CD147 Promotes Tumorigenesis via Exosome-Mediated Signaling in Rhabdomyosarcoma
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Lines and Cell Culture
2.2. Viral Transduction
2.3. Exosome Isolation
2.4. Protein Extraction and Analysis
2.5. Reverse Transcription Real-Time Polymerase Chain Reaction (RTq-PCR)
2.6. Cell Viability, Colony Formation, and Scratch Assays
2.7. Cell Proliferation, Transwell Migration, and Transwell Invasion Assays
2.8. Uptake of CFSE-Labeled RMS-Derived Exosomes by Fibroblasts
2.9. Immunohistochemical Staining of Human Tumor Samples
2.10. Statistical Analysis and Imaging
3. Results
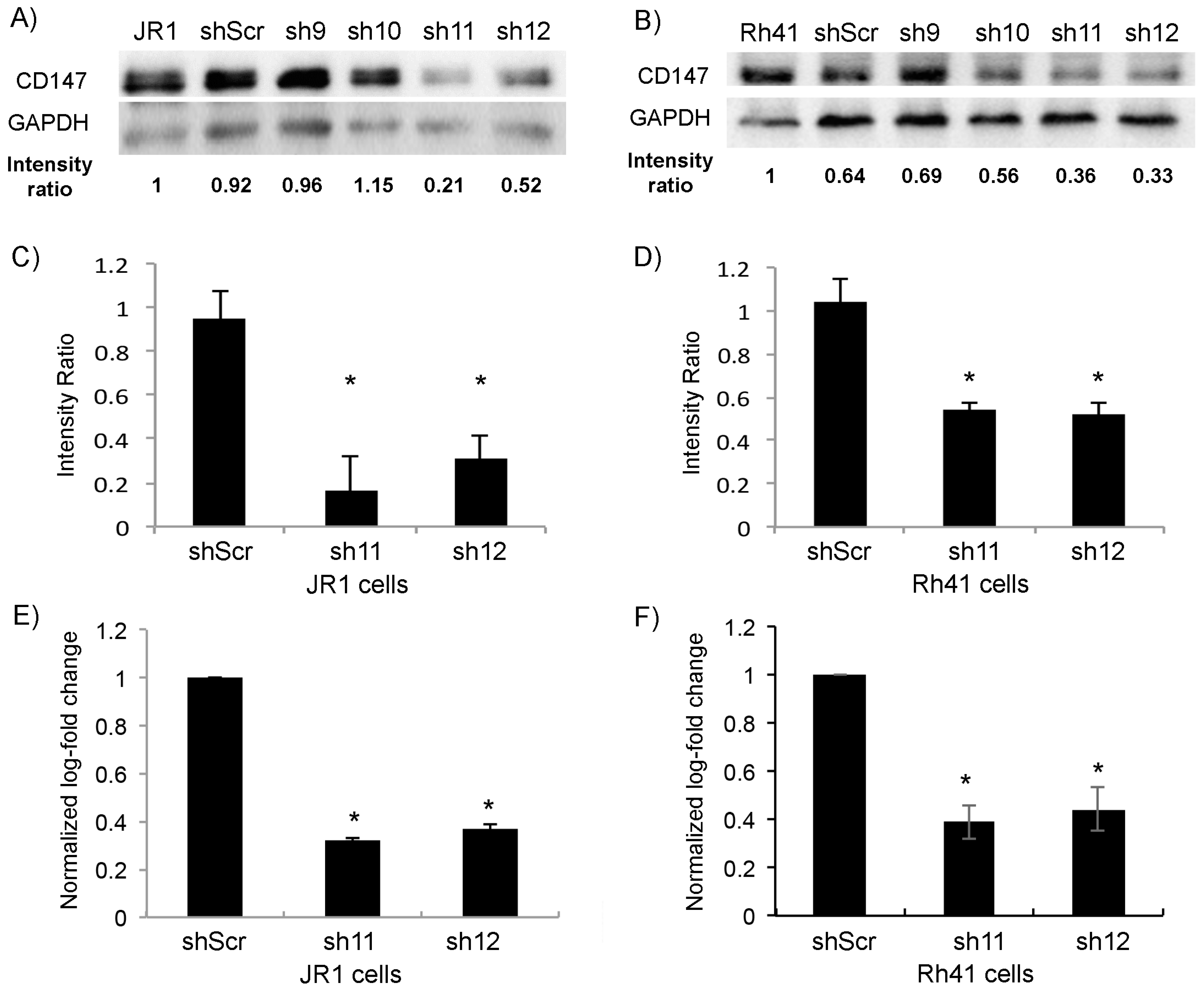
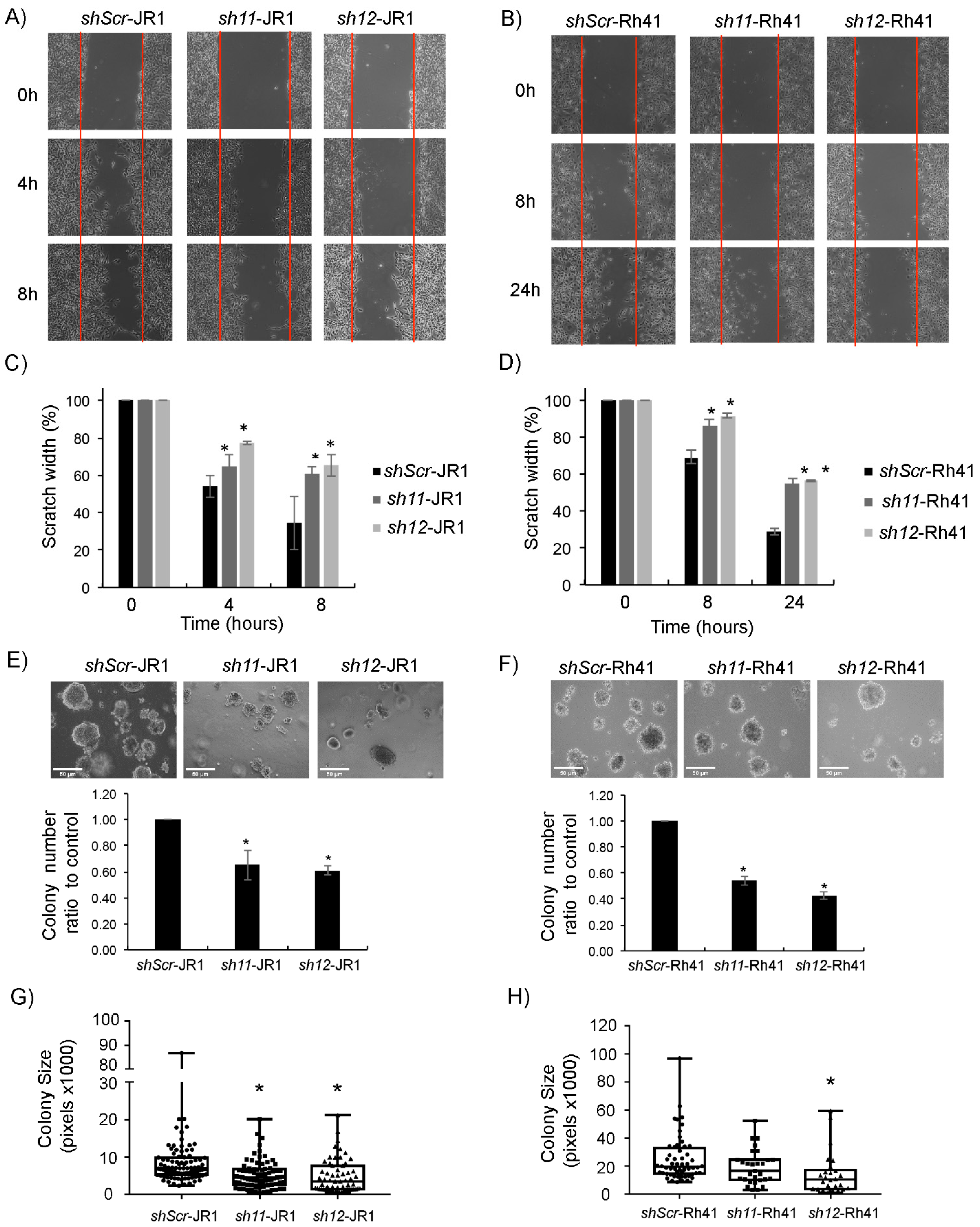
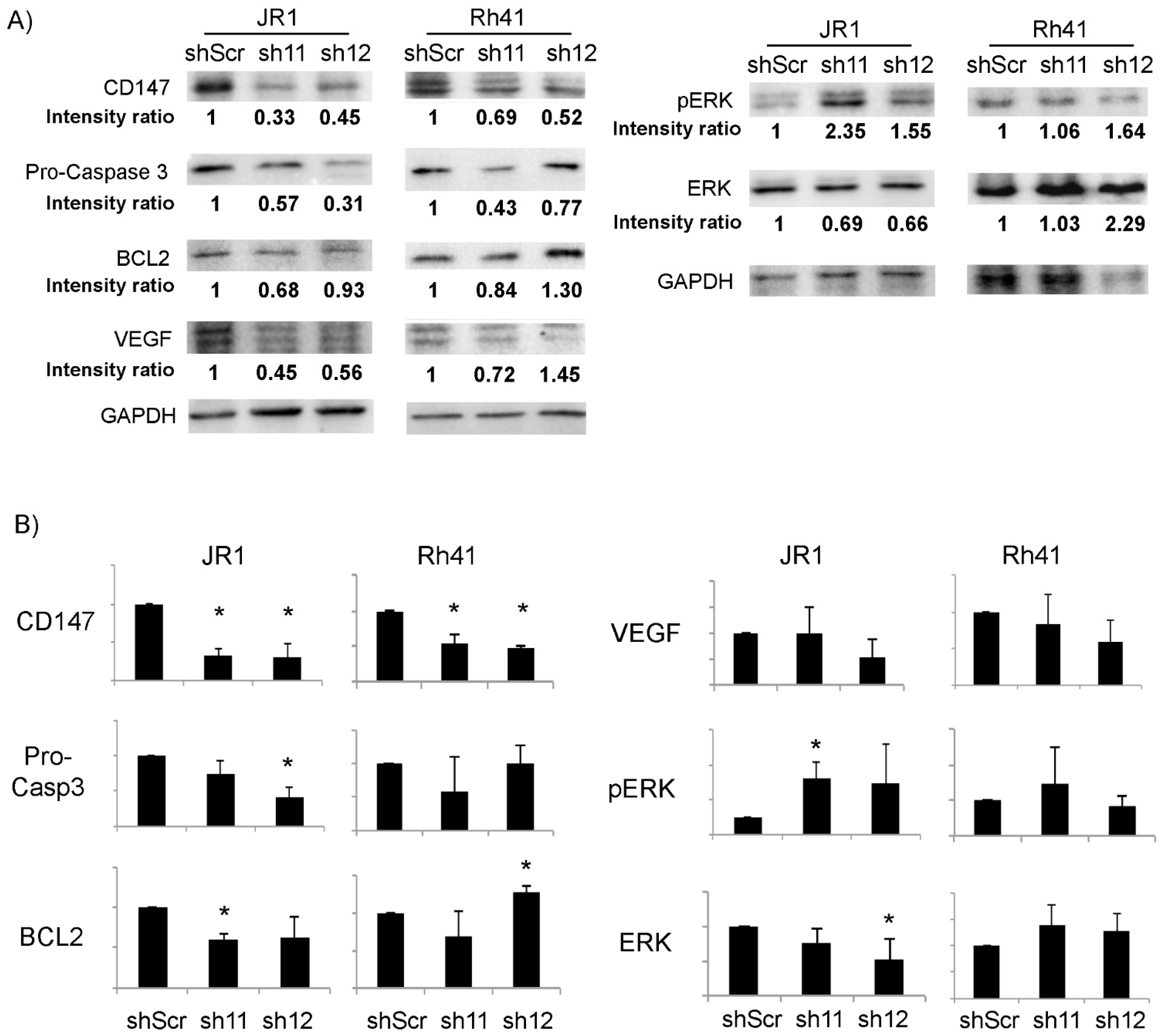
3.1. CD147 Is Expressed in RMS Cells, and Its Knockdown Diminishes RMS Cell Invasive Properties
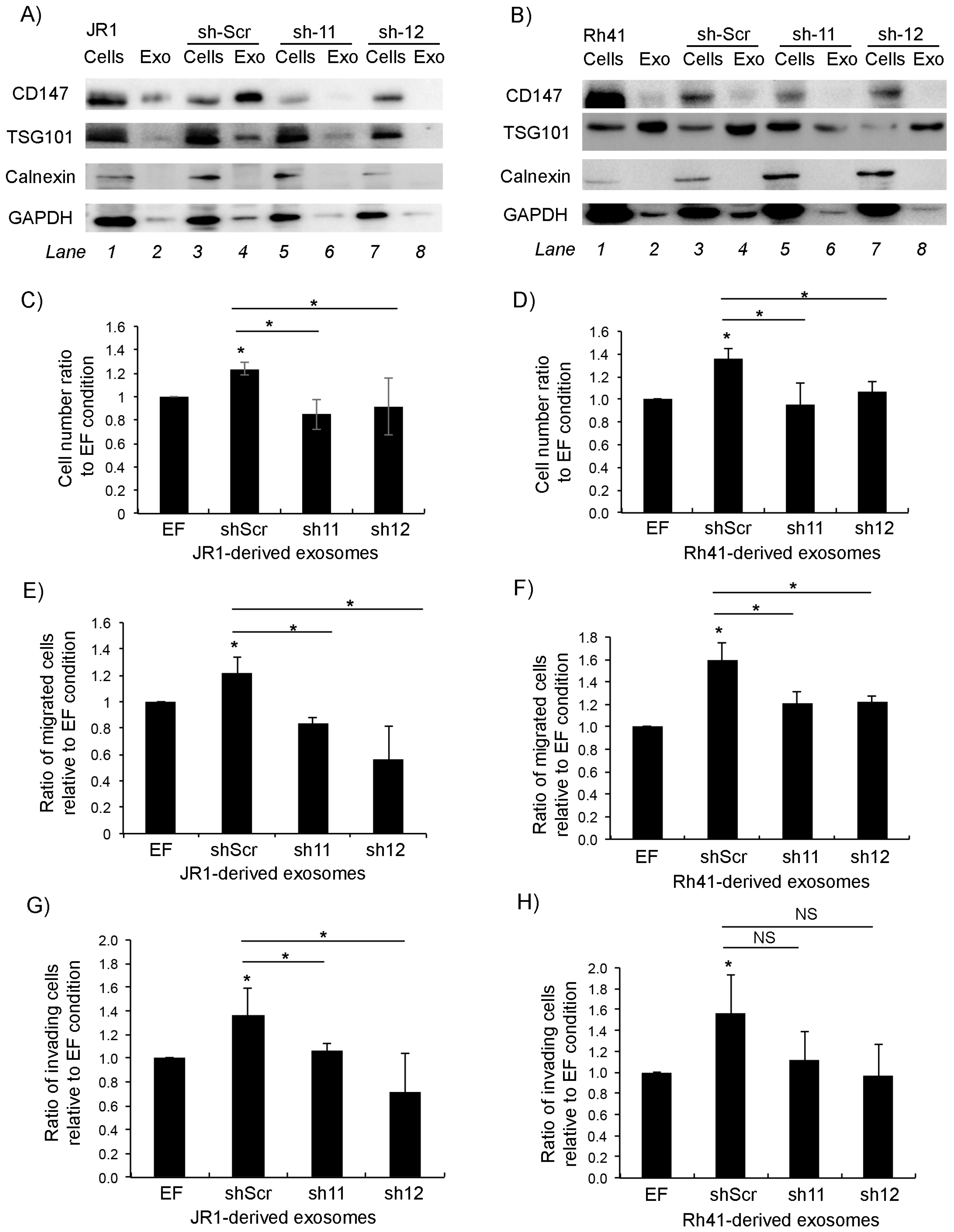
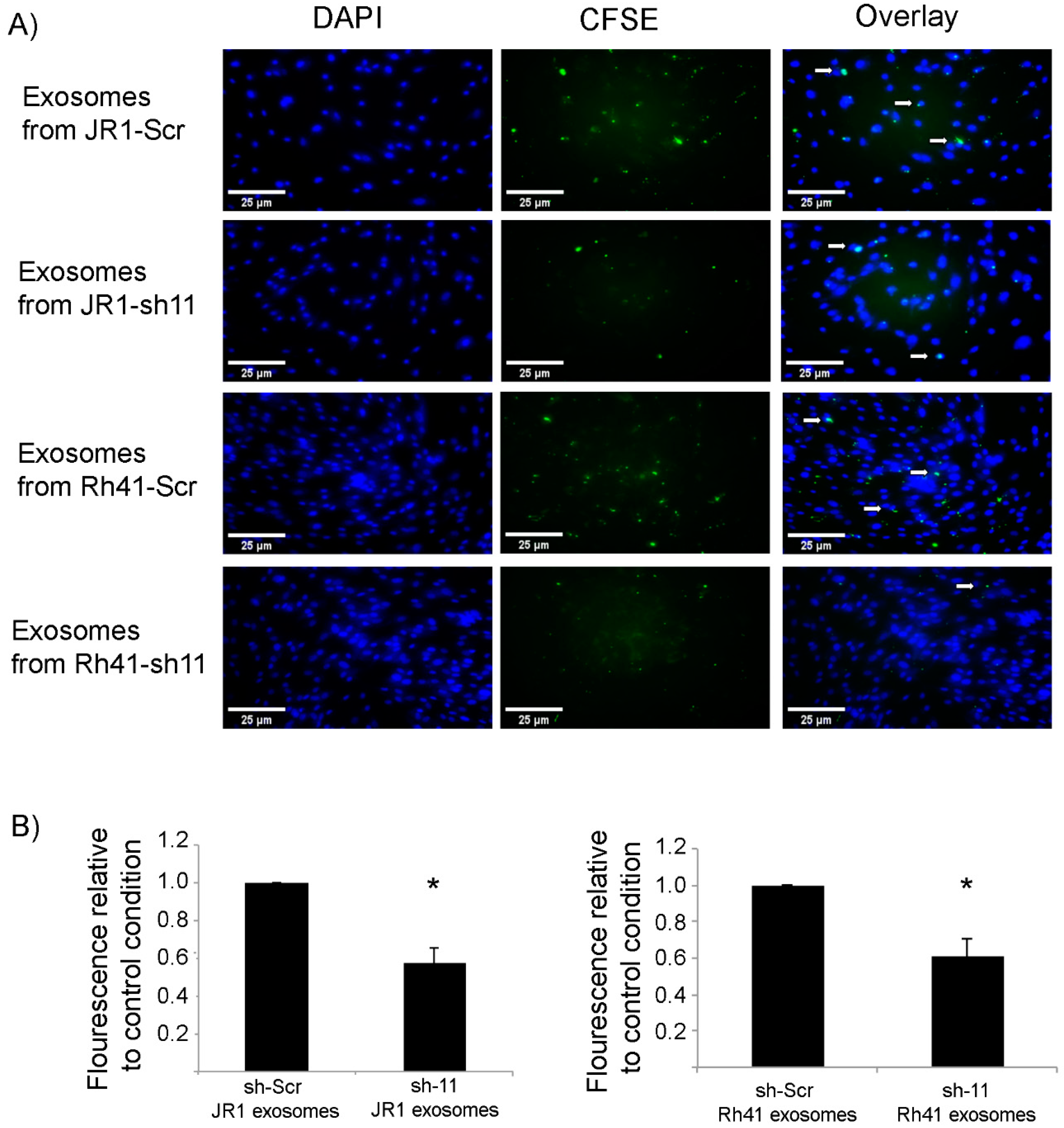
3.2. Knockdown of CD147 Modulates the Effect of RMS-Derived Exosomes on Recipient Fibroblasts
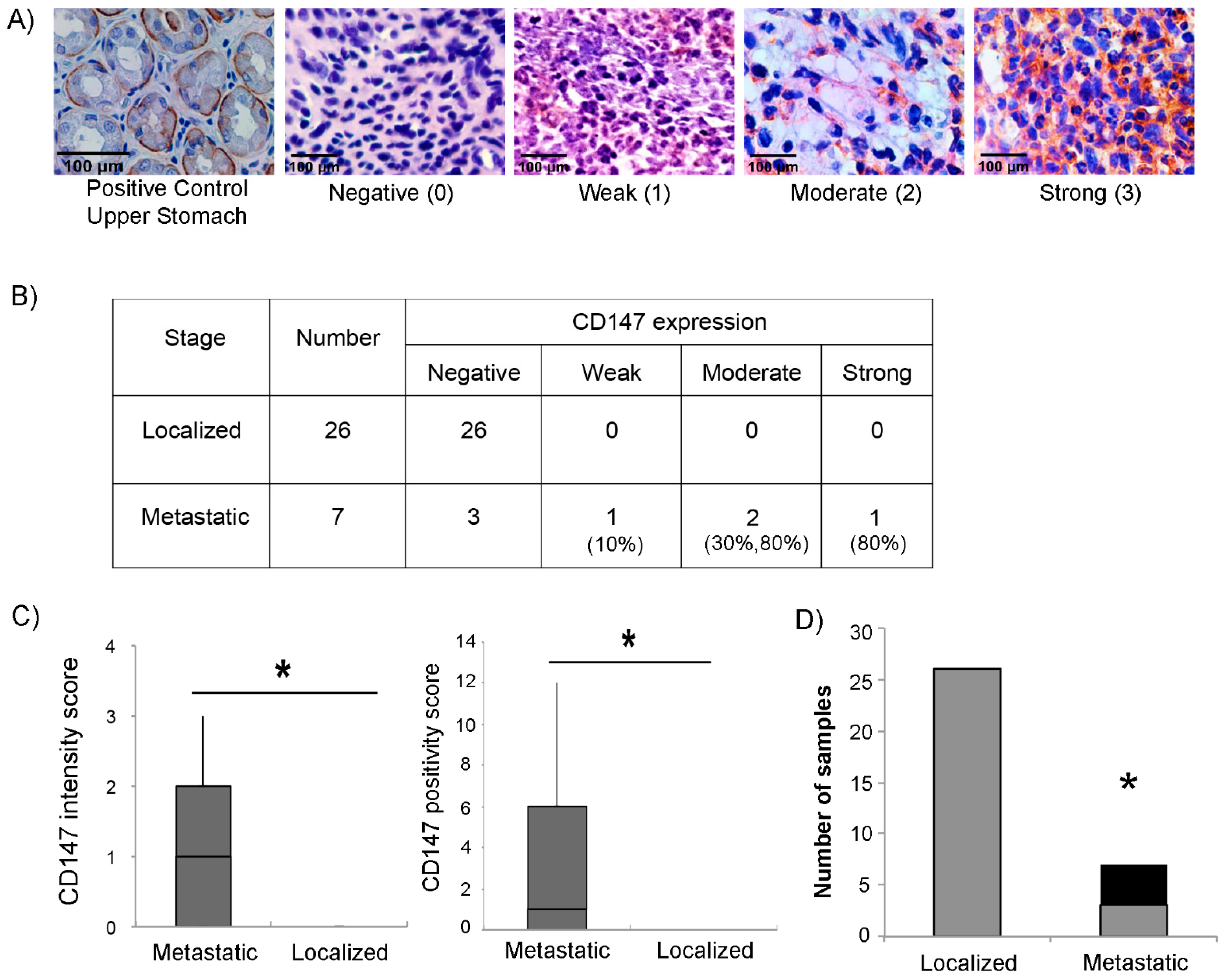
3.3. Expression of CD147 in RMS Tumors Correlates with the Metastatic Stage
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sultan, I.; Qaddoumi, I.; Yaser, S.; Rodriguez-Galindo, C.; Ferrari, A. Comparing Adult and Pediatric Rhabdomyosarcoma in the Surveillance, Epidemiology and End Results Program, 1973 to 2005: An Analysis of 2600 Patients. J. Clin. Oncol. 2009, 27, 3391–3397. [Google Scholar] [CrossRef] [PubMed]
- Saab, R.; Spunt, S.L.; Skapek, S.X. Myogenesis and Rhabdomyosarcoma: The Jekyll and Hyde of Skeletal Muscle. Curr. Top. Dev. Biol. 2011, 94, 197–234. [Google Scholar] [CrossRef] [PubMed]
- Rudzinski, E.R.; Anderson, J.R.; Hawkins, D.S.; Skapek, S.X.; Parham, D.M.; Teot, L.A. The World Health Organization Classification of Skeletal Muscle Tumors in Pediatric Rhabdomyosarcoma a Report from the Children’s Oncology Group. Arch. Pathol. Lab. Med. 2015, 139, 1281–1287. [Google Scholar] [CrossRef] [PubMed]
- Parham, D.M.; Barr, F.G. Classification of Rhabdomyosarcoma and Its Molecular Basis. Adv. Anat. Pathol. 2013, 20, 387–397. [Google Scholar] [CrossRef]
