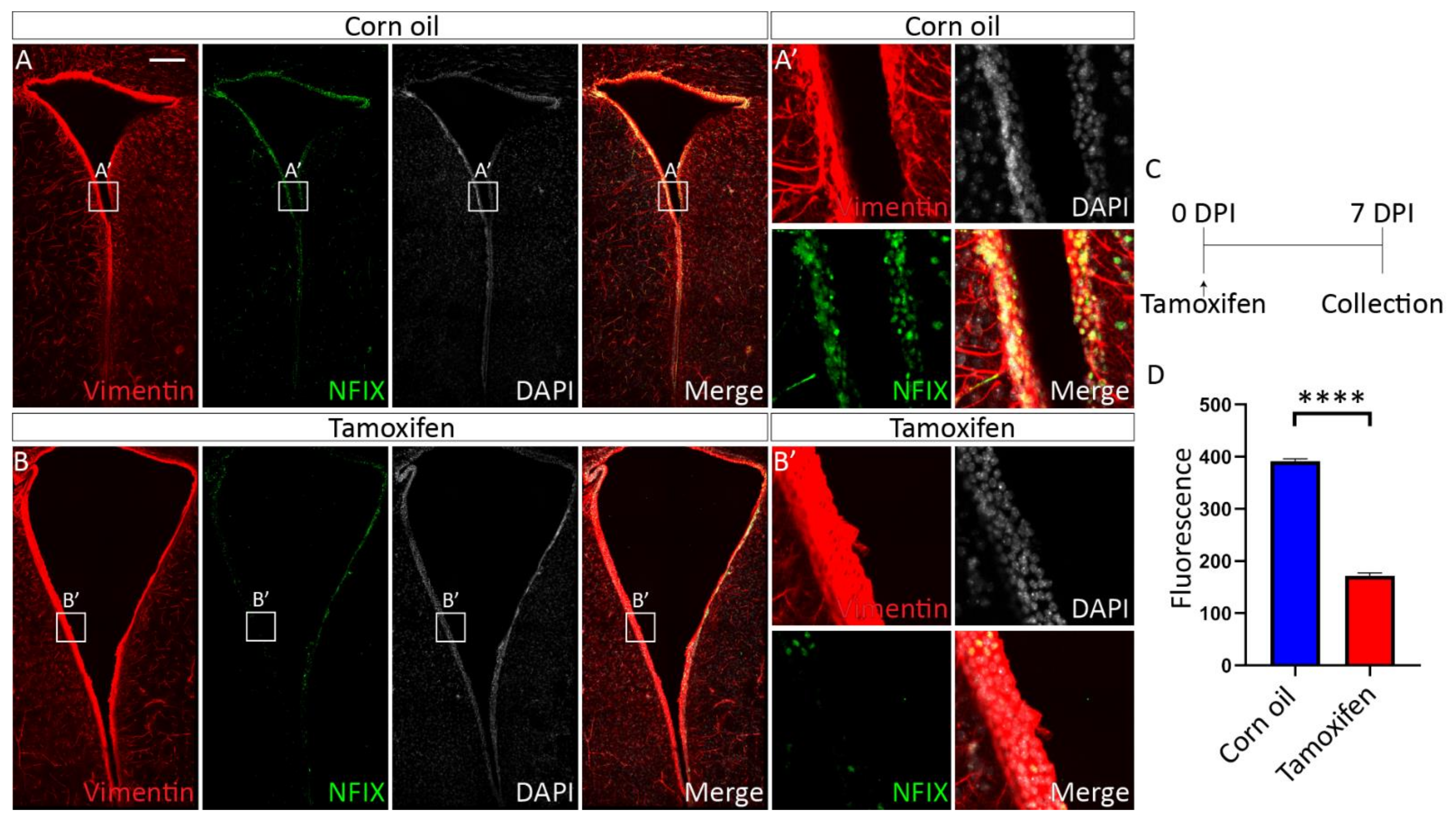
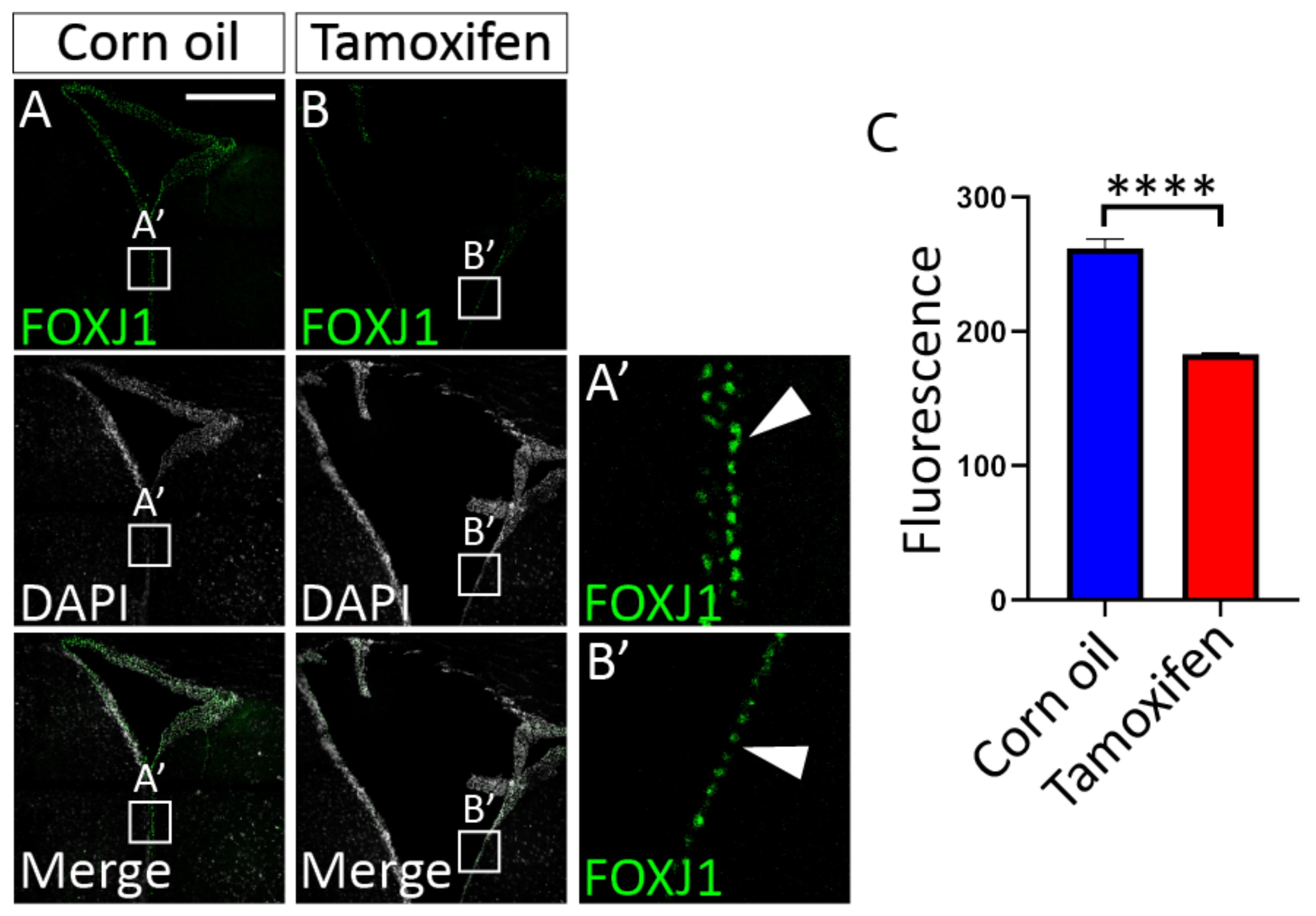
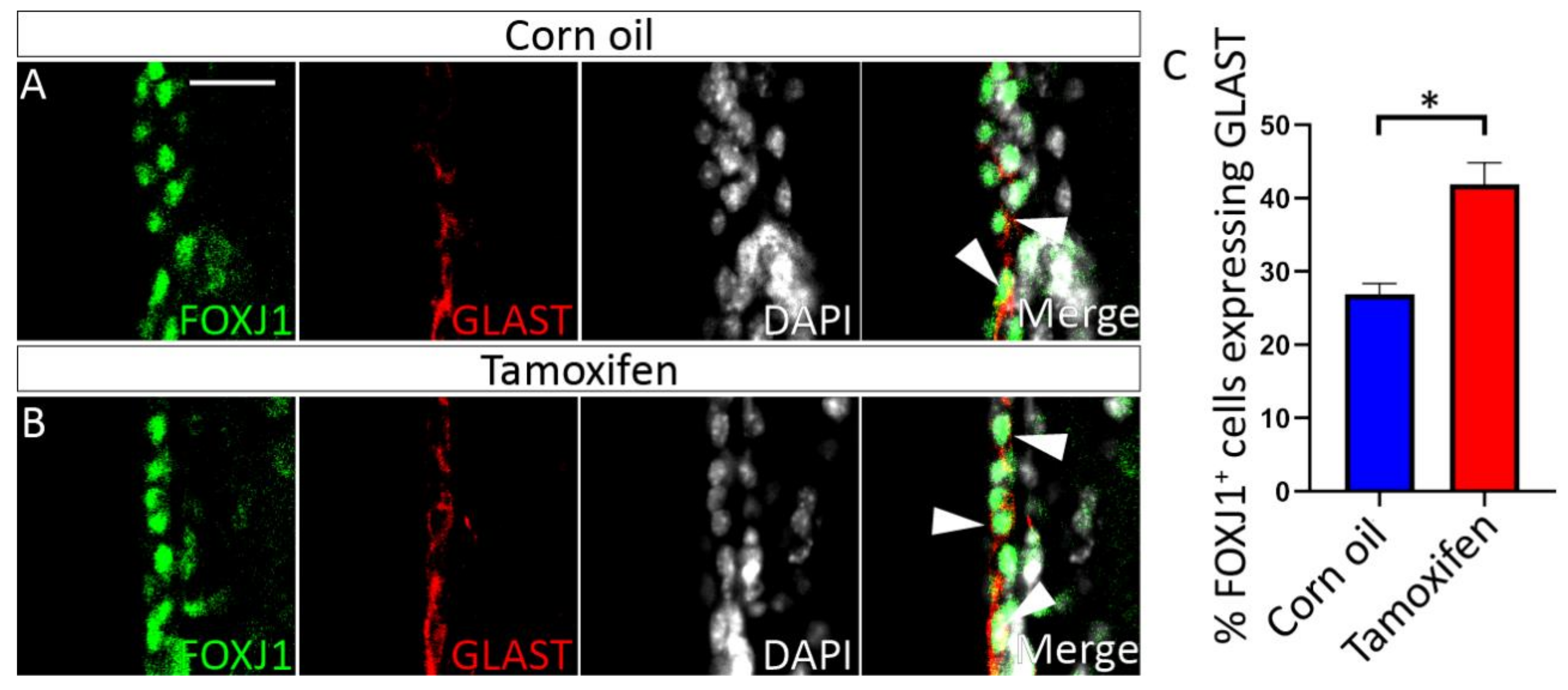
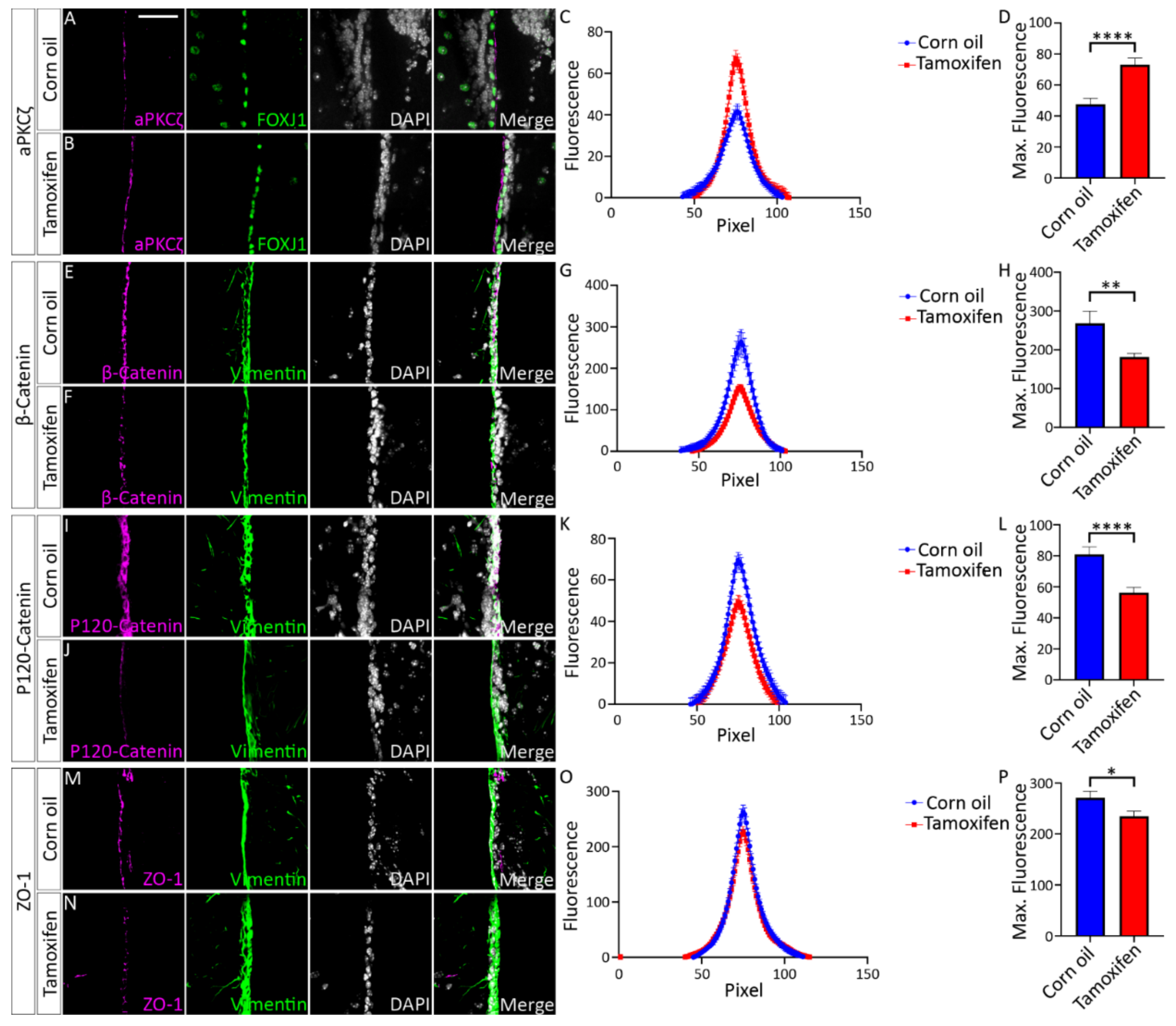
3.4.1. Tamoxifen Treatment of Adult NfixiFOXJ1−GFP Mice Results in Decreased NFIX Expression in Ependymal Cells
3.4.2. Loss of NFIX from Mature Ependymal Cells Leads to Ventricular Expansion
4. Discussion
4.1. NFIX Drives Acquisition and Maintenance of Ependymal Cell Identity Traits
4.2. Expression of NFIX Is Essential for Cell Adhesion in Ependymal Cells
4.3. Altered Cilia Structure in P5 Nfix−/− Mice May Also Contribute to Hydrocephalus
4.4. Mutations in NFIX Are Associated with Malan Syndrome in Humans
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Redmond, S.A.; Figueres-Oñate, M.; Obernier, K.; Nascimento, M.A.; Parraguez, J.I.; López-Mascaraque, L.; Fuentealba, L.C.; Alvarez-Buylla, A. Development of Ependymal and Postnatal Neural Stem Cells and Their Origin from a Common Embryonic Progenitor. Cell Rep. 2019, 27, 429–441.e423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spassky, N.; Merkle, F.; Flames, N.; Tramontin, A.; Garcia-Verdugo, J.; Alvarez-Buylla, A. Adult Ependymal Cells Are Postmitotic and Are Derived from Radial Glial Cells during Embryogenesis. J. Neurosci. 2005, 25, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Del Bigio, M. Ependymal cells: Biology and pathology. Acta Neuropathol. 2010, 119, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Whish, S.; Dziegielewska, K.M.; Møllgård, K.; Noor, N.M.; Liddelow, S.A.; Habgood, M.D.; Richardson, S.J.; Saunders, N.R. The inner CSF-brain barrier: Developmentally controlled access to the brain via intercellular junctions. Front. Neurosci. 2015, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Lee, L. Riding the wave of ependymal cilia: Genetic susceptibility to hydrocephalus in primary ciliary dyskinesia. J. Neurosci. Res. 2013, 91, 1117–1132. [Google Scholar] [CrossRef]
- Jiménez, A.J.; Domínguez-Pinos, M.-D.; Guerra, M.M.; Fernández-Llebrez, P.; Pérez-Fígares, J.-M. Structure and function of the ependymal barrier and diseases associated with ependyma disruption. Tissue Barriers 2014, 2, e28426. [Google Scholar] [CrossRef]
- Harkins, D.; Cooper, H.M.; Piper, M. The role of lipids in ependymal development and the modulation of adult neural stem cell function during aging and disease. Semin. Cell Dev. Biol. 2020, 112, 61–68. [Google Scholar] [CrossRef]
- Spector, R.; Robert Snodgrass, S.; Johanson, C.E. A balanced view of the cerebrospinal fluid composition and functions: Focus on adult humans. Exp. Neurol. 2015, 273, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Khasawneh, A.; Garling, R.; Harris, C. Cerebrospinal fluid circulation: What do we know and how do we know it? Brain Circ. 2018, 4, 14–18. [Google Scholar]
