Integrin Regulators in Neutrophils
Abstract
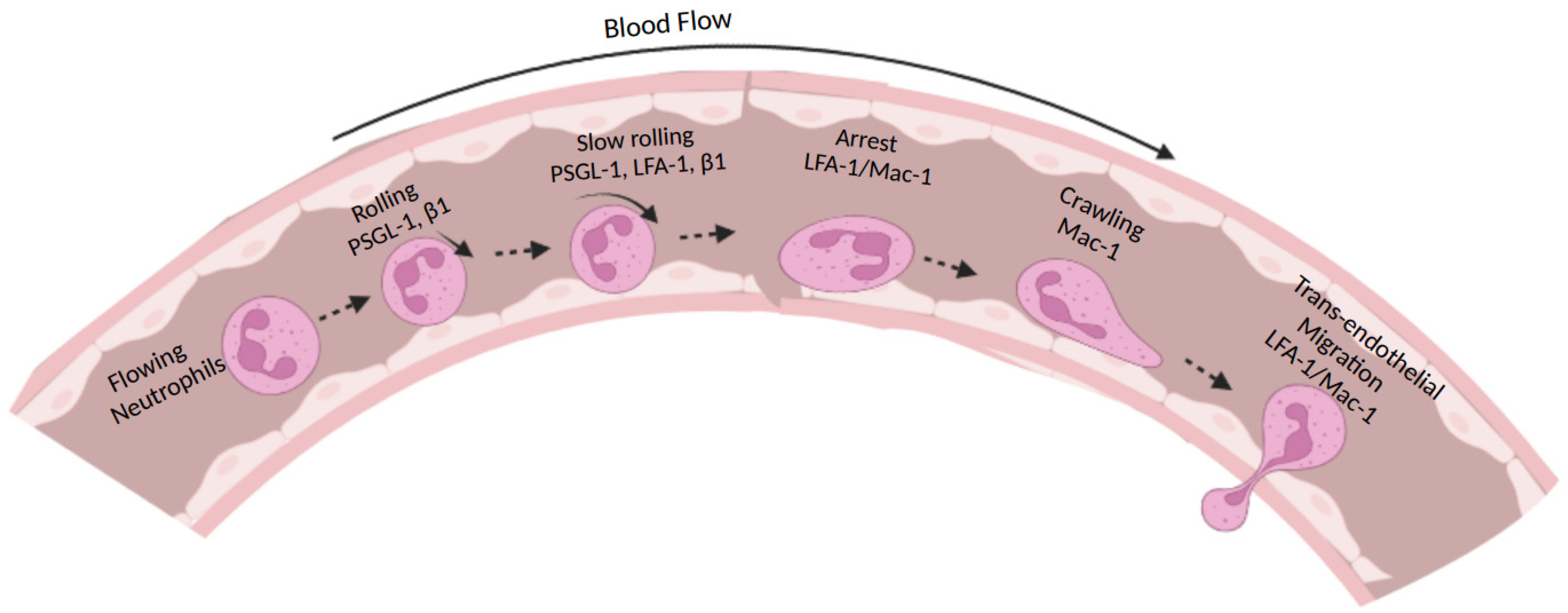
:1. Introduction
2. Types of Integrins Expressed on Neutrophils
3. Integrin Conformational Changes during Activation
4. Integrin Activation Modulators
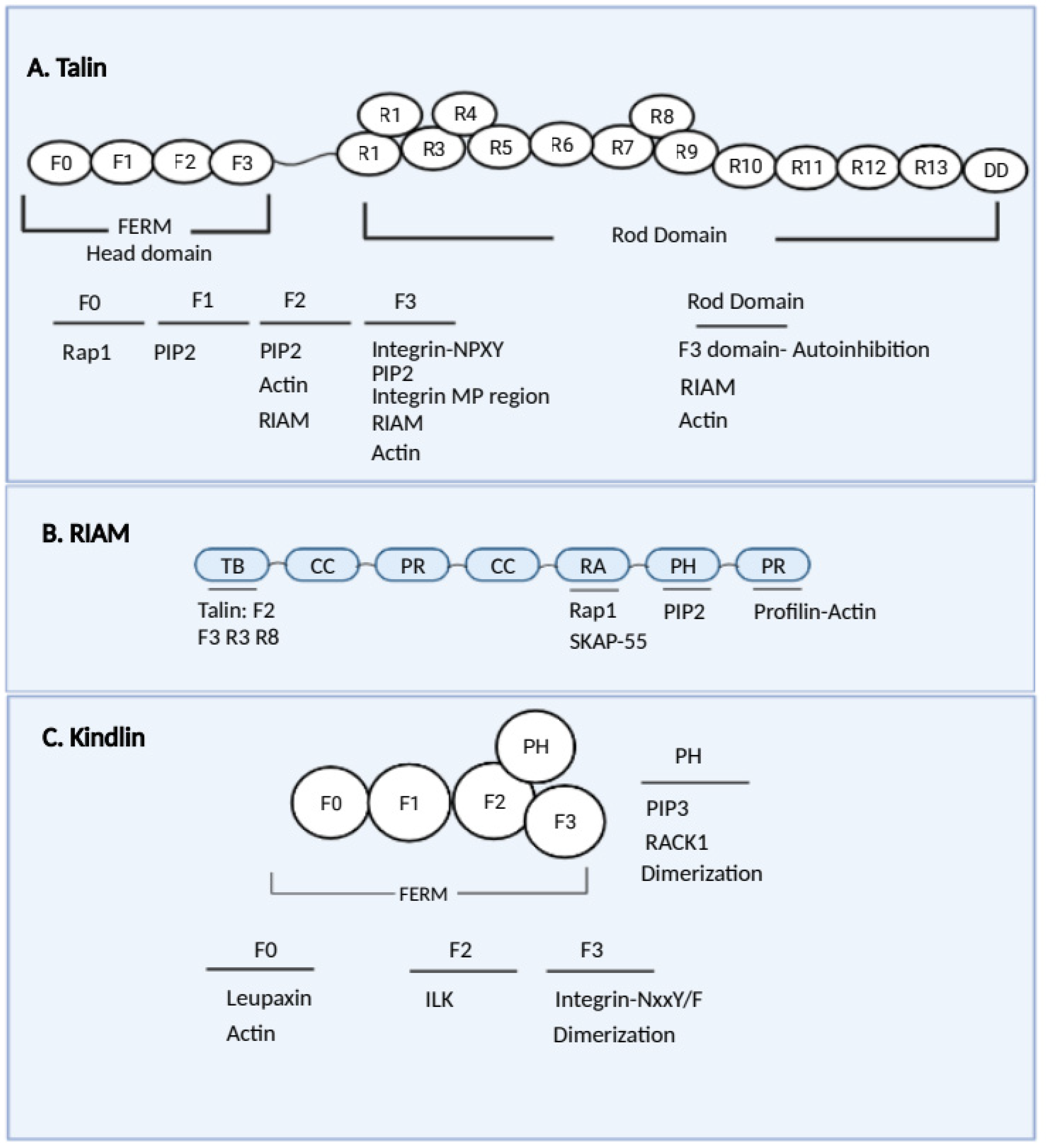
4.1. Talins
4.2. RIAM
4.3. Rap1
4.4. Kindlin
4.5. Linking of Integrins to the Actin Cytoskeleton
4.6. Other Molecules That Regulate Neutrophil Integrins
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Witter, A.R.; Okunnu, B.M.; Berg, R.E. The Essential Role of Neutrophils During Infection with the Intracellular Bacterial Pathogen Listeria Monocytogenes. J. Immunol. 2016, 197, 1557–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, R.A.; Collar, A.L.; Swamydas, M.; Rodriguez, C.A.; Lim, J.K.; Mendez, L.M.; Fink, D.L.; Hsu, A.P.; Zhai, B.; Karauzum, H.; et al. CARD9-Dependent Neutrophil Recruitment Protects against Fungal Invasion of the Central Nervous System. PLoS Pathog. 2015, 11, e1005293. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Komano, J.; Saitoh, Y.; Misawa, T.; Takahama, M.; Kozaki, T.; Uehata, T.; Iwasaki, H.; Omori, H.; Yamaoka, S.; et al. Neutrophil Extracellular Traps Mediate a Host Defense Response to Human Immunodeficiency Virus-1. Cell Host Microbe 2012, 12, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Jenne, C.N.; Wong, C.H.Y.; Zemp, F.J.; McDonald, B.; Rahman, M.M.; Forsyth, P.A.; McFadden, G.; Kubes, P. Neutrophils Recruited to Sites of Infection Protect from Virus Challenge by Releasing Neutrophil Extracellular Traps. Cell Host Microbe 2013, 13, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, K.; Hoffman, H.M.; Kubes, P.; Cassatella, M.A.; Zychlinsky, A.; Hedrick, C.C.; Catz, S.D. Neutrophils: New Insights and Open Questions. Sci. Immunol. 2018, 3, eaat4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mócsai, A. Diverse Novel Functions of Neutrophils in Immunity, Inflammation, and Beyond. J. Exp. Med. 2013, 210, 1283–1299. [Google Scholar] [CrossRef] [Green Version]
- Bonaventura, A.; Montecucco, F.; Dallegri, F.; Carbone, F.; Lüscher, T.F.; Camici, G.G.; Liberale, L. Novel Findings in Neutrophil Biology and Their Impact on Cardiovascular Disease. Cardiovasc. Res. 2019, 115, 1266–1285. [Google Scholar] [CrossRef]
- Scapini, P.; Cassatella, M.A. Social Networking of Human Neutrophils within the Immune System. Blood 2014, 124, 710–719. [Google Scholar] [CrossRef]
- Filippi, M.-D. Neutrophil Transendothelial Migration: Updates and New Perspectives. Blood 2019, 133, 2149–2158. [Google Scholar] [CrossRef]
- Liew, P.X.; Kubes, P. The Neutrophil’s Role During Health and Disease. Physiol. Rev. 2019, 99, 1223–1248. [Google Scholar] [CrossRef]
- Margraf, A.; Ley, K.; Zarbock, A. Neutrophil Recruitment: From Model Systems to Tissue-Specific Patterns. Trends Immunol. 2019, 40, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Marki, A.; Esko, J.D.; Pries, A.R.; Ley, K. Role of the Endothelial Surface Layer in Neutrophil Recruitment. J. Leukoc. Biol. 2015, 98, 503–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morikis, V.A.; Simon, S.I. Neutrophil Mechanosignaling Promotes Integrin Engagement with Endothelial Cells and Motility Within Inflamed Vessels. Front. Immunol. 2018, 9, 2774. [Google Scholar] [CrossRef] [PubMed]
