Reprogramming of Amino Acid Metabolism Differs between Community-Acquired Pneumonia and Infection-Associated Exacerbation of Chronic Obstructive Pulmonary Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Study Population
2.2. Mass Spectrometry
2.3. S. aureus Infection
2.4. Statistical Analysis
3. Results
3.1. Description of the Study Population
3.2. Metabolite Detection
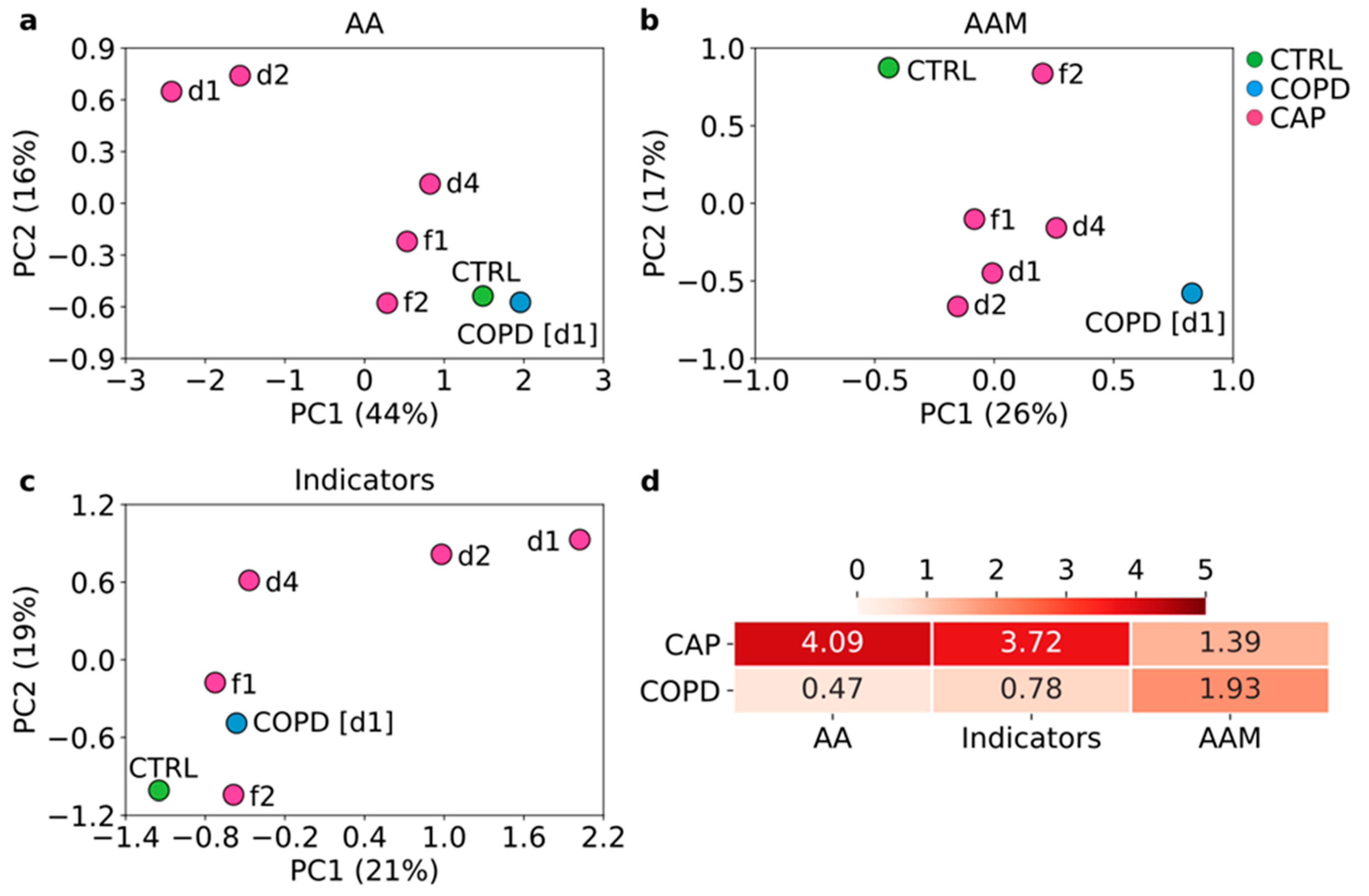
3.3. Reprogramming of Amino Acid Metabolism Differs between CAP and COPD and Tends to Normalize with Clinical Improvement of CAP
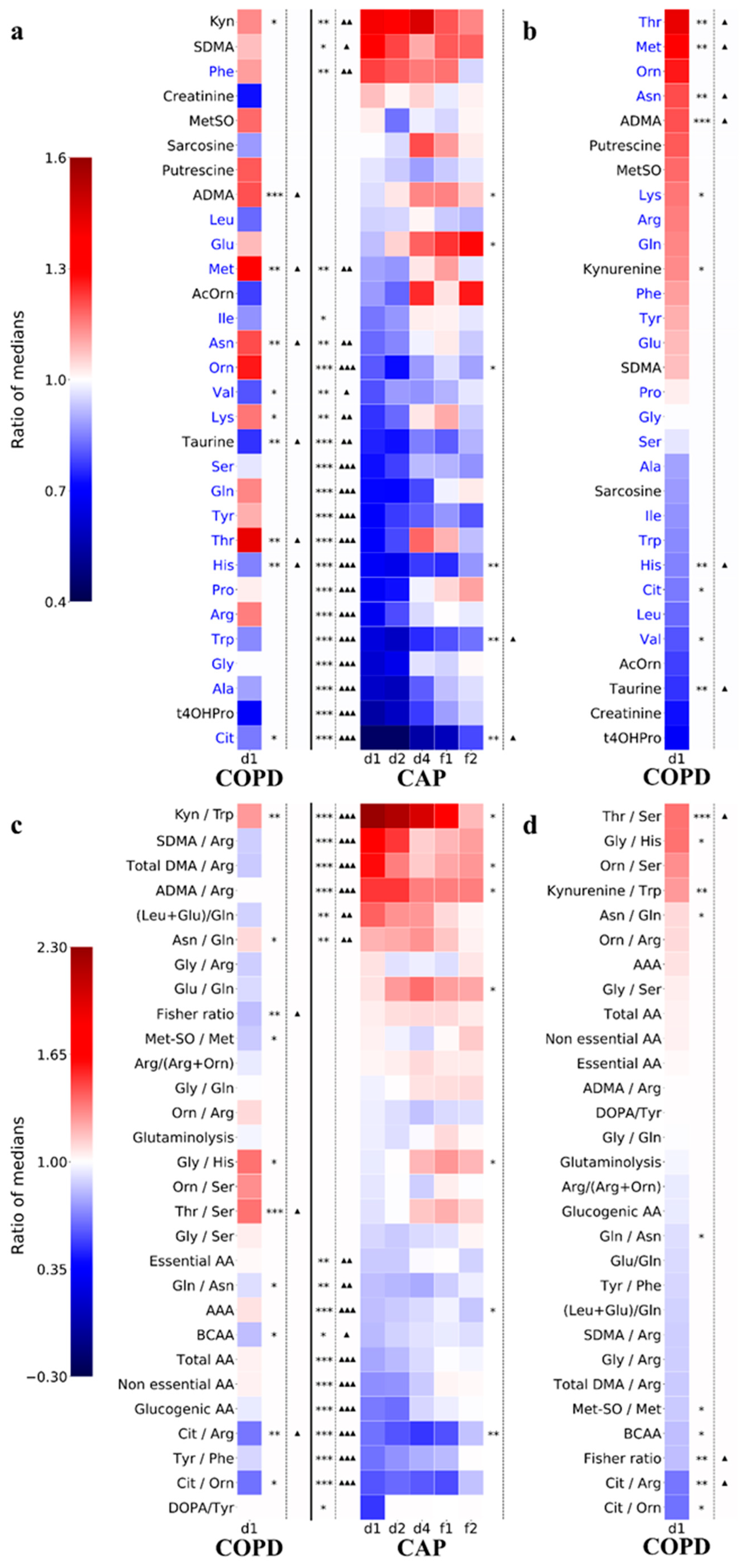
3.4. Decreased Amino Acid Concentrations Are the Hallmark of Acute CAP
3.5. Persistence of Abnormalities in Amino Acid Metabolism during Resolution of CAP
3.6. Comparisons with Amino Acid Metabolic Changes in S. aureus-Infected Human Cell Lines
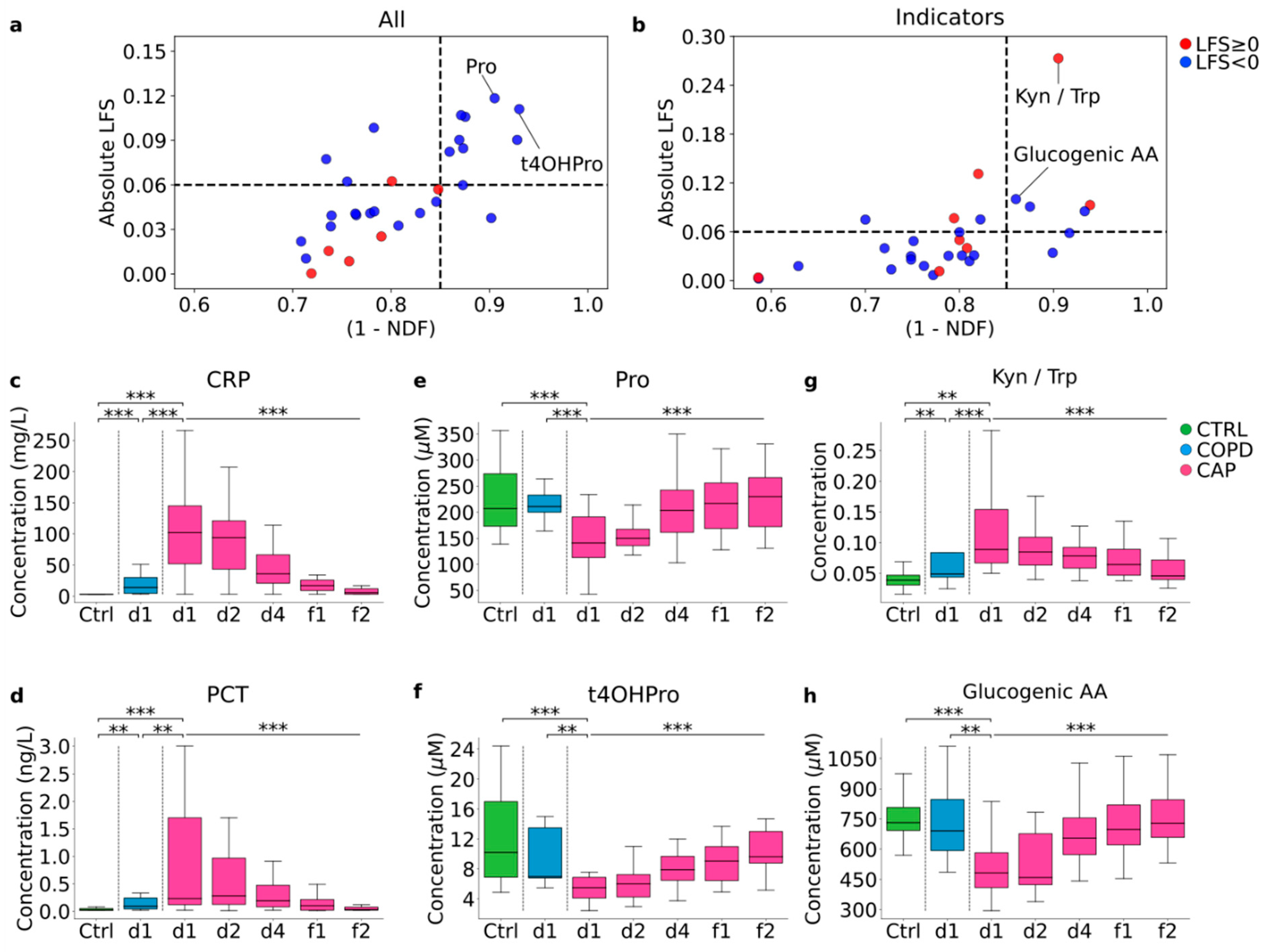
3.7. Identification of Diagnostic Biomarker Candidates
3.8. Changes in Amino Acid Metabolism That Correlate with Resolution of CAP
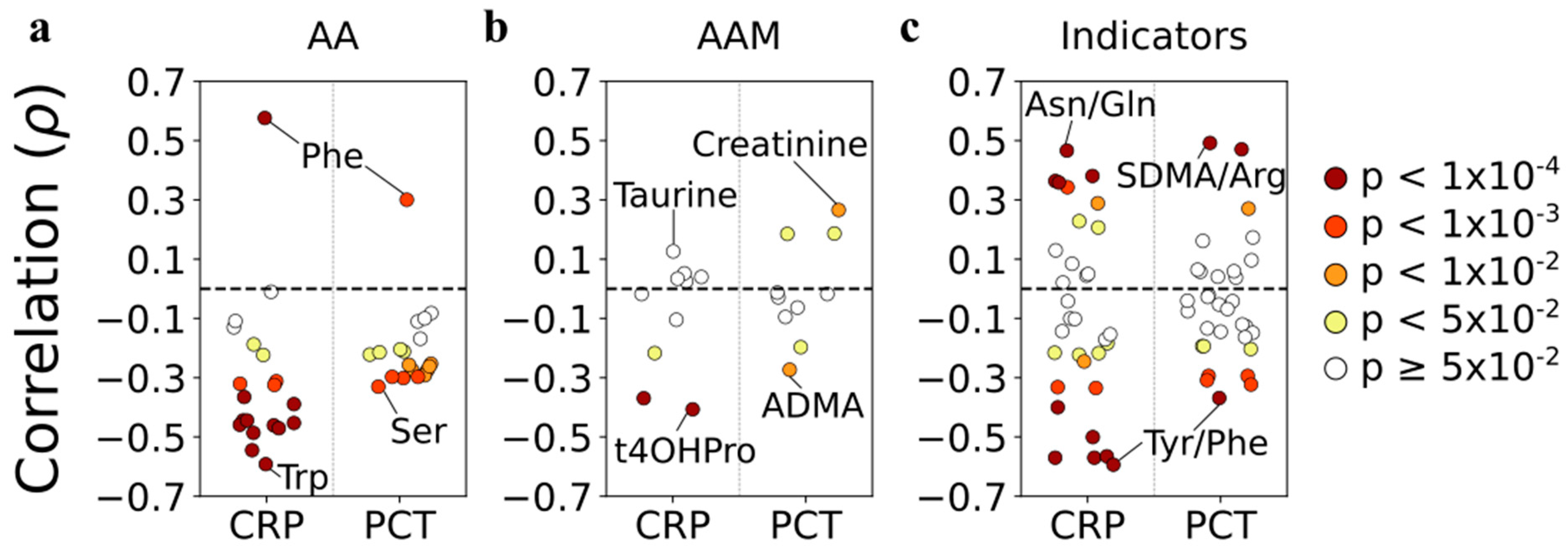