- Johanson, C.; Stopa, E.; McMillan, P.; Roth, D.; Funk, J.; Krinke, G. The Distributional Nexus of Choroid Plexus to Cerebrospinal Fluid, Ependyma and Brain: Toxicologic/Pathologic Phenomena, Periventricular Destabilization, and Lesion Spread. Toxicol. Pathol. 2011, 39, 186–212. [Google Scholar] [CrossRef]
- Veeraval, L.; O’Leary, C.J.; Cooper, H.M. Adherens Junctions: Guardians of Cortical Development. Front. Cell Dev. Biol. 2020, 8, 6. [Google Scholar] [CrossRef]
- Jacquet, B.; Salinas-Mondragon, R.; Liang, H.; Therit, B.; Buie, J.D.; Dykstra, M.; Campbell, K.; Ostrowski, L.; Brody, S.; Ghashghaei, H. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 2009, 136, 4021–4031. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Álvarez, G.; Daclin, M.; Shihavuddin, A.; Lansade, P.; Fortoul, A.; Faucourt, M.; Clavreul, S.; Lalioti, M.-E.; Taraviras, S.; Hippenmeyer, S.; et al. Adult Neural Stem Cells and Multiciliated Ependymal Cells Share a Common Lineage Regulated by the Geminin Family Members. Neuron 2019, 102, 159–172.e157. [Google Scholar] [CrossRef] [Green Version]
- Vidovic, D.; Davila, R.A.; Gronostajski, R.M.; Harvey, T.J.; Piper, M. Transcriptional regulation of ependymal cell maturation within the postnatal brain. Neural Dev. 2018, 13, 2. [Google Scholar] [CrossRef] [Green Version]
- Lavado, A.; Oliver, G. Six3 is required for ependymal cell maturation. Development 2011, 138, 5291–5300. [Google Scholar] [CrossRef] [Green Version]
- Vidovic, D.; Harris, L.; Harvey, T.J.; Evelyn Heng, Y.H.; Smith, A.G.; Osinski, J.; Hughes, J.; Thomas, P.; Gronostajski, R.M.; Bailey, T.L.; et al. Expansion of the lateral ventricles and ependymal deficits underlie the hydrocephalus evident in mice lacking the transcription factor NFIX. Brain Res. 2015, 1616, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.E.; Piper, M.; Plachez, C.; Yeh, Y.-T.; Baizer, J.S.; Osinski, J.M.; Litwack, E.D.; Richards, L.J.; Gronostajski, R.M. The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 2008, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, E.M.; Guerra, M.M. Neural Stem Cells and Fetal-Onset Hydrocephalus. Pediatr. Neurosurg. 2017, 52, 446–461. [Google Scholar] [CrossRef]
- Rekate, H.L. A Contemporary Definition and Classification of Hydrocephalus. Semin. Pediatr. Neurol. 2009, 16, 9–15. [Google Scholar] [CrossRef]
- Orešković, D.; Radoš, M.; Klarica, M. Role of choroid plexus in cerebrospinal fluid hydrodynamics. Neuroscience 2017, 354, 69–87. [Google Scholar] [CrossRef]
- Oliver, C.; González, C.A.; Alvial, G.; Flores, C.A.; Rodríguez, E.M.; Bátiz, L.F. Disruption of CDH2/N-Cadherin-Based Adherens Junctions Leads to Apoptosis of Ependymal Cells and Denudation of Brain Ventricular Walls. J. Neuropathol. Exp. Neurol. 2013, 72, 846–860. [Google Scholar] [CrossRef] [Green Version]
- Ohata, S.; Nakatani, J.; Herranz-Pérez, V.; Cheng, J.; Belinson, H.; Inubushi, T.; Snider, W.D.; García-Verdugo, J.M.; Wynshaw-Boris, A.; Álvarez-Buylla, A. Loss of Dishevelleds Disrupts Planar Polarity in Ependymal Motile Cilia and Results in Hydrocephalus. Neuron 2014, 83, 558–571. [Google Scholar] [CrossRef] [Green Version]