- Rudzinski, E.R.; Anderson, J.R.; Chi, Y.Y.; Gastier-Foster, J.M.; Astbury, C.; Barr, F.G.; Skapek, S.X.; Hawkins, D.S.; Weigel, B.J.; Pappo, A.; et al. Histology, Fusion Status, and Outcome in Metastatic Rhabdomyosarcoma: A Report from the Children’s Oncology Group. Pediatr. Blood Cancer 2017, 64. [Google Scholar] [CrossRef]
- Mercado, G.E.; Barr, F.G. Fusions Involving PAX and FOX Genes in the Molecular Pathogenesis of Alveolar Rhabdomyosarcoma: Recent Advances. Curr. Mol. Med. 2007, 7, 47–61. [Google Scholar] [CrossRef]
- Liu, J.; Guzman, M.A.; Pezanowski, D.; Patel, D.; Hauptman, J.; Keisling, M.; Hou, S.J.; Papenhausen, P.R.; Pascasio, J.M.; Punnett, H.H.; et al. FOXO1-FGFR1 Fusion and Amplification in a Solid Variant of Alveolar Rhabdomyosarcoma. Mod. Pathol. 2011, 24, 1327–1335. [Google Scholar] [CrossRef]
- Wachtel, M.; Dettling, M.; Koscielniak, E.; Stegmaier, S.; Treuner, J.; Simon-Klingenstein, K.; Bühlmann, P.; Niggli, F.K.; Schäfer, B.W. Gene Expression Signatures Identify Rhabdomyosarcoma Subtypes and Detect a Novel t(2;2)(Q35;P23) Translocation Fusing PAX3 to NCOA1. Cancer Res. 2004, 64, 5539–5545. [Google Scholar] [CrossRef]
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive Genomic Analysis of Rhabdomyosarcoma Reveals a Landscape of Alterations Affecting a Common Genetic Axis in Fusion-Positive and Fusion-Negative Tumors. Cancer Discov. 2014, 4, 216–231. [Google Scholar] [CrossRef]
- Williamson, D.; Missiaglia, E.; De Reyniès, A.; Pierron, G.; Thuille, B.; Palenzuela, G.; Thway, K.; Orbach, D.; Laé, M.; Fréneaux, P.; et al. Fusion Gene-Negative Alveolar Rhabdomyosarcoma Is Clinically and Molecularly Indistinguishable from Embryonal Rhabdomyosarcoma. J. Clin. Oncol. 2010, 28, 2151–2158. [Google Scholar] [CrossRef]
- Haduong, J.H.; Heske, C.M.; Allen-Rhoades, W.; Xue, W.; Teot, L.A.; Rodeberg, D.A.; Donaldson, S.S.; Weiss, A.; Hawkins, D.S.; Venkatramani, R. An Update on Rhabdomyosarcoma Risk Stratification and the Rationale for Current and Future Children’s Oncology Group Clinical Trials. Pediatr. Blood Cancer 2022, 69, e29511. [Google Scholar] [CrossRef] [PubMed]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.A.; Barr, F.G. Molecular Diagnostics in the Management of Rhabdomyosarcoma. Expert. Rev. Mol. Diagn. 