- Morikis, V.A.; Hernandez, A.A.; Magnani, J.L.; Sperandio, M.; Simon, S.I. Targeting Neutrophil Adhesive Events to Address Vaso-Occlusive Crisis in Sickle Cell Patients. Front. Immunol. 2021, 12, 1256. [Google Scholar] [CrossRef]
- Maas, S.L.; Soehnlein, O.; Viola, J.R. Organ-Specific Mechanisms of Transendothelial Neutrophil Migration in the Lung, Liver, Kidney, and Aorta. Front. Immunol. 2018, 9, 2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadik, C.D.; Kim, N.D.; Luster, A.D. Neutrophils Cascading Their Way to Inflammation. Trends Immunol. 2011, 32, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Herter, J.; Zarbock, A. Integrin Regulation during Leukocyte Recruitment. J. Immunol. 2013. [Google Scholar] [CrossRef] [Green Version]
- Kourtzelis, I.; Mitroulis, I.; von Renesse, J.; Hajishengallis, G.; Chavakis, T. From Leukocyte Recruitment to Resolution of Inflammation: The Cardinal Role of Integrins. J. Leukoc. Biol. 2017, 102, 677–683. [Google Scholar] [CrossRef] [Green Version]
- Sökeland, G.; Schumacher, U. The Functional Role of Integrins during Intra- and Extravasation within the Metastatic Cascade. Mol. Cancer 2019, 18, 12. [Google Scholar] [CrossRef]
- Fan, Z.; Ley, K. Leukocyte Arrest: Biomechanics and Molecular Mechanisms of Β2 Integrin Activation. Biorheology 2015, 52, 353–377. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Hu, L.; Fan, Z. Β2 Integrin Activation and Signal Transduction in Leukocyte Recruitment. Am. J. Physiol. Cell Physiol. 2021, 321, C308–C316. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the Site of Inflammation: The Leukocyte Adhesion Cascade Updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Arias-Mejias, S.M.; Warda, K.Y.; Quattrocchi, E.; Alonso-Quinones, H.; Sominidi-Damodaran, S.; Meves, A. The Role of Integrins in Melanoma: A Review. Int. J. Derm. 2020, 59, 525–534. [Google Scholar] [CrossRef]
- Kadry, Y.A.; Calderwood, D.A. Chapter 22: Structural and Signaling Functions of Integrins. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183206. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.-H.; Carman, C.V.; Springer, T.A. Structural Basis of Integrin Regulation and Signaling. Annu. Rev. Immunol. 2007, 25, 619–647. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Layseca, P.; Icha, J.; Hamidi, H.; Ivaska, J. Integrin Trafficking in Cells and Tissues. Nat. Cell Biol. 2019, 21, 122–132. [Google Scholar] [CrossRef]
- Ou, Z.; Dolmatova, E.; Lassègue, B.; Griendling, K.K. Β1- and Β2-Integrins: Central Players in Regulating Vascular Permeability and Leukocyte Recruitment during Acute Inflammation. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H734–H739. [Google Scholar] [CrossRef]
- Arnaout, M.A.; Mahalingam, B.; Xiong, J.-P. Integrin Structure, Allostery, and Bidirectional Signaling. Annu. Rev. Cell Dev. Biol. 2005, 21, 381–410. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Luo, B.-H. Integrin Bi-Directional Signaling across the Plasma Membrane. J. Cell. Physiol. 2013, 228, 306–312. [Google Scholar] [CrossRef]
- Stadtmann, A.; Zarbock, A. The Role of Kindlin in Neutrophil Recruitment to Inflammatory Sites. Curr. Opin. Hematol. 2017, 24, 38–45. [Google Scholar] [CrossRef]