- Kousi, M.; Katsanis, N. The Genetic Basis of Hydrocephalus. Annu. Rev. Neurosci. 2016, 39, 409–435. [Google Scholar] [CrossRef]
- Stauber, M.; Weidemann, M.; Dittrich-Breiholz, O.; Lobschat, K.; Alten, L.; Mai, M.; Beckers, A.; Kracht, M.; Gossler, A. Identification of FOXJ1 effectors during ciliogenesis in the foetal respiratory epithelium and embryonic left-right organiser of the mouse. Dev. Biol. 2017, 423, 170–188. [Google Scholar] [CrossRef]
- Muthusamy, N.; Vijayakumar, A.; Cheng, G.; Ghashghaei, H.T. A Knock-in Foxj1 CreERT2::GFP mouse for recombination in epithelial cells with motile cilia. Genesis 2014, 52, 350–358. [Google Scholar] [CrossRef] [Green Version]
- Moazen, M.; Alazmani, A.; Rafferty, K.; Liu, Z.-J.; Gustafson, J.; Cunningham, M.L.; Fagan, M.J.; Herring, S.W. Intracranial pressure changes during mouse development. J. Biomech. 2016, 49, 123–126. [Google Scholar] [CrossRef]
- Jones, H.C.; Deane, R.; Bucknall, R.M. Developmental changes in cerebrospinal fluid pressure and resistance to absorption in rats. Dev. Brain Res. 1987, 33, 23–30. [Google Scholar] [CrossRef]
- Guerra, M.; Sival, D.; Jimenez, A.; Dominguez Pinos, M.; Den Dunnen, W.; Batiz, L.; Perez-Figares, J.; Rodriguez, E. Defects in cell-cell junctions lead to neuroepithelial/ependymal denudation in the telencephalon of human hydrocephalic foetuses. Cereb. Fluid Res. 2010, 7, S56. [Google Scholar] [CrossRef] [Green Version]
- Guerra, M.M.; Henzi, J.R.; Ortloff, D.A.; Lichtin, C.N.; Vío, A.K.; Jiménez, A.A.; Dominguez-Pinos, M.M.; González, P.C.; Jara, E.M.; Hinostroza, M.F.; et al. Cell Junction Pathology of Neural Stem Cells Is Associated With Ventricular Zone Disruption, Hydrocephalus, and Abnormal Neurogenesis. J. Neuropathol. Exp. Neurol. 2015, 74, 653–671. [Google Scholar] [CrossRef] [Green Version]
- Oishi, S.; Harkins, D.; Kurniawan, N.D.; Kasherman, M.; Harris, L.; Zalucki, O.; Gronostajski, R.M.; Burne, T.H.J.; Piper, M. Heterozygosity for Nuclear Factor One X in mice models features of Malan syndrome. EBioMedicine 2019, 39, 388–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalucki, O.; Harris, L.; Harvey, T.J.; Harkins, D.; Widagdo, J.; Oishi, S.; Matuzelski, E.; Yong, X.L.H.; Schmidt, H.; Anggono, V.; et al. NFIX-Mediated Inhibition of Neuroblast Branching Regulates Migration Within the Adult Mouse Ventricular-Subventricular Zone. Cereb. Cortex 2019, 29, 3590–3604. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.; Zalucki, O.; Clément, O.; Fraser, J.; Matuzelski, E.; Oishi, S.; Harvey, T.J.; Burne, T.H.J.; Heng, J.I.-T.; Gronostajski, R.M.; et al. Neurogenic differentiation by hippocampal neural stem and progenitor cells is biased by NFIX expression. Development 2018, 145, dev155689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiscox, S.; Jiang, W.G.; Obermeier, K.; Taylor, K.; Morgan, L.; Burmi, R.; Barrow, D.; Nicholson, R.I. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of β-catenin phosphorylation. Int. J. Cancer 2006, 118, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, E.M.; Guerra, M.M.; Vío, K.; González, C.; Ortloff, A.; Bátiz, L.F.; Rodríguez, S.; Jara, M.C.; Muñoz, R.I.; Ortega, E.; et al. A cell junction pathology of neural stem cells leads to abnormal neurogenesis and hydrocephalus. Biol. Res. 2012, 45, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Zaman, M.; Yael, K.; Maria, D.-M.; Elaine, C.; Sara, G.-P.; Christian, O.; Bin, C.; Jose Manuel, G.-V.; Nader, S.; Arturo, A.-B. Bi- and uniciliated ependymal cells define continuous floor-plate-derived tanycytic territories. Nat. Commun. 2017, 8, 13759. [Google Scholar]
- Yoshida, S.; Matsumoto, S.; Kanchika, T.; Hagiwara, T.; Minami, T. The organic mercury compounds, methylmercury and ethylmercury, inhibited ciliary movement of ventricular ependymal cells in the mouse brain around the concentrations reported for human poisoning. Neurotoxicology 2016, 57, 69–74. [Google Scholar] [CrossRef]
- Worthington, W.C.; Cathcart, R.S. Ependymal Cilia: Distribution and Activity in the Adult Human Brain. Science 1963, 139, 221–222. [Google Scholar] [CrossRef]
- Worthington, W.C.; Cathcart, R.S. Ciliary Currents on Ependymal Surfaces. Ann. New York Acad. Sci. 1966, 130, 944–950. [Google Scholar] [CrossRef]
- Harris, L.; Zalucki, O.; Gobius, I.; McDonald, H.; Osinki, J.; Harvey, T.J.; Essebier, A.; Vidovic, D.; Gladwyn-Ng, I.; Burne, T.H.; et al. Transcriptional regulation of intermediate progenitor cell generation during hippocampal development. Development 2016, 143, 4620. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.; Essebier, A.; Gronostajski, R.M.; Boden, M.; Wainwright, B.J.; Harvey, T.J.; Piper, M. Cell-type-specific expression of NFIX in the developing and adult cerebellum. Brain Struct. Funct. 2016, 222, 2251–2270. [Google Scholar] [CrossRef]
- Piper, M.; Harris, L.; Barry, G.; Heng, Y.H.E.; Plachez, C.; Gronostajski, R.M.; Richards, L.J. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J. Comp. Neurol. 2011, 519, 3532–3548. [Google Scholar] [CrossRef] [Green Version]
- Heng, Y.H.E.; Zhou, B.; Harris, L.; Harvey, T.; Smith, A.; Horne, E.; Martynoga, B.; Andersen, J.; Achimastou, A.; Cato, K.; et al. NFIX Regulates Proliferation and Migration Within the Murine SVZ Neurogenic Niche. Cereb. Cortex 2015, 25, 3758–3778. [Google Scholar] [CrossRef]
- Niessen, C.M. Tight Junctions/Adherens Junctions: Basic Structure and Function. J. Investig. Dermatol. 2007, 127, 2525–2532. [Google Scholar] [CrossRef] [Green Version]
- Piper, M.; Barry, G.; Hawkins, J.; Mason, S.; Lindwall, C.; Little, E.; Sarkar, A.; Smith, A.G.; Moldrich, R.X.; Boyle, G.M.; et al. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 9127–9139. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.S.; Harris, L.; Lim, J.W.C.; Harvey, T.J.; Piper, M.; Gronostajski, R.M.; Richards, L.J.; Bunt, J. Differential neuronal and glial expression of nuclear factor I proteins in the cerebral cortex of adult mice. J. Comp. Neurol. 2017, 525, 2465–2483. [Google Scholar] [CrossRef]