2017, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Linardic, C.M. PAX3-FOXO1 Fusion Gene in Rhabdomyosarcoma. Cancer Lett. 2008, 270, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Ghayad, S.E.; Rammal, G.; Ghamloush, F.; Basma, H.; Nasr, R.; Diab-Assaf, M.; Chelala, C.; Saab, R. Exosomes Derived from Embryonal and Alveolar Rhabdomyosarcoma Carry Differential MiRNA Cargo and Promote Invasion of Recipient Fibroblasts. Sci. Rep. 2016, 6, 37088. [Google Scholar] [CrossRef] [PubMed]
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Dillon, S.T.; Reeves, R.K.; Manickam, C.; Morgello, S.; Kirk, G.D.; Mehta, S.H.; Gabuzda, D. Exosome Markers Associated with Immune Activation and Oxidative Stress in HIV Patients on Antiretroviral Therapy. Sci. Rep. 2018, 8, 7227. [Google Scholar] [CrossRef]
- Han, L.; Lam, E.W.F.; Sun, Y. Extracellular Vesicles in the Tumor Microenvironment: Old Stories, but New Tales. Mol. Cancer 2019, 18, 59. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular Vesicles in Cancer — Implications for Future Improvements in Cancer Care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Whiteside, T.L. The Tumor Microenvironment and Its Role in Promoting Tumor Growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef]
- Rammal, G.; Fahs, A.; Kobeissy, F.; Mechref, Y.; Zhao, J.; Zhu, R.; Diab-Assaf, M.; Saab, R.; Ghayad, S.E. Proteomic Profiling of Rhabdomyosarcoma-Derived Exosomes Yield Insights into Their Functional Role in Paracrine Signaling. J. Proteome Res. 2019, 18, 3567–3579. [Google Scholar] [CrossRef]
- Ghamloush, F.; Ghayad, S.E.; Rammal, G.; Fahs, A.; Ayoub, A.J.; Merabi, Z.; Harajly, M.; Zalzali, H.; Saab, R. The PAX3-FOXO1 Oncogene Alters Exosome MiRNA Content and Leads to Paracrine Effects Mediated by Exosomal MiR-486. Sci. Rep. 2019, 9, 14242. [Google Scholar] [CrossRef] [PubMed]