- Xiong, J.P.; Stehle, T.; Diefenbach, B.; Zhang, R.; Dunker, R.; Scott, D.L.; Joachimiak, A.; Goodman, S.L.; Arnaout, M.A. Crystal Structure of the Extracellular Segment of Integrin Alpha Vbeta3. Science 2001, 294, 339–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, T.-L.; Kim, C.; Ginsberg, M.H.; Ulmer, T.S. The Structure of the Integrin AIIbβ3 Transmembrane Complex Explains Integrin Transmembrane Signalling. EMBO J. 2009, 28, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Morse, E.M.; Brahme, N.N.; Calderwood, D.A. Integrin Cytoplasmic Tail Interactions. Biochemistry 2014, 53, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Rocco, M.; Rosano, C.; Weisel, J.W.; Horita, D.A.; Hantgan, R.R. Integrin Conformational Regulation: Uncoupling Extension/Tail Separation from Changes in the Head Region by a Multiresolution Approach. Structure 2008, 16, 954–964. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Xu, Z.; Peng, Y.; Wang, J.; Xiang, Y. Integrin Β4 as a Potential Diagnostic and Therapeutic Tumor Marker. Biomolecules 2021, 11, 1197. [Google Scholar] [CrossRef]
- Goksoy, E.; Ma, Y.-Q.; Wang, X.; Kong, X.; Perera, D.; Plow, E.F.; Qin, J. Structural Basis for the Autoinhibition of Talin in Regulating Integrin Activation. Mol. Cell 2008, 31, 124–133. [Google Scholar] [CrossRef] [Green Version]
- Harburger, D.S.; Bouaouina, M.; Calderwood, D.A. Kindlin-1 and -2 Directly Bind the C-Terminal Region of β Integrin Cytoplasmic Tails and Exert Integrin-Specific Activation Effects. J. Biol. Chem. 2009, 284, 11485–11497. [Google Scholar] [CrossRef] [Green Version]
- Moser, M.; Nieswandt, B.; Ussar, S.; Pozgajova, M.; Fässler, R. Kindlin-3 Is Essential for Integrin Activation and Platelet Aggregation. Nat. Med. 2008, 14, 325–330. [Google Scholar] [CrossRef]
- .Calderwood, D.A.; Fujioka, Y.; de Pereda, J.M.; García-Alvarez, B.; Nakamoto, T.; Margolis, B.; McGlade, C.J.; Liddington, R.C.; Ginsberg, M.H. Integrin Beta Cytoplasmic Domain Interactions with Phosphotyrosine-Binding Domains: A Structural Prototype for Diversity in Integrin Signaling. Proc. Natl. Acad. Sci. USA 2003, 100, 2272–2277. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, S.; Grinstein, S. Phagocytosis and Innate Immunity. Curr. Opin. Immunol. 2002, 14, 136–145. [Google Scholar] [CrossRef]
- Torres-Gomez, A.; Cabañas, C.; Lafuente, E.M. Phagocytic Integrins: Activation and Signaling. Front. Immunol. 2020, 11, 738. [Google Scholar] [CrossRef] [PubMed]
- Blythe, E.N.; Weaver, L.C.; Brown, A.; Dekaban, G.A. Β2 Integrin CD11d/CD18: From Expression to an Emerging Role in Staged Leukocyte Migration. Front. Immunol. 2021, 12, 775447. [Google Scholar] [CrossRef] [PubMed]
- Madamanchi, A.; Santoro, S.A.; Zutter, M.M. A2β1 Integrin. Adv. Exp. Med. Biol. 2014, 819, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Kirveskari, J.; Bono, P.; Granfors, K.; Leirisalo-Repo, M.; Jalkanen, S.; Salmi, M. Expression of Alpha4-Integrins on Human Neutrophils. J. Leukoc. Biol. 2000, 68, 243–250. [Google Scholar]
- Pierini, L.M.; Lawson, M.A.; Eddy, R.J.; Hendey, B.; Maxfield, F.R. Oriented Endocytic Recycling of Alpha5beta1 in Motile Neutrophils. Blood 2000, 95, 2471–2480. [Google Scholar] [CrossRef]