- Paez-Gonzalez, P.; Lay, M.; Pyun, J.; Kuo, C. Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus. Nat. Commun. 2018, 9, 1655. [Google Scholar]
- Waki, H.; Nakamura, M.; Yamauchi, T.; Wakabayashi, K.-I.; Yu, J.; Hirose-Yotsuya, L.; Take, K.; Sun, W.; Iwabu, M.; Okada-Iwabu, M.; et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet. 2011, 7, e1002311. [Google Scholar] [CrossRef]
- Pjanic, M.; Schmid, C.D.; Gaussin, A.; Ambrosini, G.; Adamcik, J.; Pjanic, P.; Plasari, G.; Kerschgens, J.; Dietler, G.; Bucher, P.; et al. Nuclear Factor I genomic binding associates with chromatin boundaries. BMC Genom. 2013, 14, 99. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.; Essebier, A.; Brown, A.S.; Davila, R.A.; Harkins, D.; Zalucki, O.; Shapiro, L.P.; Penzes, P.; Wainwright, B.J.; Scott, M.P.; et al. Common Regulatory Targets of NFIA, NFIX and NFIB during Postnatal Cerebellar Development. Cerebellum 2020, 19, 89–101. [Google Scholar] [CrossRef]
- Chaand, M.; Fiore, C.; Johnston, B.T.; Moon, D.H.; Carulli, J.P.; Shearstone, J.R. Chromatin Accessibility Mapping of Primary Erythroid Cell Populations Leads to Identification and Validation of Nuclear Factor I X (NFIX) As a Novel Fetal Hemoglobin (HbF) Repressor. Blood 2019, 134 (Suppl. S1), 812. [Google Scholar] [CrossRef]
- Adam, R.C.; Yang, H.; Ge, Y.; Infarinato, N.R.; Gur-Cohen, S.; Miao, Y.; Wang, P.; Zhao, Y.; Lu, C.P.; Kim, J.E.; et al. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices. Nat. Cell Biol. 2020, 22, 640–650. [Google Scholar] [CrossRef]
- Matuzelski, E.; Bunt, J.; Harkins, D.; Lim, J.W.C.; Gronostajski, R.M.; Richards, L.J.; Harris, L.; Piper, M. Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord. Dev. Biol. 2017, 432, 286–297. [Google Scholar] [CrossRef]
- Fraser, J.; Essebier, A.; Brown, A.S.; Davila, R.A.; Sengar, A.S.; Tu, Y.; Ensbey, K.S.; Day, B.W.; Scott, M.P.; Gronostajski, R.M.; et al. Granule neuron precursor cell proliferation is regulated by NFIX and intersectin 1 during postnatal cerebellar development. Brain Struct. Funct. 2018, 224, 811–827. [Google Scholar] [CrossRef]
- Walker, M.; Stabley, D.; McKinney-Freeman, S. NFIX is a putative novel regulator of hematopoietic stem and progenitor cell adhesion. Exp. Hematol. 2016, 44, S106–S107. [Google Scholar] [CrossRef]
- Walker, M.; Brown, S.; Cheng, Y.; Li, Y.; McKinney-Freeman, S. 3143—Characterization of Genome-Wide Binding of Nfix in Hematopoietic Cells. Exp. Hematol. 2020, 88, S82. [Google Scholar] [CrossRef]