- Paolillo, M.; Schinelli, S. Integrins and Exosomes, a Dangerous Liaison in Cancer Progression. Cancers 2017, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.D.; Toole, B.P. How, with Whom and When: An Overview of CD147-Mediated Regulatory Networks Influencing Matrix Metalloproteinase Activity. Biosci. Rep. 2016, 36, e00283. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Zeng, X.; Gu, H.; Li, M.; Tan, H.; Jin, Z.; Hua, T.; Shi, R.; Wang, H. CD147/EMMPRIN Overexpression and Prognosis in Cancer: A Systematic Review and Meta-Analysis. Sci. Rep. 2016, 6, 32804. [Google Scholar] [CrossRef]
- Xu, J.; Lu, Y.; Qiu, S.; Chen, Z.N.; Fan, Z. A Novel Role of EMMPRIN/CD147 in Transformation of Quiescent Fibroblasts to Cancer-Associated Fibroblasts by Breast Cancer Cells. Cancer Lett. 2013, 335, 280–286. [Google Scholar] [CrossRef]
- Caudron, A.; Battistella, M.; Feugeas, J.P.; Pagès, C.; Basset-Seguin, N.; Sadoux, A.; Podgorniak, M.P.; Janin, A.; Lebbe, C.; Mourah, S. EMMPRIN/CD147 as a Novel Independent Prognostic Biomarker in Melanoma. J. Clin. Oncol. 2015, 25, 618–622. [Google Scholar] [CrossRef]
- Voigt, H.; Vetter-Kauczok, C.S.; Schrama, D.; Hofmann, U.B.; Becker, J.C.; Houben, R. CD147 Impacts Angiogenesis and Metastasis Formation. Cancer Invest. 2009, 27, 329–333. [Google Scholar] [CrossRef]
- Yang, S.; Qi, F.; Tang, C.; Wang, H.; Qin, H.; Li, X.; Li, J.; Wang, W.; Zhao, C.; Gao, H. CD147 Promotes the Proliferation, Invasiveness, Migration and Angiogenesis of Human Lung Carcinoma Cells. Oncol. Lett. 2017, 13, 898–904. [Google Scholar] [CrossRef]
- Li, L.; Dong, X.; Peng, F.; Shen, L. Integrin Β1 Regulates the Invasion and Radioresistance of Laryngeal Cancer Cells by Targeting CD147. Cancer Cell Int. 2018, 18, 80. [Google Scholar] [CrossRef]
- Li, Y.; Wu, J.; Song, F.; Tang, J.; Wang, S.J.; Yu, X.L.; Chen, Z.N.; Jiang, J.L. Extracellular Membrane-Proximal Domain of HAb18G/CD147 Binds to Metal Ion-Dependent Adhesion Site (MIDAS) Motif of Integrin Β1 to Modulate Malignant Properties of Hepatoma Cells. J. Biol. Chem. 2012, 287, 4759–4772. [Google Scholar] [CrossRef]