- Bohnsack, J.F. CD11/CD18-Independent Neutrophil Adherence to Laminin Is Mediated by the Integrin VLA-6. Blood 1992, 79, 1545–1552. [Google Scholar] [CrossRef] [Green Version]
- Mambole, A.; Bigot, S.; Baruch, D.; Lesavre, P.; Halbwachs-Mecarelli, L. Human Neutrophil Integrin A9β1: Up-Regulation by Cell Activation and Synergy with Β2 Integrins during Adhesion to Endothelium under Flow. J. Leukoc. Biol. 2010, 88, 321–327. [Google Scholar] [CrossRef]
- Langereis, J.D. Neutrophil Integrin Affinity Regulation in Adhesion, Migration, and Bacterial Clearance. Cell Adhes. Migr. 2013, 7, 476–481. [Google Scholar] [CrossRef] [Green Version]
- Lefort, C.; Ley, K. Neutrophil Arrest by LFA-1 Activation. Front. Immunol. 2012, 3, 157. [Google Scholar] [CrossRef] [Green Version]
- Salas, A.; Shimaoka, M.; Kogan, A.N.; Harwood, C.; von Andrian, U.H.; Springer, T.A. Rolling Adhesion through an Extended Conformation of Integrin AlphaLbeta2 and Relation to Alpha I and Beta I-like Domain Interaction. Immunity 2004, 20, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Chesnutt, B.C.; Smith, D.F.; Raffler, N.A.; Smith, M.L.; White, E.J.; Ley, K. Induction of LFA-1-Dependent Neutrophil Rolling on ICAM-1 by Engagement of E-Selectin. Microcirculation 2006, 13, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, Y.; Spelten, O.; Zhang, H.; Ley, K.; Zarbock, A. Rolling on E- or P-Selectin Induces the Extended but Not High-Affinity Conformation of LFA-1 in Neutrophils. Blood 2010, 116, 617–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiVietro, J.A.; Smith, M.J.; Smith, B.R.; Petruzzelli, L.; Larson, R.S.; Lawrence, M.B. Immobilized IL-8 Triggers Progressive Activation of Neutrophils Rolling in Vitro on P-Selectin and Intercellular Adhesion Molecule-1. J. Immunol. 2001, 167, 4017–4025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, W.; Wang, Q.; Deng, Z.; Li, T.; Xiao, H.; Wu, Z. TRAF1 Suppresses Antifungal Immunity through CXCL1-Mediated Neutrophil Recruitment during Candida Albicans Intradermal Infection. Cell Commun. Signal. 2020, 18, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopalan, P.K.; Smith, C.W.; Lu, H.; Berg, E.L.; McIntire, L.V.; Simon, S.I. Neutrophil CD18-Dependent Arrest on Intercellular Adhesion Molecule 1 (ICAM-1) in Shear Flow Can Be Activated through L-Selectin. J. Immunol. 1997, 158, 367–375. [Google Scholar]
- Lyck, R.; Enzmann, G. The Physiological Roles of ICAM-1 and ICAM-2 in Neutrophil Migration into Tissues. Curr. Opin. Hematol 2015, 22, 53–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillipson, M.; Heit, B.; Colarusso, P.; Liu, L.; Ballantyne, C.M.; Kubes, P. Intraluminal Crawling of Neutrophils to Emigration Sites: A Molecularly Distinct Process from Adhesion in the Recruitment Cascade. J. Exp. Med. 2006, 203, 2569–2575. [Google Scholar] [CrossRef]
- Buffone, A.; Anderson, N.R.; Hammer, D.A. Human Neutrophils Will Crawl Upstream on ICAM-1 If Mac-1 Is Blocked. Biophys. J. 2019, 117, 1393–1404. [Google Scholar] [CrossRef]
- Proebstl, D.; Voisin, M.-B.; Woodfin, A.; Whiteford, J.; D’Acquisto, F.; Jones, G.E.; Rowe, D.; Nourshargh, S. Pericytes Support Neutrophil Subendothelial Cell Crawling and Breaching of Venular Walls in Vivo. J. Exp. Med. 2012, 209, 1219–1234. [Google Scholar] [CrossRef] [Green Version]