- Denny, S.K.; Yang, D.; Chuang, C.-H.; Brady, J.J.; Lim, J.S.; Grüner, B.M.; Chiou, S.-H.; Schep, A.N.; Baral, J.; Hamard, C.; et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 2016, 166, 328–342. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Chen, J.; Yuan, W.; Ruan, H.; Shu, Y.; Ji, J.; Wu, L.; Tang, Q.; Zhou, Z.; Zhang, X.; et al. Nuclear factor I/B promotes colorectal cancer cell proliferation, epithelial-mesenchymal transition and 5-fluorouracil resistance. Cancer Sci. 2019, 110, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Zhu, X.; Liu, W.; Ruan, T.; Wan, W.; Tao, K. NFIB promotes cell growth, aggressiveness, metastasis and EMT of gastric cancer through the Akt/Stat3 signaling pathway. Oncol. Rep. 2018, 40, 1565–1573. [Google Scholar] [CrossRef]
- Xiao, K.; Oas, R.G.; Chiasson, C.M.; Kowalczyk, A.P. Role of p120-catenin in cadherin trafficking. Biochim. Biophys. Acta. Mol. Cell Res. 2007, 1773, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheelock, M.J.; Shintani, Y.; Maeda, M.; Fukumoto, Y.; Johnson, K.R. Cadherin switching. J. Cell Sci. 2008, 121, 727–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.A.; Ireton, R.C.; Reynolds, A.B. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003, 163, 525–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belkin, A.M.; Stepp, M.A. Integrins as receptors for laminins. Microsc. Res. Tech. 2000, 51, 280–301. [Google Scholar] [CrossRef]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2009, 339, 269–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klemke, M.; Weschenfelder, T.; Konstandin, M.H.; Samstag, Y. High affinity interaction of integrin α4β1 (VLA-4) and vascular cell adhesion molecule 1 (VCAM-1) enhances migration of human melanoma cells across activated endothelial cell layers. J. Cell. Physiol. 2007, 212, 368–374. [Google Scholar] [CrossRef]
- Fisch, C.; Dupuis-Williams, P. Ultrastructure of cilia and flagella-back to the future. Biol. Cell 2011, 103, 249–270. [Google Scholar] [CrossRef]
- Feistel, K.; Blum, M. Three types of cilia including a novel 9+4 axoneme on the notochordal plate of the rabbit embryo. Dev. Dyn. 2006, 235, 3348–3358. [Google Scholar] [CrossRef]
- Hao, K.; Chen, Y.; Yan, X.; Zhu, X. Cilia locally synthesize proteins to sustain their ultrastructure and functions. Nat. Commun. 2021, 12, 6971. [Google Scholar] [CrossRef]
- Priolo, M.; Schanze, D.; Tatton-Brown, K.; Mulder, P.A.; Tenorio, J.; Kooblall, K.; Acero, I.H.; Alkuraya, F.S.; Arias, P.; Bernardini, L.; et al. Further delineation of Malan syndrome. Hum. Mutat. 2018, 39, 1226–1237. [Google Scholar] [CrossRef]
- Mulder, P.A.; van Balkom, I.D.C.; Landlust, A.M.; Priolo, M.; Menke, L.A.; Acero, I.H.; Alkuraya, F.S.; Arias, P.; Bernardini, L.; Bijlsma, E.K.; et al. Development, behaviour and sensory processing in Marshall–Smith syndrome and Malan syndrome: Phenotype comparison in two related syndromes. J. Intellect. Disabil. Res. 2020, 64, 956–969. [Google Scholar] [CrossRef]
- Francisco, M.; Purificación, M.-R.; Amparo, S.-C.; Antonio, P.-A.; Silvestre, O.; Mónica, R.; Sonia, M.; Sandra, M.; Jorge, P.; Carmen, O. Novel mutations of NFIX gene causing Marshall-Smith syndrome or Sotos-like syndrome: One gene, two phenotypes. Pediatr. Res. 2015, 78, 533–539. [Google Scholar]