- Landras, A.; de Moura, C.R.; Jouenne, F.; Lebbe, C.; Menashi, S.; Mourah, S. CD147 Is a Promising Target of Tumor Progression and a Prognostic Biomarker. Cancers 2019, 11, 1803. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Khanna, S.; Giles, P.J.; Brennan, P.; Brewis, I.A.; Staffurth, J.; Mason, M.D.; Clayton, A. Proteomics Analysis of Bladder Cancer Exosomes. Mol. Cell. Proteomics 2010, 9, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.H.; Liu, Y.J.; Tang, L.L.; Wang, S.M.; Yan, G.J.; Liao, L.Q. Plasma Soluble Cluster of Differentiation 147 Levels Are Increased in Breast Cancer Patients and Associated with Lymph Node Metastasis and Chemoresistance. Hong Kong Med. J. 2018, 24, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; König, A.K.; Marmé, F.; Runz, S.; Wolterink, S.; Koensgen, D.; Mustea, A.; Sehouli, J.; Altevogt, P. Systemic Presence and Tumor-Growth Promoting Effect of Ovarian Carcinoma Released Exosomes. Cancer Lett. 2009, 278, 73–81. [Google Scholar] [CrossRef]
- Clayton, J.; Pincott, J.R.; Van Den Berghe, J.A.; Kemshead, J.T. Comparative Studies between a New Human Rhabdomyosarcoma Cell Line, JR-1 and Its Tumour of Origin. Br. J. Cancer 1986, 54, 83–90. [Google Scholar] [CrossRef]
- Hinson, A.R.P.; Jones, R.; Lisa, L.E.; Belyea, B.C.; Barr, F.G.; Linardic, C.M. Human Rhabdomyosarcoma Cell Lines for Rhabdomyosarcoma Research: Utility and Pitfalls. Front. Oncol. 2013, 3, 183. [Google Scholar] [CrossRef]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In Vitro Scratch Assay: A Convenient and Inexpensive Method for Analysis of Cell Migration in Vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Yin, H.; Shao, Y.; Chen, X. The Effects of CD147 on the Cell Proliferation, Apoptosis, Invasion, and Angiogenesis in Glioma. Neurol. Sci. 2017, 38, 129–136. [Google Scholar] [CrossRef]
- Chen, L.; Pan, Y.; Gu, L.; Nie, Z.; He, B.; Song, G.; Li, R.; Xu, Y.; Gao, T.; Wang, S. ERK1/2 Signalling Pathway Is Involved in CD147-Mediated Gastric Cancer Cell Line SGC7901 Proliferation and Invasion. Exp. Biol. Med. 2013, 238, 903–912. [Google Scholar] [CrossRef]
