Abstract
Lignans, as secondary metabolites synthesized within a phenylpropanoid pathway, play various roles in plants, including their involvement in growth and plant defense processes. The health and nutritional benefits of lignans are unquestionable, and many studies have been devoted to these attributes. Although the regulatory role of miRNAs in the biosynthesis of secondary metabolites has been widely reported, there is no systematic review available on the miRNA-based regulatory mechanism of lignans biosynthesis. However, the genetic background of lignan biosynthesis in plants is well characterized. We attempted to put together a regulatory mosaic based on current knowledge describing miRNA-mediated regulation of genes, enzymes, or transcription factors involved in this biosynthesis process. At the same time, we would like to underline the fact that further research is necessary to improve our understanding of the miRNAs regulating plant lignan biosynthesis by exploitation of current approaches for functional identification of miRNAs.
1. Introduction
Phenylpropanoid metabolites biosynthesis is a complex network producing various important secondary metabolites, including lignans, and its regulatory mechanism is important for plant growth, development, and (a)biotic stress protection [1]. The most recent studies are focused on unraveling the miRNAs regulatory potential in the biosynthetic pathways of major secondary metabolites such as alkaloids, terpenoids, and flavonoids [1,2,3,4]. Lignans are polyphenolic compounds that are widely distributed in plant species. They represent one of the main components of plant cell walls [5] and show a variety of biological functions, including health-promoting effects. Despite their common biochemical basis, they possess wide structural variability. It is believed that there are up to 30,000 variants of lignans, of which only about 6000 are currently known [6]. Their strong biological activity encourages biochemists to create synthetic substitutes or incorporate them into drugs [7]. The wide spectrum of their structural and functional variability made it possible to identify certain types of lignans as good candidates for natural agrochemicals with the potential to inhibit seed germination and plant growth [5].
There are two main roles attributed to the lignans: plant defense and antioxidant activity [8]. The lignan content and compositions are significantly dependent on genetic and environmental background, including some other factors such as the maturity stage of the seeds and fruits [8,9]. Environmental factors can affect secondary metabolites biosynthesis by modulation of gene expression through miRNAs and transcription factors. The list of miRNAs and related transcription factors involved in flavonoid biosynthesis mediated by environmental factors is displayed in the following study [10].
Lignans biosynthesis is spatially and temporally dependent [11,12,13,14] as well as species and genotype specific [15,16].
MicroRNAs (miRNAs), as non-coding RNA molecules that bind to highly complementary sites in target mRNAs, play a key role in the post-transcriptional regulation of genes. They are highly conserved and have formed part of the genomes of viruses, plants, animals, and humans [17,18]. In addition to highly conserved miRNAs, less conserved miRNAs are also found in plants and interfere in several aspects of plant regulation such as response to biotic and abiotic stresses, regulation of developmental processes—leaf morphogenesis, physiology of cellular structures, alteration of vegetative phase, and flowering time [19,20].
Phenylpropanoids represent the largest group of plant secondary metabolites, involved in many biological and developmental processes, which at the same time points to their importance and meaning for plants. It is known that miRNAs also participate in phenylpropanoid biosynthesis by targeting transcription factors [1]. Therefore, the knowledge of the miRNA-based regulatory mechanisms of their biosynthesis is extremely important [2,3]. Currently, there is an effort to increase the production of secondary metabolites, which are produced by medicinal plants in small quantities and are used as insecticides, dyes, drugs, and toxins in agriculture, medicine, and industry. One of the ways how to increase their production is exploitation of RNA technologies that target small non-coding RNAs, which requires the knowledge of miRNAs involved in secondary metabolite biosynthesis [21].
While the genetic regulation of lignans biosynthesis is handled at a very good level, the direct links between this biosynthesis and miRNA-based regulation do not reach such a level of complexity [22,23]. In the review, we focused on the individual steps of lignan biosynthesis and searched for available information on the molecular regulation of these processes. Given the need for a deeper knowledge of the stress adaptive, nutritional, and medicinal potential of lignans, we would like to contribute to the understanding of the regulatory role of miRNAs in these perfect metabolic mechanisms and provide an overview of current approaches for functional miRNA assays in secondary metabolites biosynthesis.
2. Plant Lignan Biosynthesis and Tissue Compartmentation
Lignans, belonging to the group of phenylpropane derivatives (phenylpropanoids), are biosynthesized in the cell cytoplasm through the action of phenylalanine ammonia lyase (PAL) and pinoresinol-lariciresinol reductase (PRL), which catalyze the lignans biosynthesis [5]. They are synthesized by the shikimate pathway using the aromatic amino acid phenylalanine [24]. The product of phenylalanine deamination is cinnamic acid, which is further reduced by the ester of coenzyme A to an intermediate aldehyde. Subsequent hydroxylation by the P450 enzyme leads to the formation of hydroxycinnamic acid (p-coumaric acid) and/or its polyhydroxyl analogs. Their methylation by O-methyltransferase results in monomeric phenylpropanoid subunits—cinnamic acid derivatives, with subsequent reduction by the NADPH enzyme to alcohol. These compounds—hydroxycinnamic acid derivatives, aldehydes, and alcohols—represent monomeric units in the biosynthesis of lignans or neolignans. Dimerizations of these monomers are mediated by laccases, or peroxidases, followed by postdimerization transformations by methylation and/or hydroxylation processes [24,25]. A schematic of the synthesis of lignans is illustrated in Figure 1.
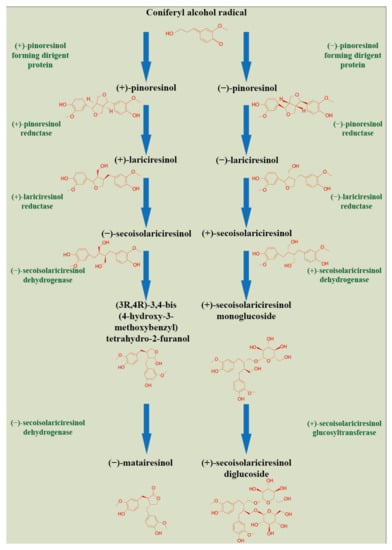
Figure 1.
Plant lignans biosynthesis. (Harenčár, modified based on [26]).
As was already mentioned, more information has been available on the action of miRNAs in lignin biosynthesis. Since the biosynthesis of lignan and lignin monolignols shares common steps, it is possible to apply this knowledge to both, including subsequent dimerization (lignans) or polymerization (lignin) of monolignols [27]. Both lignans and lignins are polyphenols derived from the shikimate pathway, sharing common phenylpropanoid precursors—monolignols. Subsequently, the biosynthesis of lignans and lignins branches off, either to the process of dimerization (lignans) or polymerization (lignin) of monolignols (Figure 2). The biosynthesis pathway of these polyphenols is also sharing common enzymes, including the monolignol dimerization or polymerization process, where laccases and peroxidases are actively present [28,29,30]. The regulation of monolignols production and polymerization, as well as lignan production, includes NAC (NAM, ATAF1/2, CUC2) and MYB transcription factors [31].
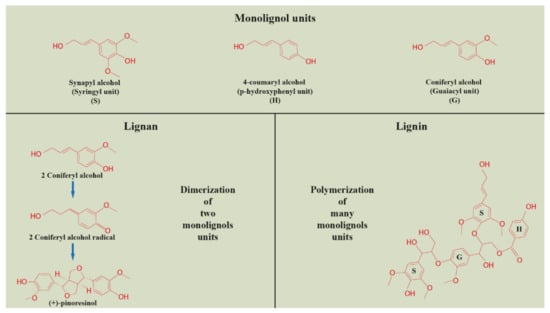
Figure 2.
Dimerization and polymerization of monolignols units in the process of lignan and lignin biosynthesis, respectively (Harenčár, modified based on [26]).
The biosynthetic pathway of lignin is divided into two branches: the phenylpropanoid pathway from phenylalanine to hydroxycinnamic acid and the monolignol pathway from hydroxycinnamic acid to monomeric phenylpropanoid subunits (monolignols) [32]. The structure of lignin is a heteropolymer composed of three hydroxycinnamyl alcohol monomers, coniferyl alcohol, p-coumaryl alcohol, and sinapyl alcohol, which differ in their degree of methoxylation [33]. These monolignols produce guaiacyl (G), p-hydroxyphenyl (H), and syringyl (S) phenylpropanoid units when incorporated into the lignin polymer [27]. As with lignans, the amount and composition of lignin vary depending on the plant species, developmental phase, tissue type, and environmental conditions [34]. The lignins of dicotyledonous plants are generally composed of a combination of G and S (with less H) units in comparison to monocotyledonous plants, where these phenylpropanoid units are almost evenly represented [33].