- Chong, D.L.W.; Rebeyrol, C.; José, R.J.; Williams, A.E.; Brown, J.S.; Scotton, C.J.; Porter, J.C. ICAM-1 and ICAM-2 Are Differentially Expressed and Up-Regulated on Inflamed Pulmonary Epithelium, but Neither ICAM-2 nor LFA-1: ICAM-1 Are Required for Neutrophil Migration Into the Airways In Vivo. Front. Immunol. 2021, 12, 691957. [Google Scholar] [CrossRef]
- Kinoshita, K.; Leung, A.; Simon, S.; Evans, E. Long-Lived, High-Strength States of ICAM-1 Bonds to Beta2 Integrin, II: Lifetimes of LFA-1 Bonds under Force in Leukocyte Signaling. Biophys. J. 2010, 98, 1467–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarantos, M.R.; Lum, A.F.H.; Staunton, D.E.; Simon, S.I. Kinetics of LFA-1 Binding to ICAM-1 Studied in a Cell-Free System. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2004, 2004, 4974–4977. [Google Scholar] [CrossRef] [PubMed]
- Gorina, R.; Lyck, R.; Vestweber, D.; Engelhardt, B. Β2 Integrin-Mediated Crawling on Endothelial ICAM-1 and ICAM-2 Is a Prerequisite for Transcellular Neutrophil Diapedesis across the Inflamed Blood-Brain Barrier. J. Immunol. 2014, 192, 324–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Yang, H.; Wang, M.; Lü, S.; Zhang, Y.; Long, M. Ligand-Specific Binding Forces of LFA-1 and Mac-1 in Neutrophil Adhesion and Crawling. Mol. Biol. Cell 2018, 29, 408–418. [Google Scholar] [CrossRef]
- Ostermann, G.; Weber, K.S.C.; Zernecke, A.; Schröder, A.; Weber, C. JAM-1 Is a Ligand of the Beta(2) Integrin LFA-1 Involved in Transendothelial Migration of Leukocytes. Nat. Immunol. 2002, 3, 151–158. [Google Scholar] [CrossRef]
- Yakubenko, V.P.; Cui, K.; Ardell, C.L.; Brown, K.E.; West, X.Z.; Gao, D.; Stefl, S.; Salomon, R.G.; Podrez, E.A.; Byzova, T.V. Oxidative Modifications of Extracellular Matrix Promote the Second Wave of Inflammation via Β2 Integrins. Blood 2018, 132, 78–88. [Google Scholar] [CrossRef]
- Chen, J.; Zhong, M.-C.; Guo, H.; Davidson, D.; Mishel, S.; Lu, Y.; Rhee, I.; Pérez-Quintero, L.-A.; Zhang, S.; Cruz-Munoz, M.-E.; et al. SLAMF7 Is Critical for Phagocytosis of Haematopoietic Tumour Cells via Mac-1 Integrin. Nature 2017, 544, 493–497. [Google Scholar] [CrossRef] [Green Version]
- Bose, T.O.; Colpitts, S.L.; Pham, Q.-M.; Puddington, L.; Lefrançois, L. CD11a Is Essential for Normal Development of Hematopoietic Intermediates. J. Immunol. 2014, 193, 2863–2872. [Google Scholar] [CrossRef] [Green Version]
- Lampiasi, N.; Russo, R.; Zito, F. The Alternative Faces of Macrophage Generate Osteoclasts. Biomed Res. Int. 2016, 2016, 9089610. [Google Scholar] [CrossRef] [Green Version]
- Herrero-Turrión, M.J.; Calafat, J.; Janssen, H.; Fukuda, M.; Mollinedo, F. Rab27a Regulates Exocytosis of Tertiary and Specific Granules in Human Neutrophils. J. Immunol. 2008, 181, 3793–3803. [Google Scholar] [CrossRef] [Green Version]
- Martín-Martín, B.; Nabokina, S.M.; Blasi, J.; Lazo, P.A.; Mollinedo, F. Involvement of SNAP-23 and Syntaxin 6 in Human Neutrophil Exocytosis. Blood 2000, 96, 2574–2583. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F.; Calafat, J.; Janssen, H.; Martín-Martín, B.; Canchado, J.; Nabokina, S.M.; Gajate, C. Combinatorial SNARE Complexes Modulate the Secretion of Cytoplasmic Granules in Human Neutrophils. J. Immunol. 2006, 177, 2831–2841. [Google Scholar] [CrossRef] [Green Version]