- Amer, K.M.; Thomson, J.E.; Congiusta, D.; Dobitsch, A.; Chaudhry, A.; Li, M.; Chaudhry, A.; Bozzo, A.; Siracuse, B.; Aytekin, M.N.; et al. Epidemiology, Incidence, and Survival of Rhabdomyosarcoma Subtypes: SEER and ICES Database Analysis. J. Orthop. Res. 2019, 37, 2226–2230. [Google Scholar] [CrossRef]
- Li, H.; Jiang, C.; Wu, D.; Shi, S.; Liao, M.; Wang, J.; Li, Y.; Xu, Z. The Prognostic and Clinicopathologic Characteristics of CD147 and Esophagus Cancer: A Meta-Analysis. PLoS ONE 2017, 12, e0180271. [Google Scholar] [CrossRef]
- Peng, J.; Jiang, H.; Guo, J.; Huang, J.; Yuan, Q.; Xie, J.; Xiao, K. CD147 Expression Is Associated with Tumor Proliferation in Bladder Cancer via GSDMD. Biomed. Res. Int. 2020, 7638975. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, C.; Niu, R.; Hu, G.; Gu, Z.; Zhuang, Z. MiR-890 Inhibits Proliferation and Invasion and Induces Apoptosis in Triple-Negative Breast Cancer Cells by Targeting CD147. BMC Cancer 2019, 19, 577. [Google Scholar] [CrossRef]
- Millimaggi, D.; Mari, M.; D’Ascenzo, S.; Carosa, E.; Jannini, E.A.; Zucker, S.; Carta, G.; Pavan, A.; Dolo, V. Tumor Vesicle-Associated CD147 Modulates the Angiogenic Capability of Endothelial Cells. Neoplasia 2007, 9, 349–357. [Google Scholar] [CrossRef]
- Schneiderhan, W.; Scheler, M.; Holzmann, K.H.; Marx, M.; Gschwend, J.E.; Bucholz, M.; Gress, T.M.; Seufferlein, T.; Adler, G.; Oswald, F. CD147 Silencing Inhibits Lactate Transport and Reduces Malignant Potential of Pancreatic Cancer Cells in in Vivo and in Vitro Models. Gut 2009, 58, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.H.; Wang, Y.; Wen, L.; Zhai, Z.B.; Ai, Z.H.; Yao, N.L.; Wang, L.; Liu, W.C.; Chen, B.L.; Li, Y.; et al. Basigin-2 Is the Predominant Basigin Isoform That Promotes Tumor Cell Migration and Invasion and Correlates with Poor Prognosis in Epithelial Ovarian Cancer. J. Transl. Med. 2013, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Kulyar, M.F.e.A.; Yao, W.; Ding, Y.; Du, H.; Li, K.; Zhang, L.; Li, A.; Huachun, P.; Waqas, M.; Mehmood, K.; et al. Cluster of Differentiation 147 (CD147) Expression Is Linked with Thiram Induced Chondrocyte’s Apoptosis via Bcl-2/Bax/Caspase-3 Signalling in Tibial Growth Plate under Chlorogenic Acid Repercussion. Ecotoxicol. Environ. Saf. 2021, 213, 112059. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, H.; Jeong, K.; Jung, M.Y.; Hahn, B.S.; Yoon, K.S.; Kwan Jin, B.; Jahng, G.H.; Kang, I.; Ha, J.; et al. Release of Overexpressed CypB Activates ERK Signaling through CD147 Binding for Hepatoma Cell Resistance to Oxidative Stress. Apoptosis 2012, 17, 784–796. [Google Scholar] [CrossRef]
- Li, F.; Zhang, J.; Guo, J.; Jia, Y.; Han, Y.; Wang, Z. RNA Interference Targeting CD147 Inhibits Metastasis and Invasion of Human Breast Cancer MCF-7 Cells by Downregulating MMP-9/VEGF Expression. Acta Biochim. Biophys. Sin. 2018, 50, 676–684. [Google Scholar] [CrossRef]
- Hu, X.; Su, J.; Zhou, Y.; Xie, X.; Peng, C.; Yuan, Z.; Chen, X. Repressing CD147 Is a Novel Therapeutic Strategy for Malignant Melanoma. Oncotarget 2017, 8, 25806–25813. [Google Scholar] [CrossRef]
- Fu, J.; Fu, J.; Chen, X.; Zhang, Y.; Gu, H.; Bai, Y. CD147 and VEGF Co-Expression Predicts Prognosis in Patients with Acute Myeloid Leukemia. Jpn. J. Clin. Oncol. 2010, 40, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, C.M.; Woenne, E.C.; Brix, G.; Radzwill, N.; Ilg, M.; Bachert, P.; Peschke, P.; Kirsch, S.; Kauczor, H.U.; Delorme, S.; et al. Impact of Stroma on the Growth, Microcirculation, and Metabolism of Experimental Prostate Tumors. Neoplasia 2007, 9, 57–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Said, N.A.; Najwer, I.; Socha, M.J.; Fulton, D.J.; Mok, S.C.; Motamed, K. SPARC Inhibits LPA-Mediated Mesothelial-Ovarian Cancer Cell Crosstalk. Neoplasia 2007, 9, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Kesavan, P.; Nakada, M.T.; Yan, L. Tumor-Stroma Interaction: Positive Feedback Regulation of Extracellular Matrix Metalloproteinase Inducer (EMMPRIN) Expression and Matrix Metalloproteinase-Dependent Generation of Soluble EMMPRIN. Mol. Cancer Res. 2004, 2, 73–80. [Google Scholar] [CrossRef]
- Sidhu, S.S.; Mengistab, A.T.; Tauscher, A.N.; LaVail, J.; Basbaum, C. The Microvesicle as a Vehicle for EMMPRin in Tumor-Stromal Interactions. Oncogene 2004, 23, 956–963. [Google Scholar] [CrossRef]