In lignin biosynthesis, 15 genes were identified, and the binding sites for lignification-associated transcription factors (AC-I, AC-II, AC-III, and AC-IV) were identified in their promoter regions [33]. These factors bind to the promoter regions through cis regulatory elements related to hormone response (to ABA, salicylic acid, and methyl jasmonate), environmental stress, and development stage [33]. The analysis also indicated that the genes involved in monolignol biosynthesis may represent a putative target for members of the following miRNA families miR160, miR164, miR166, miR167, miR169, miR171, miR 5384, and miR6223 [33]. Several miRNA families such as miR156, miR164, and miR397 are involved in the lignin metabolism, content, and composition [35]. The regulatory network of microRNA-Long Non-coding RNA-transcription factors showed that specifically the regulation patterns of lnc6873-MYB2, lnc4458-MYB330, and lnc6437-MYB308 may be involved in lignin biosynthesis [1]. The crucial role of miR397 in the direct regulation of 12 laccase genes and 4 peroxidase genes involved in lignin polymerization was verified, including MYB (MYB021, MYB52) and NAC (SND1-A2, VND6-C2) transcription factors, directly activated by these genes [36]. The specific features of lignans and lignin are shown in Table 1.

Table 1.
Different properties of plant lignans and lignins.
The enantiomeric composition of some lignans was already determined, for example, in trees, medicinal plants, and some plant foods (linseeds, sesame seeds). It is known that etantiomers of the same type of lignan may show different biological properties and elicit different biological effects [9]. Stereochemical characterization of lignans showed varied etantiomeric composition in the whole berries compared to the seeds [9].
The composition of polyphenols in the lignan macromolecule varies during the developmental stages of the flax seed [8]. Monolignol glucosides accumulated at the early stages of seed development in a free form, whereas SDG was bounded inside the lignan macromolecule and accumulated mainly in the later stages. One of the explanations for the lignan macromolecule formation is their role in lipid protection against oxidation. The oilseed lignans are concentrated in the hull and those of berries or fruits in the seeds or kernels [9]. However, in the case of sesame seeds, some of the lignans are concentrated in the hull (lariciresinol, cyclolariciresinol), whereas others (sesamin) are evenly distributed in the seed. The lignan—podophyllotoxin (PTOX) shows phytotoxicity; therefore, it must be stored in the vacuoles as glucosides of the PTOX-producing plant cells [37]. Lignans have been identified except seeds in flowering aerial parts [38], leaves, roots [39], fruits and vegetables [40], and wooden parts [41]. The immunohistochemical labeling allowed to localize the lignans mainly in the secondary wall of the sclerite cells of the outer integument of the seed, and very light labeling was also observed in cytoplasmic inclusions of the endosperm [42].
3. Genetic Regulation of Plant Lignan Biosynthesis
Dirigent (DIR) proteins are considered mediators of lignans and their biosynthesis in the plant response to abiotic stress [43]. The expression of selected DIR genes has been shown to be involved in the formation of (−)-pinoresinol in the seed coat and (+)-pinoresinol in vegetative organs [44]. The synthesis of lignans involves the dimerization of two coniferyl alcohols. This step, directed by a dirigent protein (DIR), leads to the formation of pinoresinol (PINO) [44]. Consequently, the pinoresinol is converted to lariciresinol (LARI) and then to secoisolariciresinol (SECO) by the pinoresinol-lariciresinol reductase (PLR) [28,45]. The conversion of the (−)-pinoresinol to (−)-lariciresinol is catalyzed by pinoresinol reductase (PrR), which is coded by two genes, PrR1 and PrR2. The promoter of the PrR1 gene is regulated by transcription factors SND1 and MYB46 [46]. The transcription activity of the key gene in lignan synthesis, pinoresinol-lariciresinol reductase (PLR), was studied by RT-PCR and promoter-reporter transgenesis during the flax seed development. LuPLR gene was expressed in the seed coat, and consequently, the synthesis of PLR enzyme in mature seeds confirmed its involvement in SDG synthesis [47]. To better understand the function of the PLR enzyme in lignan biosynthesis, the RNAi strategy using hairpin dsRNA structures was applied for functional analysis of the LuPLR1 gene [48]. Transgenic RNAi plants with silenced LuPLR1 gene failed to accumulate SDG. HPLC analysis of lignan and neolignane content displayed a dramatic decrease in SDG in transgenic seeds.
The glycosylation of secoisolariciresinol (SECO) into SDG (secoisolariciresinol diglucoside) is achieved by uridine glycosyltransferases (UGTs). It has been shown [49] that the UGT74S1 gene is the only one able to glycosylate SECO into SDG despite the identification of two duplicated genes (UGT74S4 and UGT74S3) closely related to UGT74S1 but incapable glycosylation.
Regulatory proteins known as transcription factors (TFs) are responsible for spatio-temporal synthesis and accumulation of secondary metabolites. TFs regulate the expression of target genes by binding to promoter-specific sequences, cis elements, in response to phytohormones and environmental conditions [50]. Investigation of spatio-temporal expression of DIR multigene family in flax genome revealed the function of specific DIRs in (−)-pinoresinol formation in seed-coats and (+)-pinoresinol in vegetative organs and/or specific responses to stress [44]. Phylogeny analysis grouped flax DIRs genes into six distinct clusters, where representatives of cluster DIR-a were connected to lignan biosynthesis, and those in clusters DIR-b/d and DIR-g have their putative function in lignin biosynthesis. A genome-wide analysis of the flax dirigent protein family records an extensive study by authors [44].
The enantiomeric composition of lignans is not only species-specific but also shows spatial and temporal specification. Similarly, pinoresinol-lariciresinol reductase (PLR) genes display spatio-temporal and organ-specific expression patterns [28,51]. Pinoresinol-lariciresinol reductases with opposite enantiospecificity determine the enantiomeric composition of lignans in different plant organs [51]. These enantiomers may represent the final products of lignans biosynthesis or are precursors for the synthesis of other groups of lignans [28]. Detailed characteristics of pinoresinol-lariciresinol reductases are provided in this review [28]. This suggests that the regulation of PRLs genes expression is crucial for lignans synthesis, and this statement is supported by the modification of PRLs gene expression patterns under in vitro induced stress conditions [47]. Stimulated expression of several lignan biosynthesis genes (DIR, PLR, and UGT) was observed under the elicitation effect of chitosan in addition to a significant increase in biomass of flax cell cultures [52].
4. The Involvement of miRNA in Plant Lignan Biosynthesis
Current trends in understanding the regulatory mechanisms of secondary metabolite biosynthesis point to a transcriptional level of regulation mediated by transcription factors, as well as to a post-transcriptional level of regulation with the involvement of interfering RNA molecules (miRNAs) [50,53]. There are several miRNA-mediated or miRNA-targeted modules of secondary metabolites regulation: miRNA-targeted biosynthesis-related genes, miRNA-targeted transcription factors (TFs), and miRNA-targeted noncoding RNAs (ncRNAs) [23]. The current observations pointed out that not only do miRNAs directly regulate lncRNAs but that they also indirectly regulate TFs through targeting of lncRNAs [1]. Regulatory machinery of gene expression is triggered through a combinatorial action of miRNAs and TFs [1,54]; additionally, they control the expression of each other. Most of the miRNA targets are transcription factors (TFs) which regulate plant growth, development, secondary metabolites biosynthesis, and stress tolerance [22]. The list of miRNA-targeted transcription factors involved in regulating flavonoid biosynthesis in plants is stated in the current study [55].
The phenylpropanoid pathway and the role of miRNAs in flavonoid biosynthesis is one of the most extensively studied [52,56]; however, the miRNA-based regulation of lignans is not characterized sufficiently. The schematic representation of the shikimate pathway leading to lignans precursors production followed by lignans biosynthesis and their interactions with known miRNAs is depicted in Figure 3.
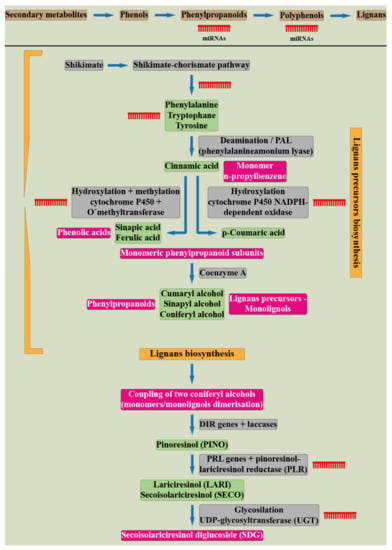
Figure 3.
The schematic representation of the lignans biosynthesis indicating the known points of the pathway in which the miRNAs participate (Ražná, Harenčár, elaborated based on [55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73].