- Mollinedo, F.; Martín-Martín, B.; Calafat, J.; Nabokina, S.M.; Lazo, P.A. Role of Vesicle-Associated Membrane Protein-2, Through Q-Soluble N-Ethylmaleimide-Sensitive Factor Attachment Protein Receptor/R-Soluble N-Ethylmaleimide-Sensitive Factor Attachment Protein Receptor Interaction, in the Exocytosis of Specific and Tertiary Granules of Human Neutrophils. J. Immunol. 2003, 170, 1034–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramadass, M.; Johnson, J.L.; Catz, S.D. Rab27a Regulates GM-CSF-Dependent Priming of Neutrophil Exocytosis. J. Leukoc. Biol. 2017, 101, 693–702. [Google Scholar] [CrossRef]
- Masuda, S.; Nakazawa, D.; Shida, H.; Miyoshi, A.; Kusunoki, Y.; Tomaru, U.; Ishizu, A. NETosis Markers: Quest for Specific, Objective, and Quantitative Markers. Clin. Chim. Acta 2016, 459, 89–93. [Google Scholar] [CrossRef]
- Werr, J.; Xie, X.; Hedqvist, P.; Ruoslahti, E.; Lindbom, L. Beta1 Integrins Are Critically Involved in Neutrophil Locomotion in Extravascular Tissue In Vivo. J. Exp. Med. 1998, 187, 2091–2096. [Google Scholar] [CrossRef] [Green Version]
- Davenpeck, K.L.; Sterbinsky, S.A.; Bochner, B.S. Rat Neutrophils Express Alpha4 and Beta1 Integrins and Bind to Vascular Cell Adhesion Molecule-1 (VCAM-1) and Mucosal Addressin Cell Adhesion Molecule-1 (MAdCAM-1). Blood 1998, 91, 2341–2346. [Google Scholar] [CrossRef]
- Henderson, R.B.; Lim, L.H.; Tessier, P.A.; Gavins, F.N.; Mathies, M.; Perretti, M.; Hogg, N. The Use of Lymphocyte Function-Associated Antigen (LFA)-1-Deficient Mice to Determine the Role of LFA-1, Mac-1, and Alpha4 Integrin in the Inflammatory Response of Neutrophils. J. Exp. Med. 2001, 194, 219–226. [Google Scholar] [CrossRef] [Green Version]
- Issekutz, T.B.; Miyasaka, M.; Issekutz, A.C. Rat Blood Neutrophils Express Very Late Antigen 4 and It Mediates Migration to Arthritic Joint and Dermal Inflammation. J. Exp. Med. 1996, 183, 2175–2184. [Google Scholar] [CrossRef] [Green Version]
- Canalli, A.A.; Proença, R.F.; Franco-Penteado, C.F.; Traina, F.; Sakamoto, T.M.; Saad, S.T.O.; Conran, N.; Costa, F.F. Participation of Mac-1, LFA-1 and VLA-4 Integrins in the in Vitro Adhesion of Sickle Cell Disease Neutrophils to Endothelial Layers, and Reversal of Adhesion by Simvastatin. Haematologica 2011, 96, 526–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibbotson, G.C.; Doig, C.; Kaur, J.; Gill, V.; Ostrovsky, L.; Fairhead, T.; Kubes, P. Functional Alpha4-Integrin: A Newly Identified Pathway of Neutrophil Recruitment in Critically Ill Septic Patients. Nat. Med. 2001, 7, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Werr, J.; Johansson, J.; Eriksson, E.E.; Hedqvist, P.; Ruoslahti, E.; Lindbom, L. Integrin Alpha(2)Beta(1) (VLA-2) Is a Principal Receptor Used by Neutrophils for Locomotion in Extravascular Tissue. Blood 2000, 95, 1804–1809. [Google Scholar] [CrossRef]
- Dangerfield, J.; Larbi, K.Y.; Huang, M.-T.; Dewar, A.; Nourshargh, S. PECAM-1 (CD31) Homophilic Interaction up-Regulates Alpha6beta1 on Transmigrated Neutrophils in Vivo and Plays a Functional Role in the Ability of Alpha6 Integrins to Mediate Leukocyte Migration through the Perivascular Basement Membrane. J. Exp. Med. 2002, 196, 1201–1211. [Google Scholar] [CrossRef]