- Lu, T.M.; Houghton, S.; Magdeldin, T.; Barcia Durán, J.G.; Minotti, A.P.; Snead, A.; Sproul, A.; Nguyen, D.-H.T.; Xiang, J.; Fine, H.A.; et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. USA 2021, 118, e2016950118. [Google Scholar] [CrossRef]


| Antibody Name/Target | Catalogue Number | Host Species | Manufacturer | Concentration |
|---|---|---|---|---|
| Primary Antibodies | ||||
| aPKCζ | Sc-216 | Rabbit | Santa Cruz | 1:300 |
| β-Catenin | 05-655 | Mouse | Millipore | 1:300 |
| P120-Catenin | 33-9600 | Rabbit | Abcam | 1:300 |
| ZO-1 | 40-2300 | Mouse | Invitrogen | 1:300 |
| NFIX | SAB1401263 | Mouse | Sigma-Aldrich | 1:400 |
| FOXJ1 | MA5-31419 | Mouse | eBioscience | 1:500 |
| Vimentin | ab92547 | Rabbit | Abcam | 1:500 |
| PAX6 | ab78545 | Mouse | Abcam | 1:500 |
| Secondary Antibodies | ||||
| Donkey α Rabbit Cy3 | AB_2307443 | Donkey | Jackson ImmunoResearch | 1:300 |
| Donkey α Mouse 647 | AB_2340863 | Donkey | Jackson ImmunoResearch | 1:300 |
| DAPI | - | Thermofisher | 1:1000 | |
| Antibody Name/Target | Catalogue Number | Host Species | Manufacturer | Concentration |
|---|---|---|---|---|
| Primary Antibodies | ||||
| PAX6 | ab78545 | Rabbit | Abcam | 1:500 |
| aPKCζ | Sc-216 | Rabbit | Santa Cruz | 1:300 |
| β-Catenin | 05-655 | Mouse | Millipore | 1:300 |
| E-Cadherin | 24E10 | Rabbit | Cell Signalling | 1:300 |
| PAR3 | 07-033 | Rabbit | Millipore | 1:300 |
| P120-Catenin | 33-9600 | Rabbit | Abcam | 1:300 |
| ZO-1 | 40-2300 | Mouse | Invitrogen | 1:300 |
| Secondary Antibodies | ||||
| Donkey α Rabbit Cy3 | AB_2307443 | Donkey | Jackson ImmunoResearch | 1:300 |
| Donkey α Mouse 647 | AB_2340863 | Donkey | Jackson ImmunoResearch | 1:300 |
| DAPI | - | Thermofisher | 1:1000 | |
| Average Raw CT | ||||||
|---|---|---|---|---|---|---|
| Gene | Protein | Fold Change | Student t-Test p-Value | Scramble | Knockdown | Predicted NFI Binding Site |
| CDH1 | E-Cadherin | 1.08 | 0.449571 | 1.96 | 1.84 | −1806, −51 |
| CTNNB1 | Β-Catenin | 0.94 | 0.608607 | 5.31 | 5.4 | −2418, −1040, −862, −335, |
| CTNND1 | P120-Catenin | 0.72 | 0.040615 | 3.99 | 4.47 | −268 |
| TJP1 | ZO-1 | 1.13 | 0.019167 | 4.05 | 3.87 | −628, −341 |
| Average Raw CT | |||||
|---|---|---|---|---|---|
| Gene | Fold Change | Student t-Test p-Value | Scramble | Knockdown | Predicted NFI Binding Site |
| TNC | 3.36 | 0.37651 | 1.60E-01 | 9.50E-02 | −98, −288, −2010 |
| ACTN3 | 2.28 | 0.96452 | 1.00E−04 | 4.50E−05 | −881, −1407, −1565, −1796, −476, −605, −704 |
| COL11A1 | 2.14 | 0.20736 | 6.80E−06 | 1.50E−05 | −2190, −1940, −888, −829 |
| CDSN | 1.4 | 0.04696 | 3.20E−01 | 3.80E−01 | −1679, −1029, −248, +174 |
| NME1 | 1.31 | 0.0055 | 1.60E−03 | 1.20E−03 | − |
| PKP2 | 1.19 | 0.04353 | 5.20E−03 | 6.70E−03 | − |
| DSP | 1.16 | 0.02935 | 2.80E−06 | 6.00E−06 | − |
| WASF1 | 1.16 | 0.0032 | 4.50E−02 | 6.30E−02 | − |