A recent study [1] identified 80 differentially expressed genes (DEGs) involved in the phenylpropanoid biosynthesis pathway, which belong to 14 gene families (PAL, phenylalanine ammonia-lyase; 4CL, 4-coumarate-CoA ligase; C4H, cannamate-4-hydroxylase; CCR, cinnamoyl-CoA reductase; F5H, ferulate-5-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; CCoAOMT, caffeoyl-CoA O-methyltransferase; HCT, hydroxyl cinnamoyl transferase; CAD, cinnamyl-alcohol dehydrogenase; POD, peroxidase; C3H, p-coumaroyl ester 3-hydroxylase; CALDH, coniferyl-aldehyde dehydrogenase; β-G, beta-glucosidase; and CGT, coniferyl-alcohol glucosyltransferase. These DEGs were regulated by 23 miRNAs, whereas five 4CL genes were targeted by miR393a-5p, four C4H genes by Novel325/353/418/424 miRNAs, and C3H was targeted by Novel234 miRNA, which points to a significant effect of 4CL, C4H, and C3H genes on phenylpropanoid metabolism. In addition to this, the study [1] revealed that miRNAs and long non-coding RNAs (lncRNAs) indirectly regulate phenylpropanoid biosynthesis by targeting MYBs transcription factors, which act as repressors (MYB4, MYB39, MYB44, and MYB308) or activators (MYB2, MYB5, MYB12, MYB330, and MYB340) of phenylpropanoid metabolite pathway. This miRNAs-lncRNas-TFs co-regulation pattern contributes to tissue-specific variability of phenylpropanoid metabolites biosynthesis and is essential for regulating plant organ development [1,57].
Lignans, together with other polyphenolic metabolites (which are categorized into flavonoids), are synthesized by a central phenylpropanoid pathway, sharing common enzymes and substrates. Several miRNAs were recognized to regulate flavonoid biosynthesis by targeting the mRNAs encoding key enzymes [58] or transcription factors of the phenylpropanoid pathway [55]. O-methyltransferase in phenylalanine metabolism is targeted by miR1438. Activation of 4-coumarate and 4-hydroxycinnamates to the respective thiol esters is regulated by miR172i. Activated thiol esters are then used as a precursor of monolignols.
Cytochromes P450 (CYPs, CYP450, and P450) are heme-containing enzymes. They are known as monooxygenases catalyzing oxygenation/hydroxylation reactions in the biosynthesis of many secondary metabolites [59,60]. The microarray analysis of 326 CYP genes in the rice genome displayed tissue-specific and zone-specific expression patterns. At the same time, differential expression of several CYP genes was observed in response to drought stress. It has been confirmed that the expression of CYP genes can be post-transcriptionally regulated by miRNAs families, such as miR440, miR531, miR812, and by species-specific families miR1848, miR5523 (rice genome) [59]. It has been elucidated that CYPs involved in the phenylpropanoid pathway are primarily regulated and targeted by miR413 [60]. A Cytochrome P450 has been found to be a target for miR5035, miR2275d [61], and for lus-miR168b [62].
Phenylalanine ammonia-lyase (PAL) is the key enzyme of the phenylpropanoid pathway, participating in the precursor production of important secondary metabolites. PAL encoding genes are generally well studied [63]; however, the knowledge of the underlying regulatory mechanism of miRNAs involved in the PAL gene regulation does not reach the same level. Twelve common walnut PAL genes were targeted by several miRNAs belonging to miR172, miR399, miR408, miR477, miR830, and miR7729 families, pointing to tight regulation of PAL genes at the pre- and post-transcriptional level [63]. Inhibition of PAL gene expression by miR477 was observed as a response to pathogen infection [64]. The phenylalanine ammonia lyase (PAL) converting L-phenylalanine to ammonia and trans cinnamic acid is a target for miR1873 [65].
Laccases (LACs) belong to oxidase family enzymes (multicopper oxidoreductases). Due to containing four copper (Cu) ions, they can catalyze the oxidation of various aromatic and nonaromatic substrates, including oxidative polymerization of monolignols [66]. Laccase genes are responsible for the polymerization of monolignols, and post-transcriptional regulation of these genes by miRNAs was identified [29]. Almost 82% of soybean laccases are potential targets of miRNAs, such as miR397a/b, miR408d, and miR5671a, whereas maize laccase genes were potentially targeted by miR397a/b or miR528a/b. Expression analyses of soybean and maize laccase genes revealed tissue- and developmental-specific activity profile, including their specific response to abiotic and biotic stress factors [66]. Transcripts of six flax (Linum usitatissimum L.) laccase genes were targeted by lus-miR397 [67]. Almost 59% of identified laccase genes in Populus trichocarpa Torr. & Gray were predicted to be target of ptr-miR397a. Transgenics overexpressing in Ptr-MIR397a resulted in reduction of laccase genes expression as well as in lignin reduction, indicating that the biosynthesis of lignin is regulated by miRNA [68]. In the P. trichocarpa Torr. & Gray genome, three miR397 (miR397a, b, and c) were identified; however, only ptr-miR397a was sufficiently abundant to be detected [67].
The glycosylation of SECO into SDG is regulated by UDP-glycosyltransferases (UGTs, or uridine diphosphate glycosyltransferases). Flax UGT74S1 gene coding UDP-glycosyltransferases was reported as the key enzyme controlling SECO glycosylation in flax [69,70], using SECO as substrate to form SDG.
As lignans and lignins are phenylpropanoid substances sharing common enzymes and substrates [53], some of the knowledge of miRNA-mediated lignins regulation might be implemented in understanding miRNA-mediated lignans biosynthesis. Transcripts belonging to the blue copper protein (BCP) family are targeted by miR408. The functions of these proteins include oxygenase and oxidase activities and monolignol coupling catalysed by laccases within the cell wall [71]. In Arabidopsis, miR408 has been shown to regulate laccase genes involved in cell wall lignification [72]. Together with another two miRNAs (miR397 and miR857), it has been shown to target the transcripts for the members of the lignin-related laccase copper protein family [72]. The LIM1 gene, which may act as a transcriptional activator of lignin biosynthesis, is targeted by miR167h [73].
To obtain insight into the regulation of lignan biosynthesis during Schisandra chinensis (Turcz.) Baill. fruit development, long-read transcriptome sequencing was conducted. More than 21,800 assembled transcripts (or unigenes) were classified into 58 TF families, amongst which were identified as most abundant: bHLH, NAC, WRKY, MYB/−related, FAR1, C3H, B3, C2H2, ERF, GRAS, bZIP, HB-other, TALE, G2-like, YABBY, and Trihelix families [74]. Almost 13,000 unigenes were assigned to 20 primary pathways (according to KEGG pathway assignments), where the most represented were ‘metabolic pathways’ and ‘biosynthesis of secondary metabolites’. Subsequent analysis for transcripts related to ‘biosynthesis of secondary metabolites’ besides other pathways confirmed those related to ‘phenylpropanoid biosynthesis’ [74].
Several types of transcription factors have been implicated in the regulation of lignan biosynthesis. MYB (myeloblastosis) is a large family of proteins having different numbers of MYB domain repeats; for example, MYB-related or R2R3-MYB. MYB TF regulates, among others, the biosynthesis of secondary metabolites [9]. MYB-related transcription factors regulate flavonoid metabolism in plants and are incorporated in the regulation of phenylpropanoid metabolism [75]. MYB12 transcription factor belonging to the group of R2R3-MYB transcription factors represents a flavonol-specific regulator of phenylpropanoid biosynthesis [50,76] and is putatively targeted by miRNA858 [77]. The expression of several MYB transcription factors was downregulated by overexpression of miR858. Higher expression of MYBs in Arabidopsis thaliana (L.) Heynh. transgenic lines expressing artificial miRNA target mimics (MIM858) resulted in supported flavonoids synthesis at the cost of lignin synthesis [77].
The APETALA 2 (AP2)/ethylene-responsive factor (ERF) belongs to transcription factors (TFs) families, well-documented in plant responses to a wide range of biotic and abiotic stresses [78], in the control of primary and secondary metabolism and plant growth and development [79]. Positive regulation of plant lignans biosynthesis by APETALA2/ethylene response factor (AP2/ERF) was suggested. Knocking down the expression of li049, one novel AP2/ERF gene from Isatis indigotica Fort., caused a remarkable reduction of lignans/lignins contents [80]. AP2-domain-containing TF are also targets for lus-miR172e [81].