3.9. Correlations with Systemic Inflammation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferreira-Coimbra, J.; Sarda, C.; Rello, J. Burden of Community-Acquired Pneumonia and Unmet Clinical Needs. Adv. Ther. 2020, 37, 1302–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musher, D.M.; Thorner, A.R. Community-acquired pneumonia. N. Engl. J. Med. 2014, 371, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Ishida, T. Diagnostic markers for community-acquired pneumonia. Ann. Transl. Med. 2020, 8, 609. [Google Scholar] [CrossRef] [PubMed]
- Ewig, S.; Torres, A. Severe community-acquired pneumonia. Clin. Chest Med. 1999, 20, 575–587. [Google Scholar] [CrossRef]
- Faulk, W.P.; Mata, L.J.; Edsall, G. Effects of malnutrition on the immune response in humans: A review. Trop Dis. Bull. 1975, 72, 89–103. [Google Scholar]
- Yoneda, J.; Andou, A.; Takehana, K. Regulatory roles of amino acids in immune response. Curr. Rheum. Rev. 2009, 5, 252–258. [Google Scholar] [CrossRef]
- Zhao, H.; Raines, L.N.; Huang, S.C. Carbohydrate and Amino Acid Metabolism as Hallmarks for Innate Immune Cell Activation and Function. Cells 2020, 9, 562. [Google Scholar] [CrossRef] [Green Version]