| TJP1 | 1.13 | 0.01917 | 9.70E−06 | 3.00E−05 | −341, −628 |
| ACTN4 | 0.83 | 0.02028 | 3.00E−06 | 6.20E−06 | −206, −1756, −2120 |
| ITGB1 | 0.82 | 0.0338 | 3.40E−04 | 9.80E−05 | −2797, −2933, −2671, −2807 |
| LAMC1 | 0.77 | 0.03058 | 8.90E−03 | 5.20E−03 | −176, −205, −629, −2467, −2913 |
| CSNK2A2 | 0.77 | 0.0431 | 1.20E−01 | 1.10E−01 | −169, −1669 |
| CTNND1 | 0.72 | 0.04062 | 1.40E−01 | 1.10E−01 | −268 |
| ITGA3 | 0.71 | 0.02321 | 7.10E−03 | 3.80E−03 | −319, −619, −884, −1044, −1051, −1362, −2203, −2329, −2351, −2170, −2874, −2881, −2995 |
| SELL | 0.6 | 0.01999 | 6.20E−02 | 4.40E−02 | −308, −1285 |
| COL6A1 | 0.59 | 0.00512 | 6.90E−05 | 1.10E−05 | −1184, −1485, −2312 |
| CD44 | 0.58 | 0.01301 | 2.40E−01 | 2.00E−01 | − |
| ICAM1 | 0.54 | 0.00981 | 1.50E−04 | 7.10E−05 | −2849, −1716, −719 |
| LAMA2 | 0.49 | 0.38715 | 6.30E−02 | 2.60E−02 | −1489, −463, −1320, −2657 |
| SPP1 | 0.48 | 0.25159 | 9.70E−02 | 7.50E−02 | −1929, −3010 |
| DSG3 | 0.48 | 0.37356 | 1.70E−02 | 1.40E−02 | −741 |
| CTNNA3 | 0.46 | 0.07407 | 6.80E−02 | 6.00E−02 | −297, −1970, −1935, −2262, −5 |
| TIMP3 | 0.44 | 0.02463 | 2.00E−03 | 6.60E−04 | −2954 |
| LAMB3 | 0.41 | 0.13912 | 3.60E−04 | 1.40E−04 | −1960, −1821, −549, −473, −214, +189 |
| MMP14 | 0.41 | 0.05487 | 6.50E−03 | 3.90E−03 | −2947, −2626, −1745, −976, −243 |
| MMP12 | 0.34 | 0.5996 | 1.10E−05 | 5.10E−06 | −1065 |
| DSG1 | 0.32 | 0.23532 | 8.70E−03 | 7.50E−03 | −1833, −1805, −1420, −1489 |
| COL14A1 | 0.29 | 0.3944 | 1.40E−04 | 2.20E−05 | −326, −230, −173, |
| ITGA4 | 0.16 | 0.03347 | 1.30E−02 | 5.90E−03 | −2912, −78, +4, +10 |
| THBS2 | 0.15 | 0.00248 | 1.50E−05 | 5.10E−05 | −2354 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harkins, D.; Harvey, T.J.; Atterton, C.; Miller, I.; Currey, L.; Oishi, S.; Kasherman, M.; Davila, R.A.; Harris, L.; Green, K.; et al. Hydrocephalus in Nfix−/− Mice Is Underpinned by Changes in Ependymal Cell Physiology. Cells 2022, 11, 2377. https://doi.org/10.3390/cells11152377
Harkins D, Harvey TJ, Atterton C, Miller I, Currey L, Oishi S, Kasherman M, Davila RA, Harris L, Green K, et al. Hydrocephalus in Nfix−/− Mice Is Underpinned by Changes in Ependymal Cell Physiology. Cells. 2022; 11(15):2377. https://doi.org/10.3390/cells11152377
Chicago/Turabian StyleHarkins, Danyon, Tracey J. Harvey, Cooper Atterton, Ingrid Miller, Laura Currey, Sabrina Oishi, Maria Kasherman, Raul Ayala Davila, Lucy Harris, Kathryn Green, and et al. 2022. "Hydrocephalus in Nfix−/− Mice Is Underpinned by Changes in Ependymal Cell Physiology" Cells 11, no. 15: 2377. https://doi.org/10.3390/cells11152377
APA StyleHarkins, D., Harvey, T. J., Atterton, C., Miller, I., Currey, L., Oishi, S., Kasherman, M., Davila, R. A., Harris, L., Green, K., Piper, H., Parton, R. G., Thor, S., Cooper, H. M., & Piper, M. (2022). Hydrocephalus in Nfix−/− Mice Is Underpinned by Changes in Ependymal Cell Physiology. Cells, 11(15), 2377. https://doi.org/10.3390/cells11152377