- Dangerfield, J.P.; Wang, S.; Nourshargh, S. Blockade of Alpha6 Integrin Inhibits IL-1beta- but Not TNF-Alpha-Induced Neutrophil Transmigration in Vivo. J. Leukoc. Biol. 2005, 77, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Roussel, E.; Gingras, M.C. Transendothelial Migration Induces Rapid Expression on Neutrophils of Granule-Release VLA6 Used for Tissue Infiltration. J. Leukoc. Biol. 1997, 62, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Ridger, V.C.; Wagner, B.E.; Wallace, W.A.; Hellewell, P.G. Differential Effects of CD18, CD29, and CD49 Integrin Subunit Inhibition on Neutrophil Migration in Pulmonary Inflammation. J. Immunol. 2001, 166, 3484–3490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, I.D.; Humphries, M.J. Integrin Structure, Activation, and Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef] [Green Version]
- Arnaout, M.A. Biology and Structure of Leukocyte β 2 Integrins and Their Role in Inflammation. F1000Research 2016, 5, 2433. [Google Scholar] [CrossRef] [Green Version]
- Nishida, N.; Xie, C.; Shimaoka, M.; Cheng, Y.; Walz, T.; Springer, T.A. Activation of Leukocyte Β2 Integrins by Conversion from Bent to Extended Conformations. Immunity 2006, 25, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Shimonaka, M.; Katagiri, K.; Nakayama, T.; Fujita, N.; Tsuruo, T.; Yoshie, O.; Kinashi, T. Rap1 Translates Chemokine Signals to Integrin Activation, Cell Polarization, and Motility across Vascular Endothelium under Flow. J. Cell Biol. 2003, 161, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, S.; Shattil, S.J.; Eto, K.; Tai, V.; Liddington, R.C.; de Pereda, J.M.; Ginsberg, M.H.; Calderwood, D.A. Talin Binding to Integrin Beta Tails: A Final Common Step in Integrin Activation. Science 2003, 302, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, O.; Velyvis, A.; Velyviene, A.; Hu, B.; Haas, T.; Plow, E.; Qin, J. A Structural Mechanism of Integrin Alpha(IIb)Beta(3) “inside-out” Activation as Regulated by Its Cytoplasmic Face. Cell 2002, 110, 587–597. [Google Scholar] [CrossRef] [Green Version]
- Liddington, R.C.; Ginsberg, M.H. Integrin Activation Takes Shape. J. Cell Biol. 2002, 158, 833–839. [Google Scholar] [CrossRef]
- Shimaoka, M.; Takagi, J.; Springer, T.A. Conformational Regulation of Integrin Structure and Function. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 485–516. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.-P.; Stehle, T.; Goodman, S.L.; Arnaout, M.A. New Insights into the Structural Basis of Integrin Activation. Blood 2003, 102, 1155–1159. [Google Scholar] [CrossRef]
- Sun, H.; Lagarrigue, F.; Wang, H.; Fan, Z.; Lopez-Ramirez, M.A.; Chang, J.T.; Ginsberg, M.H. Distinct Integrin Activation Pathways for Effector and Regulatory T Cell Trafficking and Function. J. Exp. Med. 2021, 218, e20201524. [Google Scholar] [CrossRef]
- Beglova, N.; Blacklow, S.C.; Takagi, J.; Springer, T.A. Cysteine-Rich Module Structure Reveals a Fulcrum for Integrin Rearrangement upon Activation. Nat. Struct. Biol. 2002, 9, 282–287. [Google Scholar] [CrossRef]
- Adair, B.D.; Xiong, J.-P.; Maddock, C.; Goodman, S.L.; Arnaout, M.A.; Yeager, M. Three-Dimensional EM Structure of the Ectodomain of Integrin {alpha}V{beta}3 in a Complex with Fibronectin. J. Cell Biol. 2005, 168, 1109–1118. [Google Scholar] [CrossRef]