The involvement of a WRKY transcription factor in the regulation of lignans biosynthesis in flax was described. WRKY TFs bound to W box of LuPLR1 (pinoresinol-lariciresinol reductase 1) gene promoter, incorporated in secoisolariciresinol synthesis. It was also pointed out that LuWRKY36 positively regulates the LuPLR1 gene expression of lignans biosynthesis in response to biotic stress. More effective LuPLR1 gene expression was associated with higher secoisolariciresinol accumulation in the hypersensitive reaction of the resistant flax cultivar [28].
The scientific community has devoted considerable attention to the identification and characterization of miRNAs involved in the regulation of several secondary metabolites’ biosynthesis (Table 2). The progress made in this area is provided by several studies: Refs. [23,58,82]—describing microRNA-mediated regulation in flavonoid, terpenoid, alkaloid, and other N-containing metabolites biosynthesis; ref. [55]—pointed out the significance of miRNAs in the enhancement of flavonoid biosynthesis; ref. [83]—characterizes lncRNA mediated gene expression in flavonoids and terpenoids production in Citrus limon (L.) Osbeck; ref. [84]—describing miRNA role in phenylpropanoid, terpenoid, alkaloid, chlorophyll, and carotenoid biosynthesis; ref. [85]—identifies the role of miRNAs in phenylpropanoids, terpenoids, and alkaloids biosynthesis in medicinal plants; ref. [86]—is focused on lncRNAs regulatory potential in Populus trichocarpa Torr. & Gray phenylpropanoid pathway; ref. [87]—identifies miRNAs in secondary metabolites of Murraya koenigii (Linn.) Spreng; ref. [88,89] —states transcription factors and miRNAs mediated flavonol biosynthesis in Solanum tuberosum L.; ref. [90]—identifies miRNAs involved in artemisinin biosynthesis in Artemisia annua L.; ref. [91]—introduces miRNA-based regulation of secondary metabolites biosynthesis with pharmaceutical applications in medicinal plants; ref. [92]—reveals miRNA effects on secondary metabolism in Solanum tuberosum L.; and ref. [93]—characterizes the role of miRNA858 in phenylpropanoid pathway.

Table 2.
MiRNA and transcription factors involved in the phenylpropanoid biosynthesis regulation with focus on lignin/lignans target biosynthesis (in italics), elaborated based on references [1,2,3,4,31,33,35,36,46,57,68,94,95,96,97,98,99,100,101,102,103].
5. Current Approaches for Functional Identification of miRNAs in Plant Metabolites Biosynthesis
Identification and characterization of miRNAs participating in the regulatory mechanism of plant secondary metabolites is an important research task for the promising and targeted use of secondary metabolites and their effective modulation.
Many methodologies have evolved due to understanding microRNA silencing complex and their impact on plants organism. Within genetic engineering exist two more approaches besides the knock-outing of mentioned enzymes or factors. The first one lies in an over-expression of miRNAs and the second one in inhibition of miRNAs. Into the first category belongs technology AmiRNA or syn-tasiRNA (Figure 4). Based on a genomic construct composed of a cloning vector and primary microRNA with a stem-loop structure, a specific miRNA is over-expressed and silences genes effectively [104,105].
Among techniques that use a silencing of microRNAs are noncoding endogenous target mimicry (eTM), short tandem target mimic (STTM), or miRNA sponges. Mimicries are endogenous non-coding or low-coding sequences with different lengths. They are part of long non-coding RNA (lncRNA), and their epigenetic function is inhibition of microRNAs by mutual binding, which creates a mismatch loop in the middle of specific miRNA. Due to this incomplementarity, the miRNA is not able to cleavage and silence other mRNAs [106,107]. Based on this knowledge, the short tandem target mimic (STTM) technology was developed to inhibition of endogenous microRNAs. The main point again lies in a design of a specific STTM construct and a genetic transformation [108,109,110]. A MicroRNA sponge is an expanded methodology where a construct usually contains a whole family of microRNA, e.g., MIR-172a…MIR-172p (Figure 5) [111].
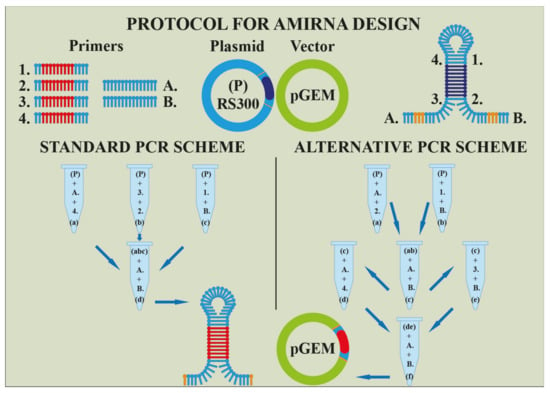
Figure 4.
An illustration of projecting artificial MicroRNA or syn-tasiRNA, respectively (Harenčár, modified based on [112,113,114]). The artificial MicroRNA is made by a sequential adding of primers (1.; 2.; 3.; 4.; A.; B.;) to the main template (P) and its intermediate products ((a), (b), (c), respectively (d)) during individual PCR reactions. The final product ((d) or (f)) is transferred to the vector pGEM.
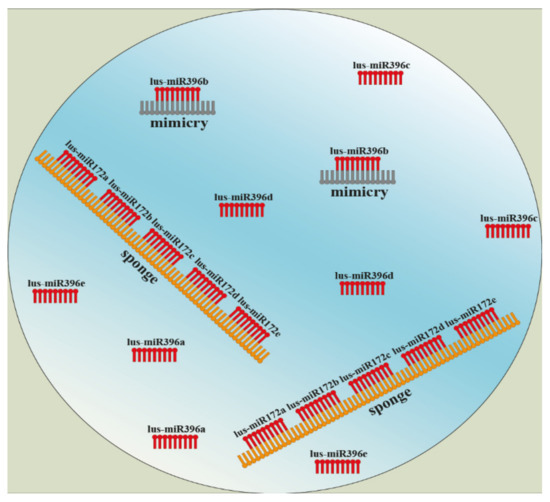
Figure 5.
Design and main principle of microRNA sponge and noncoding endogenous target mimicry (eTM) technology. (Harenčár, modified based on [111,115,116]).
For the purpose of inhibiting microRNA, it was found that small molecules can also stop a miRNA maturing by binding onto primary miRNA and preventing it from Dicer cleavage [117,118,119,120,121]. This technology is also commercially available [122,123]. Small molecules can be used as well as enhancers of microRNA cleavage [124]. Scientists identified several plant viruses that caused abnormalities by a specific microRNA upregulation [125] (Figure 6). Imaging of gene silencing in real time is also interesting [126,127].
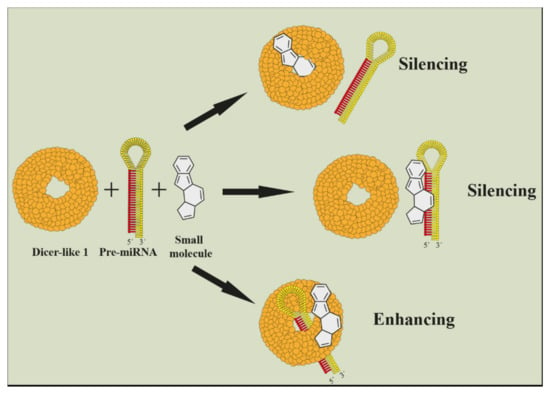
Figure 6.
Illustration of small molecule as enhancer or inhibitor of a microRNA maturation (Harenčár, modified based on [117,118,119,120,121,122,123,124]).
In silico approach represents one of the fastest developing ways how to understand the main role and big potential of microRNAs not only in plant secondary metabolites synthesis. Currently, as a primary source of information serves databases, and among the most famous belongs miRbase [118]. Plenty of software and algorithms use these data and process them according to established criteria and intention. Because microRNAs complementary pairing with their target gene sequences, algorithms used to work with computations based on permitted match and mismatch eventually on free energy [128]. Scientists usually make their algorithms accessible in the form of command lines written by a specific programming language executable in the power shell of the operating system, for example, Novomir [129]. Another has a user interface; however, they use a processor of personal computers such as p-TAREF [130], or it exists online web servers with a shared supercomputer, machine learning, adjustable criteria, and database of microRNA sequences. One of them is psRNATarget [131,132]. Many reviews and web pages follow up tools for microRNA prediction [128,133,134]. Due to the designing of constructs and genetic engineering with microRNAs, bioinformatic models were devised that help to succeed in transgenesis. Some techniques mentioned above (amirna, sponge, SSTM, syn-tasiRNA, etc.) have their own in silico design servers as well, e.g., [135,136,137,138]. RNA-induced silencing complex is quite a novel part of epigenetics applicable in medicine, nutritional science, metabolomics, and genomics and, in combination with bioinformatics, represents a big scientific potential for the future. For this reason, methods, techniques, and algorithms will be on the increase [139,140,141,142,143].