- Ning, P.; Zheng, Y.; Luo, Q.; Liu, X.; Kang, Y.; Zhang, Y.; Zhang, R.; Xu, Y.; Yang, D.; Xi, W.; et al. Metabolic profiles in community-acquired pneumonia: Developing assessment tools for disease severity. Crit. Care 2018, 22, 130. [Google Scholar] [CrossRef] [Green Version]
- Vogeli, A.; Ottiger, M.; Meier, M.A.; Steuer, C.; Bernasconi, L.; Kulkarni, P.; Huber, A.; Christ-Crain, M.; Henzen, C.; Hoess, C.; et al. Admission levels of asymmetric and symmetric dimethylarginine predict long-term outcome in patients with community-acquired pneumonia. Respir. Res. 2017, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Meier, M.A.; Ottiger, M.; Vogeli, A.; Steuer, C.; Bernasconi, L.; Thomann, R.; Christ-Crain, M.; Henzen, C.; Hoess, C.; Zimmerli, W.; et al. Activation of the tryptophan/serotonin pathway is associated with severity and predicts outcomes in pneumonia: Results of a long-term cohort study. Clin. Chem. Lab. Med. 2017, 55, 1060–1069. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Tian, T.; Liu, Y.; Li, C. The diagnostic value of homocysteine for the occurrence and acute progression of chronic obstructive pulmonary disease. BMC Pulm. Med. 2020, 20, 237. [Google Scholar] [CrossRef]
- Zinellu, A.; Fois, A.G.; Mangoni, A.A.; Paliogiannis, P.; Sotgiu, E.; Zinellu, E.; Marras, V.; Pirina, P.; Carru, C. Systemic concentrations of asymmetric dimethylarginine (ADMA) in chronic obstructive pulmonary disease (COPD): State of the art. Amino Acids 2018, 50, 1169–1176. [Google Scholar] [CrossRef]
- Zinellu, A.; Fois, A.G.; Zinellu, E.; Sotgiu, E.; Sotgia, S.; Arru, D.; Mangoni, A.A.; Carru, C.; Pirina, P. Increased kynurenine plasma concentrations and kynurenine-tryptophan ratio in mild-to-moderate chronic obstructive pulmonary disease patients. Biomark. Med. 2018, 12, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Ikeda, H. Differences in plasma amino acid levels in patients with and without bacterial infection during the early stage of acute exacerbation of COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2019, 14, 