- Hatanaka, M.; Higashi, Y.; Fukushige, T.; Baba, N.; Kawai, K.; Hashiguchi, T.; Su, J.; Zeng, W.; Chen, X.; Kanekura, T. Cleaved CD147 Shed from the Surface of Malignant Melanoma Cells Activates MMP2 Produced by Fibroblasts. Anticancer Res. 2014, 34, 7091–7096. [Google Scholar]
- Chu, D.; Zhu, S.; Li, J.; Ji, G.; Wang, W.; Wu, G.; Zheng, J. CD147 Expression in Human Gastric Cancer Is Associated with Tumor Recurrence and Prognosis. PLoS ONE 2014, 9, e101027. [Google Scholar] [CrossRef]
- Zhong, X.; Li, M.; Nie, B.; Wu, F.; Zhang, L.; Wang, E.; Han, Y. Overexpressions of RACK1 and CD147 Associated with Poor Prognosis in Stage T1 Pulmonary Adenocarcinoma. Ann. Surg. Oncol. 2013, 20, 1044–1052. [Google Scholar] [CrossRef]
- Dehner, C.A.; Armstrong, A.E.; Yohe, M.; Shern, J.F.; Hirbe, A.C. Genetic Characterization, Current Model Systems and Prognostic Stratification in PAX Fusion-Negative vs. PAX Fusion-Positive Rhabdomyosarcoma. Genes 2021, 12, 1500. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fahs, A.; Hussein, N.; Zalzali, H.; Ramadan, F.; Ghamloush, F.; Tamim, H.; El Homsi, M.; Badran, B.; Boulos, F.; Tawil, A.; et al. CD147 Promotes Tumorigenesis via Exosome-Mediated Signaling in Rhabdomyosarcoma. Cells 2022, 11, 2267. https://doi.org/10.3390/cells11152267
Fahs A, Hussein N, Zalzali H, Ramadan F, Ghamloush F, Tamim H, El Homsi M, Badran B, Boulos F, Tawil A, et al. CD147 Promotes Tumorigenesis via Exosome-Mediated Signaling in Rhabdomyosarcoma. Cells. 2022; 11(15):2267. https://doi.org/10.3390/cells11152267
Chicago/Turabian StyleFahs, Assil, Nader Hussein, Hasan Zalzali, Farah Ramadan, Farah Ghamloush, Hani Tamim, Mahmoud El Homsi, Bassam Badran, Fouad Boulos, Ayman Tawil, and et al. 2022. "CD147 Promotes Tumorigenesis via Exosome-Mediated Signaling in Rhabdomyosarcoma" Cells 11, no. 15: 2267. https://doi.org/10.3390/cells11152267
APA StyleFahs, A., Hussein, N., Zalzali, H., Ramadan, F., Ghamloush, F., Tamim, H., El Homsi, M., Badran, B., Boulos, F., Tawil, A., Ghayad, S. E., & Saab, R. (2022). CD147 Promotes Tumorigenesis via Exosome-Mediated Signaling in Rhabdomyosarcoma. Cells, 11(15), 2267. https://doi.org/10.3390/cells11152267