6. Conclusions
Due to the biological importance of secondary metabolites, understanding the regulatory mechanisms of their biosynthesis is crucial [36]. Plant lignans hold significant biological potential, not only in terms of nutritional and medicinal use, but also in terms of plant adaptive potential to environmental stress. Lignans biosynthesis pathway is known in detail; however, the knowledge of the involvement of miRNAs in regulatory processes of the biosynthesis is partial. The current understanding is limited to identification of miRNA-mediated regulation or to characterization of involved transcription factors of phenylpropanoid pathway and its individual groups of metabolites (alkaloids, flavonoids, and terpenoids). The review is focused on the individual steps of lignans biosynthesis and matching identified and characterized miRNAs and transcription factors (as primary target sequences for miRNAs) to make a mosaic of miRNA-modulated regulation of plant lignans biosynthesis. Whereas understanding the role of miRNAs in the regulation of plant lignan biosynthesis represents an important research task, the review presents several current approaches to miRNA functional analysis. We believe that the summarized information will create a knowledge platform for further screening of miRNA functions and thus contribute to a more detailed understanding of miRNA-based regulation of lignans biosynthesis.
Author Contributions
Conceptualization, K.R. and Ľ.H.; writing—original draft preparation, K.R., Ľ.H. and M.K.; writing—review and editing, K.R., Ľ.H. and M.K.; visualization, Ľ.H. and K.R.; supervision, K.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This publication was supported by the Operational program Integrated Infrastructure within the project: Demand-driven research for sustainable and innovative food, Drive4SIFood 313011V336, co-financed by the European Regional Development Fund.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tu, Z.; Xia, H.; Yang, L.; Zhai, X.; Shen, Y.; Li, H. The Roles of microRNA-Long Non-coding RNA-mRNA Networks in the Regulation of Leaf and Flower Development in Liriodendron chinense. Front. Plant Sci. 2022, 13, 816875. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Kumar, P.; Sanyal, R.; Mane, D.B.; Prasanth, D.A.; Patil, M.; Dey, A. Unravelling the regulatory role of miRNAs in secondary metabolite production in medicinal crops. Plant Gene 2021, 27, 100303. [Google Scholar] [CrossRef]
- Adjei, M.O.; Zhou, X.; Mao, M.; Rafique, F.; Ma, J. MicroRNAs Roles in Plants Secondary Metabolism. Plant Signal. Behav. 2021, 16, 1915590. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.; Chen, L.; Xiao, Y.; Wu, J.; Zeng, L.; Li, H.; Wu, Q.; Hu, Z. Recent Advanced Metabolic and Genetic Engineering of Phenylpropanoid Biosynthetic Pathways. Int. J. Mol. Sci. 2021, 22, 9544. [Google Scholar] [CrossRef]
- DellaGreca, M.; Zuppolini, S.; Zarrelli, A. Isolation of lignans as seed germination and plant growth inhibitors from Mediterranean plants and chemical synthesis of some analogues. Phytochem. Rev. 2013, 12, 717–731. [Google Scholar] [CrossRef]
- Zálešák, F.; Bon, D.J.Y.D.; Pospíšil, J. Plant secondary metabolites as a reservoir of biologically active substances. Pharmacol. Res. 2019, 146, 104284. [Google Scholar] [CrossRef]
- Barker, D. Lignans. Molecules 2019, 24, 1424. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, A.; Fliniaux, O.; Quéro, A.; Molinié, R.; Demailly, H.; Hano, C.; Paetz, C.; Roscher, A.; Grand, E.; Kovensky, J.; et al. Kinetics of the incorporation of the main phenolic compounds into the lignan macromolecule during flaxseed development. Food Chem. 2017, 15, 1–8. [Google Scholar] [CrossRef]
- Smeds, A.I.; Eklund, P.C.; Willför, S.M. Content, composition, and stereochemical characterization of lignans in berries and seeds. Food Chem. 2012, 134, 1991–1998. [Google Scholar] [CrossRef]
- Yang, K.; Han, H.; Li, Y.; Ye, J.; Xu, F. Significance of miRNA in enhancement of flavonoid biosynthesis. Plant Biol. J. 2022, 24, 217–226. [Google Scholar] [CrossRef]
- Schmidt, T.J.; Hemmati, S.; Klaes, M.; Konuklugil, B.; Mo-Hagheghzadeh, A.; Ionkova, I.; Fuss, E.; Wilhelm Alfer-mann, A. Lignans in flowering aerial parts of Linum species--chemodiversity in the light of systematics andphylogeny. Phytochemistry 2010, 71, 1714–1728. [Google Scholar] [CrossRef]
- Schmidt, T.J.; Klaes, M.; Sendker, J. Lignans in seeds of Linum species. Phytochemistry 2012, 82, 89–99. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Kim, K.W.; Lee, C.S.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: Integrated omics and MALDI mass spectrometry imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef]
- Kasote, D.M. Flaxseed phenolics as natural antioxidants. Int. Food. Res. J. 2013, 20, 27. [Google Scholar]
- Bjelková, M.; Filip, V.; Kyselka, J.; Ševčík, R.; Větrovcová, M. Výběr a charakteristika lněného semene jako vstupní suroviny (Selection and Characteristics of Flaxseed as an Input Raw Material); Agritec Plant Research Ltd.: Šumperk, Czech Republic, 2017; ISBN 978-80-87360-57-6. [Google Scholar]
- Garros, L.; Drouet, S.; Corbin, C.; Decourtil, C.; Fidel, T.; Lebas de Lacour, J.; Leclerc, E.A.; Renouard, S.; Tungmunnithum, D.; Doussot, J.; et al. Insight into the Influence of Cultivar Type, Cultivation Year, and Site on the Lignans and Related Phenolic Profiles, and the Health-Promoting Antioxidant Potential of Flax (Linum usitatissimum L.) Seeds. Molecules 2018, 23, 2636. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wu, G.; Yuan, H.; Cheng, L.; Zhao, D.; Huang, W.; Zhang, S.; Zhang, L.; Chen, H.; Zhang, J.; et al. Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. 2016, 16, 124. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Khare, T.; Kumar, V. Recent trends and advances in identification and functional characterization of plant miRNAs. Acta Physiol. Plant 2020, 42, 25. [Google Scholar] [CrossRef]
- Millar, A.A. The function of miRNAs in plants. Plants 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Sand, M. The pathway of miRNA maturation. miRNA Maturation 2014, 1095, 3–10. [Google Scholar]
- Sabzehzari, M.; Naghavi, M.R. Phyto-miRNAs-based regulation of metabolites biosynthesis in medicinal plants. Gene 2019, 682, 13–24. [Google Scholar] [CrossRef]
- Samad, A.F.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Cui, J.L.; Li, X.K. MicroRNA-Mediated Gene Regulation of Secondary Metabolism in Plants. Crit Rev. Plant Sci. 2021, 40, 459–478. [Google Scholar] [CrossRef]
- Teponno, R.B.; Kusari, S.; Spiteller, M. Recent advances in research on lignans and neolignans. Nat. Prod. Rep. 2016, 33, 1044–1092. [Google Scholar] [CrossRef] [Green Version]
- Touré, A.; Xueming, X. Flaxseed lignans: Source, biosynthesis, metabolism, antioxidant activity, bio-active components, and health benefits. Compr. Rev. Food. Sci. Food Saf. 2010, 9, 261–269. [Google Scholar] [CrossRef]
- MetaCyc. Available online: https://metacyc.org/ (accessed on 28 March 2022).