575–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogeli, A.; Ottiger, M.; Meier, M.A.; Steuer, C.; Bernasconi, L.; Huber, A.; Christ-Crain, M.; Henzen, C.; Hoess, C.; Thomann, R.; et al. Asymmetric Dimethylarginine Predicts Long-Term Outcome in Patients with Acute Exacerbation of Chronic Obstructive Pulmonary Disease. Lung 2017, 195, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Arshad, H.; Alfonso, J.C.L.; Franke, R.; Michaelis, K.; Araujo, L.; Habib, A.; Zboromyrska, Y.; Lucke, E.; Strungaru, E.; Akmatov, M.K.; et al. Decreased plasma phospholipid concentrations and increased acid sphingomyelinase activity are accurate biomarkers for community-acquired pneumonia. J. Transl. Med. 2019, 17, 365. [Google Scholar] [CrossRef]
- Biocrates_Life_Sciences. Factsheet MetIDQTM RatioExplorer. 2019. Available online: http://www.biocrates.com/images/FS_RatioExplorer%203.pdf (accessed on 15 March 2022).
- MNE-Python software package v0.18.1. Available online: http://mne-tools.github.io/dev/index.html (accessed on 15 March 2022).
- R Foundation for Statistical Computing, version 3.3.2. Available online: https://www.r-project.org (accessed on 15 March 2022).