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Markulin, L.; Corbin, C.; Renouard, S.; Drouet, S.; Durpoix, C.; Mathieu, C.; Lopez, T.; Augin, D.; Hano, C.; Lainé, É. Characterization of LuWRKY36, a flax transcription factor promoting secoisolariciresinol biosynthesis in response to Fusarium oxysporum elicitors in Linum usitatissimum L. hairy roots. Planta 2019, 250, 347–366. [Google Scholar] [CrossRef]
- Le Roy, J.; Blervacq, A.S.; Créach, A.; Huss, B.; Hawkins, S.; Neutelings, G. Spatial regulation of monolignol biosynthesis and laccase genes control developmental and stress-related lignin in flax. BMC Plant Biol. 2017, 17, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, N.G.; Davin, L.B.; Sarkanen, S. Lignin and Lignan Biosynthesis: Distinctions and Reconciliations. Chapter 1. In ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1998. [Google Scholar]
- Huis, R.; Morreel, K.; Fliniaux, O.; Lucau-Danila, A.; Fénart, S.; Grec, S.; Neutelings, G.; Chabbert, B.; Mesnard, F.; Boerjan, W.; et al. Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol. 2012, 158, 1893–1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.L.; Bhoo, S.H.; Kwon, M.; Lee, S.W.; Cho, M.H. Biochemical and Expression Analyses of the Rice Cinnamoyl-CoA Reductase Gene Family. Front. Plant Sci. 2017, 8, 2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jardim-Messeder, D.; Felix-Cordeiro, T.; Barzilai, L.; de Souza-Vieira, Y.; Galhego, V.; Bastos, G.A.; Valente-Almeida, G.; Aiube, Y.R.A.; Faria-Reis, A.; Corrêa, R.L.; et al. Genome-wide analysis of general phenylpropanoid and monolignol-specific metabolism genes in sugarcane. Funct. Integr. Genom. 2021, 21, 73–99. [Google Scholar] [CrossRef]
- Moura, J.C.M.S.; Bonine, C.A.V.; De Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and Biotic Stresses and Changes in the Lignin Content and Composition in Plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Trumbo, J.L.; Zhang, B.; Stewart, C.N., Jr. Manipulating microRNAs for improved biomass and biofuels from plant feedstocks. Plant Biotechnol. J. 2015, 13, 337–354. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Li, Q.; Wei, H.; Chang, M.J.; Tunlaya-Anukit, S.; Kim, H.; Liu, J.; Song, J.; Sun, Y.H.; Yuan, L.; et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2013, 25, 10848–10853. [Google Scholar] [CrossRef] [Green Version]
- Fuss, E. Lignans in plant cell and organ cultures: An overview. Phytochem. Rev. 2003, 2, 307–320. [Google Scholar] [CrossRef]
- Consonni, R.; Ottolina, G. NMR Characterization of Lignans. Molecules 2022, 27, 2340. [Google Scholar] [CrossRef]
- Willför, S.M.; Smeds, A.I.; Holmboma, B.R. Chromatographic analysis of lignans. J. Chromatogr. A 2006, 1112, 64–77. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Sánchez-Quesada, C.; Toledo, E.; Del-gado-Rodríguez, M.; Gaforio, J.J. Naturallylignan-rich foods: A dietary tool for health promotion? Molecules 2019, 24, 917. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Umezawa, T.; Shimada, M. Stereo-chemical diversity in lignan biosynthesis of Arctium lappa L. Biosci. Biotechnol. Biochem. 2002, 66, 1262–1269. [Google Scholar] [CrossRef] [Green Version]
- Attoumbré, J.; Bienaimé, C.; Dubois, F.; Fliniaux, M.A.; Chabbert, B.; Baltora-Rosseta, S. Development of antibodies against secoisolariciresinol—Application to the immunolocalization of lignans in Linum usitatissimum seeds. Phytochemistry 2010, 71, 1979–1987. [Google Scholar] [CrossRef]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef] [Green Version]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Lainé, É.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef]
- Anjum, S.; Komal, A.; Drouet, S.; Kausar, H.; Hano, C.; Abbasi, B.H. Feasible production of lignans and neolignans in root-derived in vitro cultures of flax (Linum usitatissimum L.). Plants 2020, 9, 409. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Zeng, Y.; Yin, Y.; Pu, Y.; Jackson, L.A.; Engle, N.L.; Martin, M.Z.; Tschaplinski, T.J.; Ding, S.Y.; Ragauskas, A.J.; et al. Pinoresinol reductase 1 impacts lignin distribution during secondary cell wall biosynthesis in Arabidopsis. Phytochemistry 2015, 112, 170–178. [Google Scholar] [CrossRef] [Green Version]
- Hano, C.; Martin, I.; Fliniaux, O.; Legrand, B.; Gutierrez, L.; Arroo, R.R.J.; Mesnard, F.; Lamblin, F.; Lainé, E. Pinoresinol–lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 2006, 224, 1291–1301. [Google Scholar] [CrossRef]
- Renouard, S.; Tribalatc, M.A.; Lamblin, F.; Mongelard, G.; Fliniaux, O.; Corbin, C.; Marosevic, D.; Pilard, S.; Demailly, H.; Gutierrez, L.; et al. RNAi-mediated pinoresinol lariciresinol reductase gene silencing in flax (Linum usitatissimum L.) seed coat: Consequences on lignans and neolignans accumulation. J. Plant Physiol. 2014, 171, 1372–1377. [Google Scholar] [CrossRef]
- Fofana, B.; Ghose, K.; McCallum, J.; You, F.M.; Cloutier, S. UGT74S1 is the key player in controlling secoisolariciresinol diglucoside (SDG) formation in flax. BMC Plant Biol. 2017, 17, 35. [Google Scholar] [CrossRef] [Green Version]
- Patra, B.; Schluttenhofer, C.; Wu, Y.; Pattanaik, S.; Yuan, L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta. Gene. Regul. Mech. 2013, 1829, 1236–1247. [Google Scholar] [CrossRef]
- Hemmati, S.; von Heimendahl, C.B.; Klaes, M.; Alfermann, A.W.; Schmidt, T.J.; Fuss, E. Pinoresinol-lariciresinol reductases with opposite enantiospecificity determine the enantiomeric composition of lignans in the different organs of Linum usitatissimum L. Planta Med. 2010, 76, 928–934. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, W.; Zahir, A.; Nadeem, M.; Garros, L.; Drouet, S.; Renouard, S.; Doussot, J.; Giglioli-Guivarc’h, N.; Hano, C.; Abbasi, B.H. Enhanced production of lignans and neolignans in chitosan-treated flax (Linum usitatissimum L.) cell cultures. Process. Biochem. 2019, 79, 155–165. [Google Scholar] [CrossRef]
- Gupta, O.P.; Karkute, S.G.; Banerjee, S.; Meena, N.L.; Dahuja, A. Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Front. Plant Sci. 2017, 8, 374. [Google Scholar] [CrossRef] [Green Version]
- Arora, S.; Rana, R.; Chhabra, A.; Jaiswal, A.; Rani, V. miRNA–transcription factor interactions: A combinatorial regulation of gene expression. Mol. Genet. Genom. 2013, 288, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Šamec, D.; Karalija, E.; Šola, I.; Vujčić Bok, V.; Salopek-Sondi, B. The Role of Polyphenols in Abiotic Stress Response: The Influence of Molecular Structure. Plants 2021, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezawa, T. The cinnamate/monolignol pathway. Phytochem. Rev. 2010, 9, 1–17. [Google Scholar] [CrossRef]
- Marcela, V.; Gerardo, V.; Agustín, A.C.; Antonio, G.M.; Oscar, R.; Diego, C.; Cruz-Hernández, A. MicroRNAs Associated with Secondary Metabolites Production. In Plant Physiological Aspects of Phenolic Compounds; Soto-Hernández, M., García-Mateos, R., Palma-Tenango, M., Eds.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Chen, H. Global identification, structural analysis and expression characterization of cytochrome P450 monooxygenase superfamily in rice. BMC Genom. 2018, 19, 35. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Xu, X.; Xu, X.; Li, Y.; Zhao, P.; Chen, X.; Shen, X.; Zhang, Z.; Chen, Y.; Liu, S.; et al. Genome-wide identification, evolution analysis of cytochrome P450 monooxygenase multigene family and their expression patterns during the early somatic embryogenesis in Dimocarpus longan Lour. Gene 2022, 826, 146453. [Google Scholar] [CrossRef]
- Biswas, S.; Hazra, S.; Chattopadhyay, S. Identification of conserved miRNAs and their putative target genes in Podophyllum hexandrum (Himalayan Mayapple). Plant Gene 2016, 6, 82–89. [Google Scholar] [CrossRef] [Green Version]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Qiu, S.; Rollins, M.; Datla, R.; Gupta, V.S.; Kadoo, N.Y. Genome-wide identification and characterization of microRNA genes and their targets in flax (Linum usitatissimum): Characterization of flax miRNA genes. Planta 2013, 237, 1149–1161. [Google Scholar] [CrossRef]
- Yan, F.; Li, H.; Zhao, P. Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.). Genes 2019, 10, 46. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Liu, S.; Liu, L.; Li, R.; Guo, R.; Xia, X.; Wei, C. miR477 targets the phenylalanine ammonia-lyase gene and enhances the susceptibility of the tea plant (Camellia sinensis) to disease during Pseudopestalotiopsis species infection. Planta 2020, 251, 59. [Google Scholar] [CrossRef]
- Singh, N.; Srivastava, S.; Sharma, A. Identification and analysis of miRNAs and their targets in ginger using bioinformatics approach. Gene 2016, 10, 570–576. [Google Scholar] [CrossRef]
- Liu, M.; Dong, H.; Wang, M.; Liu, Q. Evolutionary divergence of function and expression of laccase genes in plants. J. Genet. 2020, 99, 23. [Google Scholar] [CrossRef]
- Li, C.; Li, D.; Zhou, H.; Li, J.; Lu, S. Analysis of the laccase gene family and miR397-/miR408-mediated posttranscriptional regulation in Salvia miltiorrhiza. PeerJ 2019, 7, e7605. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Zhang, S.; Yu, Y.; Luo, Y.C.; Liu, Q.; Ju, C.; Zhang, Y.C.; Qu, L.H.; Lucas, W.J.; Wang, X.; et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef] [Green Version]
- Ghose, K.; Selvaraj, K.; McCallum, J.; Kirby, C.W.; Sweeney-Nixon, M.; Cloutier, S.J.; Deyholos, M.; Datla, R.; Fofana, B. Identification and functional characterization of a flax UDP-glycosyltransferase glucosylating secoisolariciresinol (SECO) into secoisolariciresinol monoglucoside (SMG) and diglucoside (SDG). BMC Plant Biol. 2014, 28, 82. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, P.; Sangwan, R.S.; Sangwas, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef]
- Neutelings, G.; Fénart, S.; Lucau-Danila, A.; Hawkins, S. Identification and characterization of miRNAs and their potential targets in flax. J. Plant Physiol. 2012, 169, 1754–1766. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 23, 15932–15945. [Google Scholar] [CrossRef] [Green Version]
- Arnaud, D.; Déjardin, A.; Leplé, J.C.; Lesage-Descauses, M.C.; Boizot, N.; Villar, M.; Bénédetti, H.; Pilate, G. Expression analysis of LIM gene family in poplar, toward an updated phylogenetic classification. BMC Res. Notes 2012, 17, 102. [Google Scholar] [CrossRef] [Green Version]
- Hong, C.P.; Kim, C.K.; Lee, D.J.; Jeong, H.J.; Lee, Y.; Park, S.G.; Kang, S.H. Long-read transcriptome sequencing provides insight into lignan biosynthesis during fruit development in Schisandra chinensis. BMC Genom. 2022, 23, 17. [Google Scholar] [CrossRef]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, D.; Tiwari, M.; Pandey, A.; Bhatia, C.; Sharma, A.; Trivedi, P.K. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 2016, 171, 944–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C.H. Ethylene Response Factor (ERF) Family Proteins in Abiotic Stresses and CRISPR–Cas9 Genome Editing of ERFs for Multiple Abiotic Stress Tolerance in Crop Plants: A Review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Ma, R.; Xiao, Y.; Lv, Z.; Tan, H.; Chen, R.; Li, Q.; Chen, J.; Wang, Y.; Yin, J.; Zhang, L.; et al. AP2/ERF transcription factor, Ii049, positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Front. Plant Sci. 2017, 8, 1361. [Google Scholar] [CrossRef] [Green Version]
- Xie, d.; Yu, Y.; Dai, Z.; Sun, J.; Su, J. Identification and characterization of miRNAs and target genes in developing flax seeds by multigroup analysis. Biotechnol. Biotechnol. Equip. 2021, 35, 538–550. [Google Scholar] [CrossRef]
- Hossain, R.; Quispe, C.; Saikat, A.S.M.; Jain, D.; Habib, A.; Janmeda, P.; Radha, M.T.I.; Daştan, S.D.; Kumar, M.; Butnariu, M.; et al. Biosynthesis of Secondary Metabolites Based on the Regulation of MicroRNAs. BioMed Res. Int. 2022, 2022, 9349897. [Google Scholar] [CrossRef]
- Bordoloi, K.S.; Baruah, P.M.; Das, M.; Agarwala, N. Unravelling lncRNA mediated gene expression as potential mechanism for regulating secondary metabolism in Citrus limon. Food Biosci. 2022, 46, 101448. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Avramenko, T.V. New opportunities for the regulation of secondary metabolism in plants: Focus on microRNAs. Biotechnol. Lett. 2015, 37, 1719–1727. [Google Scholar] [CrossRef]
- Xie, W.; Weng, A.; Melzig, M.F. MicroRNAs as new bioactive components in medicinal plants. Planta Med. 2016, 82, 1153–1162. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ge, X.; Du, J.; Cheng, X.; Peng, X.; Hu, J. Genome-Wide Identification of Long Non-Coding RNAs and Their Potential Functions in Poplar Growth and Phenylalanine Biosynthesis. Front. Genet. 2021, 12, 762678. [Google Scholar] [CrossRef]
- Gutiérrez-García, C.; Ahmed, S.S.; Ramalingam, S.; Selvaraj, D.; Srivastava, A.; Paul, S.; Sharma, A. Identification of microRNAs from Medicinal Plant Murraya koenigii by High-Throughput Sequencing and Their Functional Implications in Secondary Metabolite Biosynthesis. Plants 2021, 11, 46. [Google Scholar] [CrossRef]
- Lin, S.; Singh, R.K.; Moehninsi, M.; Navarre, D.A. R2R3-MYB transcription factors, StmiR858 and sucrose mediate potato flavonol biosynthesis. Hortic. Res. 2021, 8, 25. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhang, J.; Zhang, J.; Wang, Z.; Ran, A.; Guo, H.; Wang, D.; Zhang, J. Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Mol. Genet. Genom. 2017, 292, 37–52. [Google Scholar] [CrossRef]
- Khan, S.; Ali, A.; Saifi, M.; Saxena, P.; Ahlawat, S.; Abdin, M.Z. Identification and the potential involvement of miRNAs in the regulation of artemisinin biosynthesis in A. annua. Sci. Rep. 2020, 10, 13614. [Google Scholar] [CrossRef]
- Sabzehzari, M.; Naghavi, M.R. Phyto-miRNA: A molecule with beneficial abilities for plant biotechnology. Gene 2019, 683, 28–34. [Google Scholar] [CrossRef]
- Ding, C.; Shen, T.; Ran, N.; Zhang, H.; Pan, H.; Su, X.; Xu, M. Integrated Degradome and Srna Sequencing Revealed miRNA-mRNA Regulatory Networks between the Phloem and Developing Xylem of Poplar. Int. J. Mol. Sci. 2022, 23, 4537. [Google Scholar] [CrossRef]
- Li, Y.; Cui, W.; Qi, X.; Lin, M.; Qiao, C.; Zhong, Y.; Hu, C.; Fang, J. MicroRNA858 negatively regulates anthocyanin biosynthesis by repressing AaMYBC1 expression in kiwifruit (Actinidia arguta). Plant Sci. 2020, 296, 110476. [Google Scholar] [CrossRef]
- Ahmed, M.; Ahmed, F.; Ahmed, J.; Akhand, M.R.N.; Azim, K.F.; Imran, M.A.S.; Hoque, S.F.; Hasan, M. In silico identification of conserved miRNAs in the genome of fibre biogenesis crop Corchorus capsularis. Heliyon 2021, 7, e06705. [Google Scholar] [CrossRef]
- Zhang, L.; Xia, H.; Wu, J.; Li, M. MiRNA identification, characterization and integrated network analysis for flavonoid biosynthesis in Brassica raphanus. Hortic. Plant J. 2022, 8, 319–327. [Google Scholar] [CrossRef]
- Hassani, D.; Fu, X.; Shen, Q.; Khalid, M.; Rose, J.K.C.; Tang, K. Parallel Transcriptional Regulation of Artemisinin and Flavonoid Biosynthesis. Trends Plant Sci. 2020, 25, 466–476. [Google Scholar] [CrossRef]
- Geng, D.; Shen, X.; Xie, Y.; Yang, Y.; Bian, R.; Gao, Y.; Li, P.; Sun, L.; Feng, H.; Ma, F.; et al. Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 2020, 7, 102. [Google Scholar] [CrossRef]
- Liu, J.; Fan, H.; Wang, Y.; Han, C.; Wang, X.; Yu, J.; Li, D.; Zhang, Y. Genome-wide microRNA profiling using oligonucleotide microarray reveals regulatory networks of microRNAs in Nicotiana benthamiana during Beet necrotic yellow vein virus infection. Viruses 2020, 12, 310. [Google Scholar] [CrossRef] [Green Version]
- Jeena, G.S.; Singh, N.; Shukla, R.K. An insight into microRNA biogenesis and its regulatory role in plant secondary metabolism. Plant Cell Rep. 2022, 1–21. [Google Scholar] [CrossRef]