- Trabado, S.; Al-Salameh, A.; Croixmarie, V.; Masson, P.; Corruble, E.; Feve, B.; Colle, R.; Ripoll, L.; Walther, B.; Boursier-Neyret, C.; et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS One 2017, 12, e0173615. [Google Scholar] [CrossRef] [Green Version]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression, 2nd ed.; Wiley Interscience: Hoboken, NJ, USA, 2000. [Google Scholar]
- Beisel, W.R. Interrelated changes in host metabolism during generalized infectious illness. Am. J. Clin. Nutr. 1972, 25, 1254–1260. [Google Scholar] [CrossRef]
- Geisler, S.; Gostner, J.M.; Becker, K.; Ueberall, F.; Fuchs, D. Immune activation and inflammation increase the plasma phenylalanine-to-tyrosine ratio. Pteridines 2013, 24, 27–31. [Google Scholar] [CrossRef]
- Rapoport, M.I.; Beisel, W.R.; Hornick, R.B. Tryptophan metabolism during infectious illness in man. J. Infect. Dis. 1970, 122, 159–169. [Google Scholar] [CrossRef]
- Suhs, K.W.; Novoselova, N.; Kuhn, M.; Seegers, L.; Kaever, V.; Muller-Vahl, K.; Trebst, C.; Skripuletz, T.; Stangel, M.; Pessler, F. Kynurenine Is a Cerebrospinal Fluid Biomarker for Bacterial and Viral Central Nervous System Infections. J. Infect. Dis. 2019, 220, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Dehhaghi, M.; Kazemi Shariat Panahi, H.; Guillemin, G.J. Microorganisms, Tryptophan Metabolism, and Kynurenine Pathway: A Complex Interconnected Loop Influencing Human Health Status. Int. J. Tryptophan. Res. 2019, 12, 1178646919852996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, Y.; Suda, T.; Yokomura, K.; Suzuki, M.; Fujie, M.; Furuhashi, K.; Hahimoto, D.; Enomto, N.; Fujisawa, T.; Nakamura, Y.; et al. Serum activity of indoleamine 2,3-dioxygenase predicts prognosis of community-acquired pneumonia. J. Infect. 2011, 63, 215–222. [Google Scholar] [CrossRef]
- Csoma, B.; Bikov, A.; Nagy, L.; Toth, B.; Tabi, T.; Szucs, G.; Komlosi, Z.I.; Muller, V.; Losonczy, G.; Lazar, Z. Dysregulation of the endothelial nitric oxide pathway is associated with airway inflammation in COPD. Respir. Res. 2019, 20, 156. [Google Scholar] [CrossRef]
- Tajti, G.; Gesztelyi, R.; Pak, K.; Papp, C.; Keki, S.; Szilasi, M.E.; Mikaczo, A.; Fodor, A.; Szilasi, M.; Zsuga, J. Positive correlation of airway resistance and serum asymmetric dimethylarginine level in COPD patients with systemic markers of low-grade inflammation. Int. J. Chron. Obstruct. Pulmon. Dis. 2017, 12, 873–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzsics, I.; Nagy, L.; Keki, S.; Sarosi, V.; Illes, B.; Illes, Z.; Horvath, I.; Bogar, L.; Molnar, T. L-Arginine Pathway in COPD Patients with Acute Exacerbation: A New Potential Biomarker. COPD 2016, 13, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewicz, J.; Kontny, E. Taurine and inflammatory diseases. Amino Acids 2014, 46, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Bhavsar, T.M.; Patel, S.N.; Lau-Cam, C.A. Protective action of taurine, given as a pretreatment or as a posttreatment, against endotoxin-induced acute lung inflammation in hamsters. J. Biomed. Sci. 2010, 17 (Suppl. 1), S19. [Google Scholar] [CrossRef] [Green Version]
- Thibault, R.; Welch, S.; Mauras, N.; Sager, B.; Altomare, A.; Haymond, M.; Darmaun, D. Corticosteroids increase glutamine utilization in human splanchnic bed. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G548–G553. [Google Scholar] [CrossRef] [Green Version]
- Burian, K.; Hegyesi, H.; Buzas, E.; Endresz, V.; Kis, Z.; Falus, A.; Gonczol, E. Chlamydophila (Chlamydia) pneumoniae induces histidine decarboxylase production in the mouse lung. Immunol. Lett. 2003, 89, 229–236. [Google Scholar] [CrossRef]