- Wan, F.; Zhang, L.; Tan, M.; Wang, X.; Wang, G.L.; Qi, M.; Liu, B.; Gao, J.; Pan, Y.; Wang, Y. Genome-wide identification and characterization of laccase family members in eggplant (Solanum melongena L.). PeerJ 2022, 10, e12922. [Google Scholar] [CrossRef]
- Camargo-Ramírez, R.; Val-Torregrosa, B.; San Segundo, B. MiR858-Mediated Regulation of Flavonoid-Specific MYB Transcription Factor Genes Controls Resistance to Pathogen Infection in Arabidopsis. Plant Cell Physiol. 2018, 59, 190–204. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Li, Q.; Tan, H.; Chen, J.; Xiao, Y.; Ma, R.; Gao, S.; Zerbe, P.; Chen, W.; Zhang, L. Gene-to-metabolite network for biosynthesis of lignans in MeJA-elicited Isatis indigotica hairy root cultures. Front. Plant Sci. 2015, 3, 952. [Google Scholar] [CrossRef] [Green Version]
- Guillaumie, S.; Mzid, R.; Méchin, V.; Léon, C.; Hichri, I.; Destrac-Irvine, A.; Trossat-Magnin, C.; Delrot, S.; Lauvergeat, V. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 2010, 72, 215. [Google Scholar] [CrossRef]
- Carbonell, A.; Takeda, A.; Fahlgren, N.; Johnson, S.C.; Cuperus, J.T.; Carrington, J.C. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014, 165, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Mickiewicz, A.; Rybarczyk, A.; Sarzynska, J.; Figlerowicz, M.; Blazewicz, J. AmiRNA Designer-new method of artificial miRNA design. Acta Biochim. Pol. 2016, 63, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karakülah, G.; Kurtoğlu, K.Y.; Unver, T. PeTMbase: A database of plant endogenous target mimics (eTMs). PLoS ONE 2016, 11, e0167698. [Google Scholar] [CrossRef] [PubMed]
- Borah, P.; Das, A.; Milner, M.J.; Ali, A.; Bentley, A.R.; Pandey, R. Long non-coding RNAs as endogenous target mimics and exploration of their role in low nutrient stress tolerance in plants. Genes 2018, 9, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, W.; Zhao, N.; Xiao, B.; Cao, P.; Wu, X.; Ye, C.; Shen, E.; Qiu, J.; Zhu, Q.-H.; et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiol. 2015, 169, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Peng, T.; Qiao, M.; Liu, H.; Teotia, S.; Zhang, Z.; Zhao, Y.; Wang, B.; Zhao, D.; Shi, L.; Zhang, C.; et al. A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol. Plant 2018, 11, 1400–1417. [Google Scholar] [CrossRef] [Green Version]
- Barta, T.; Peskova, L.; Hampl, A. miRNAsong: A web-based tool for generation and testing of miRNA sponge constructs in silico. Sci. Rep. 2016, 6, 36625. [Google Scholar] [CrossRef]
- Eamens, A.L.; McHale, M.; Waterhouse, P.M. The use of artificial microRNA technology to control gene expression in Arabidopsis thaliana. Methods Mol. Biol. 2014, 1062, 211–224. [Google Scholar]
- Kotowska-Zimmer, A.; Pewinska, M.; Olejniczak, M. Artificial miRNAs as therapeutic tools: Challenges and opportunities. Wiley Interdiscip. Rev. RNA 2021, 12, e1640. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Trivedi, P.K. Artificial microRNA mediated gene silencing in plants: Progress and perspectives. Plant Mol. Biol. 2014, 86, 1–18. [Google Scholar] [CrossRef]
- Kluiver, J.; Gibcus, J.H.; Hettinga, C.; Adema, A.; Richter, M.K.; Halsema, N.; Slezak-Prochazka, I.; Ding, Y.; Kroesen, B.J.; van den Berg, A. Rapid generation of microRNA sponges for microRNA inhibition. PLoS ONE 2012, 7, e29275. [Google Scholar] [CrossRef] [Green Version]
- Kluiver, J.; Slezak-Prochazka, I.; Smigielska-Czepiel, K.; Halsema, N.; Kroesen, B.J.; van den Berg, A. Generation of miRNA sponge constructs. Methods 2012, 58, 113–117. [Google Scholar] [CrossRef]
- Qu, J.; Chen, X.; Sun, Y.Z.; Zhao, Y.; Cai, S.B.; Ming, Z.; You, Z.-H.; Li, J.Q. In Silico prediction of small molecule-miRNA associations based on the HeteSim algorithm. Mol. Ther-Nucl. Acids 2019, 14, 274–286. [Google Scholar] [CrossRef] [Green Version]
- Wen, D.; Danquah, M.; Chaudhary, A.K.; Mahato, R.I. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J. Control. Release 2015, 219, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Haga, C.L.; Velagapudi, S.P.; Strivelli, J.R.; Yang, W.Y.; Disney, M.D.; Phinney, D.G. Small molecule inhibition of miR-544 biogenesis disrupts adaptive responses to hypoxia by modulating ATM-mTOR signaling. ACS Chem. Biol. 2015, 10, 2267–2276. [Google Scholar] [CrossRef] [Green Version]
- Costales, M.G.; Haga, C.L.; Velagapudi, S.P.; Childs-Disney, J.L.; Phinney, D.G.; Disney, M.D. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J. Am. Chem. Soc. 2017, 139, 3446–3455. [Google Scholar] [CrossRef] [Green Version]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wires RNA 2020, 11, e1594. [Google Scholar] [CrossRef]
- Quiagen. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/functional-and-cell-analysis/mirna-functional-analysis/mircury-lna-mirna-power-target-site-blockers/mircury-lna-mirna-power-target-site-blockers/ (accessed on 28 March 2022).
- Louloupi, A.; Ørom, U.A.V. Inhibiting pri-miRNA processing with target site blockers. In miRNA Biogenesis; Springer: New York, NY, USA, 2018; pp. 63–68. [Google Scholar]
- Schmidt, M.F. Drug target miRNAs: Chances and challenges. Trends Biotechnol. 2014, 32, 578–585. [Google Scholar] [CrossRef]
- Othumpangat, S.; Bryan, N.B.; Beezhold, D.H.; Noti, J.D. Upregulation of miRNA-4776 in Influenza Virus Infected Bronchial Epithelial Cells Is Associated with Downregulation of NFKBIB and Increased Viral Survival. Viruses 2017, 9, 94. [Google Scholar] [CrossRef] [Green Version]
- Dalmadi, Á.; Miloro, F.; Bálint, J.; Várallyay, É.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef]
- Kobayashi, H.; Singer, R.H. Single-molecule imaging of microRNA-mediated gene silencing in cells. Nat. Commun. 2022, 13, 1435. [Google Scholar] [CrossRef]
- Peterson, S.M.; Thompson, J.A.; Ufkin, M.L.; Sathyanarayana, P.; Liaw, L.; Congdon, C.B. Common features of microRNA target prediction tools. Front. Genet. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Teune, J.H.; Steger, G. NOVOMIR: De novo prediction of microRNA-coding regions in a single plant-genome. J. Nucleic Acids 2010, 2010, 495904. [Google Scholar] [CrossRef] [Green Version]
- Jha, A.; Shankar, R. Employing machine learning for reliable miRNA target identification in plants. BMC Genom. 2011, 12, 636. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, 155–159. [Google Scholar] [CrossRef] [Green Version]
- psRNATarget: A Plant Small RNA Target Analysis Server (2017 Update). Available online: https://www.zhaolab.org/psRNATarget/ (accessed on 28 March 2022).
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. miRNA targets: From prediction tools to experimental validation. Methods Protoc. 2021, 4, 1. [Google Scholar] [CrossRef]
- Tools4MIRs. Available online: https://www.tools4mirs.org/ (accessed on 28 March 2022).
- WMD3—Web MicroRNA Designer. Available online: http://wmd3.weigelworld.org/cgi-bin/webapp.cgi/ (accessed on 28 March 2022).
- MiRNAsong. Available online: https://www2.med.muni.cz/histology/miRNAsong/index.php/ (accessed on 28 March 2022).
- PeTMbase. Available online: http://tools.ibg.deu.edu.tr/petmbase/index.php/ (accessed on 28 March 2022).
- P-SAMS. Available online: http://p-sams.carringtonlab.org/ (accessed on 28 March 2022).
- Hanna, J.; Hossain, G.S.; Kocerha, J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [Green Version]
- Neumeier, J.; Meister, G. SiRNA specificity: RNAi mechanisms and strategies to reduce off-target effects. Front. Plant Sci. 2021, 11, 2196. [Google Scholar] [CrossRef]
- Bai, X.; Bian, Z. MicroRNA-21 Is a Versatile Regulator and Potential Treatment Target in Central Nervous System Disorders. Front. Mol. Neurosci. 2022, 15, 842288. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Dicerna. Available online: https://dicerna.com/ (accessed on 28 March 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).