- Pessler, F.; Mayer, C.T.; Jung, S.M.; Behrens, E.M.; Dai, L.; Menetski, J.P.; Schumacher, H.R. Identification of novel monosodium urate crystal regulated mRNAs by transcript profiling of dissected murine air pouch membranes. Arthritis Res. Ther. 2008, 10, R64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauer, A.; Menzinger, I.; Gielow, L. Increased histidine decarboxylase activity of rat lung in endotoxin shock. Nature 1966, 212, 1249–1250. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.K.; Webley, W.C. Respiratory Chlamydia Infection Induce Release of Hepoxilin A3 and Histamine Production by Airway Neutrophils. Front. Immunol. 2018, 9, 2357. [Google Scholar] [CrossRef] [PubMed]
- Wise, J.K.; Hendler, R.; Felig, P. Influence of glucocorticoids on glucagon secretion and plasma amino acid concentrations in man. J. Clin. Investig. 1973, 52, 2774–2782. [Google Scholar] [CrossRef]


| CAP (n = 29) | Infection-Associated COPD Exacerbation (n = 13) | Controls (n = 33) | p Value | ||
|---|---|---|---|---|---|
| All Groups a | CAP vs. COPD b | ||||
| Demographics | |||||
| Female (%) | 38 | 46 | 36 | 0.30 | 0.25 |
| Male (%) | 62 | 54 | 64 | ||
| Median age (range) | 60 (24–90) | 62 (55–81) | 59 (24–90) | 0.78 | 0.31 |
| Medical history (past or current) | |||||
| Diabetes (%) | 38 | 17 | 24 | 2.0 × 10−3 | 9.0 × 10−4 |
| Cardiovascular disease (%) | 28 | 50 | 24 | 1.6 × 10−4 | 1.4 × 10−3 |
| Cancer (%) | 10 | 30 | 6 | < 1.0 × 10−5 | 4.1 × 10−4 |
| Treatment (at time of first blood sample) | |||||
| Antimicrobial (%) | 100 | 100 | 0 | NA | NA |
| Corticosteroid (%) | 22 | 62 | 3.3 | < 1.0 × 10−5 | < 1.0 × 10−5 |
| Disease severity | |||||
| PSI risk class (%) | GOLD grade (%) | ||||
| I = 21 | I = 0 | NA | NA | NA | |
| II = 20 | II = 25 | NA | NA | NA | |
| III = 14 | III = 25 | NA | NA | NA | |
| IV = 24 | IV = 50 | NA | NA | NA | |
| V = 21 | NA | NA | NA | NA | |
| Laboratory results | |||||
| CRP (mg/L, ref. < 5 mg/L) | 102 (3.1–428) | 14 (3.1–91) | 3.1 (3.1–7.1) | 2.5 × 10−12 | 2.5 × 10−5 |
| PCT (ng/l, ref. < 0.5 ng/L) | 0.23 (0.02–38) | 0.09 (0.02–1.0) | 0.02 (0.02–0.08) | 5.0 × 10−9 | 7.1 × 10−3 |
| Pathogen detected (%) | 41 | 54 | NA | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arshad, H.; Siokis, A.; Franke, R.; Habib, A.; Alfonso, J.C.L.; Poliakova, Y.; Lücke, E.; Michaelis, K.; Brönstrup, M.; Meyer-Hermann, M.; et al. Reprogramming of Amino Acid Metabolism Differs between Community-Acquired Pneumonia and Infection-Associated Exacerbation of Chronic Obstructive Pulmonary Disease. Cells 2022, 11, 2283. https://doi.org/10.3390/cells11152283
Arshad H, Siokis A, Franke R, Habib A, Alfonso JCL, Poliakova Y, Lücke E, Michaelis K, Brönstrup M, Meyer-Hermann M, et al. Reprogramming of Amino Acid Metabolism Differs between Community-Acquired Pneumonia and Infection-Associated Exacerbation of Chronic Obstructive Pulmonary Disease. Cells. 2022; 11(15):2283. https://doi.org/10.3390/cells11152283
Chicago/Turabian StyleArshad, Haroon, Anastasios Siokis, Raimo Franke, Aamna Habib, Juan Carlos López Alfonso, Yuliya Poliakova, Eva Lücke, Katina Michaelis, Mark Brönstrup, Michael Meyer-Hermann, and et al. 2022. "Reprogramming of Amino Acid Metabolism Differs between Community-Acquired Pneumonia and Infection-Associated Exacerbation of Chronic Obstructive Pulmonary Disease" Cells 11, no. 15: 2283. https://doi.org/10.3390/cells11152283
APA StyleArshad, H., Siokis, A., Franke, R., Habib, A., Alfonso, J. C. L., Poliakova, Y., Lücke, E., Michaelis, K., Brönstrup, M., Meyer-Hermann, M., Bilitewski, U., Vila, J., Abel, L., Illig, T., Schreiber, J., & Pessler, F. (2022). Reprogramming of Amino Acid Metabolism Differs between Community-Acquired Pneumonia and Infection-Associated Exacerbation of Chronic Obstructive Pulmonary Disease. Cells, 11(15), 2283. https://doi.org/10.3390/cells11152283