- Sen, M.; Yuki, K.; Springer, T.A. An Internal Ligand-Bound, Metastable State of a Leukocyte Integrin, AXβ2. J. Cell Biol. 2013, 203, 629–642. [Google Scholar] [CrossRef]
- Gupta, V.; Gylling, A.; Alonso, J.L.; Sugimori, T.; Ianakiev, P.; Xiong, J.-P.; Amin Arnaout, M. The β-Tail Domain (ΒTD) Regulates Physiologic Ligand Binding to Integrin CD11b/CD18. Blood 2007, 109, 3513–3520. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; McArdle, S.; Marki, A.; Mikulski, Z.; Gutierrez, E.; Engelhardt, B.; Deutsch, U.; Ginsberg, M.; Groisman, A.; Ley, K. Neutrophil Recruitment Limited by High-Affinity Bent Β2 Integrin Binding Ligand in Cis. Nat. Commun. 2016, 7, 12658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calderwood, D.A. Talin Controls Integrin Activation. BioChem. Soc. Trans. 2004, 32, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, D.A.; Yan, B.; de Pereda, J.M.; Alvarez, B.G.; Fujioka, Y.; Liddington, R.C.; Ginsberg, M.H. The Phosphotyrosine Binding-like Domain of Talin Activates Integrins. J. Biol. Chem. 2002, 277, 21749–21758. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Guo, Q.; Kim, C.; Hu, W.; Ye, F. Integrin AII b Tail Distal of GFFKR Participates in Inside-out AII b Β3 Activation. J. Thromb. Haemost 2014, 12, 1145–1155. [Google Scholar] [CrossRef] [Green Version]
- Oxley, C.L.; Anthis, N.J.; Lowe, E.D.; Vakonakis, I.; Campbell, I.D.; Wegener, K.L. An Integrin Phosphorylation Switch: The Effect of Beta3 Integrin Tail Phosphorylation on Dok1 and Talin Binding. J. Biol. Chem. 2008, 283, 5420–5426. [Google Scholar] [CrossRef] [Green Version]
- Kammerer, P.; Aretz, J.; Fässler, R. Lucky Kindlin: A Cloverleaf at the Integrin Tail. Proc. Natl. Acad. Sci. USA 2017, 114, 9234–9236. [Google Scholar] [CrossRef] [Green Version]
- Larjava, H.; Plow, E.F.; Wu, C. Kindlins: Essential Regulators of Integrin Signalling and Cell-Matrix Adhesion. EMBO Rep. 2008, 9, 1203–1208. [Google Scholar] [CrossRef]
- Kiema, T.; Lad, Y.; Jiang, P.; Oxley, C.L.; Baldassarre, M.; Wegener, K.L.; Campbell, I.D.; Ylänne, J.; Calderwood, D.A. The Molecular Basis of Filamin Binding to Integrins and Competition with Talin. Mol. Cell 2006, 21, 337–347. [Google Scholar] [CrossRef]
- Razinia, Z.; Mäkelä, T.; Ylänne, J.; Calderwood, D.A. Filamins in Mechanosensing and Signaling. Annu. Rev. Biophys. 2012, 41, 227–246. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Chit, J.C.-Y.; Feng, C.; Bhunia, A.; Tan, S.-M.; Bhattacharjya, S. An Alternative Phosphorylation Switch in Integrin Β2 (CD18) Tail for Dok1 Binding. Sci. Rep. 2015, 5, 11630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takala, H.; Nurminen, E.; Nurmi, S.M.; Aatonen, M.; Strandin, T.; Takatalo, M.; Kiema, T.; Gahmberg, C.G.; Ylänne, J.; Fagerholm, S.C. Beta2 Integrin Phosphorylation on Thr758 Acts as a Molecular Switch to Regulate 14-3-3 and Filamin Binding. Blood 2008, 112, 1853–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahan, F.; Madhavan, S.; Rolova, T.; Viazmina, L.; Grönholm, M.; Gahmberg, C.G. Phosphorylation of the α-Chain in the Integrin LFA-1 Enables Β2-Chain Phosphorylation and α-Actinin Binding Required for Cell Adhesion. J. Biol. Chem. 2018, 293, 12318–12330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bromberger, T.; Klapproth, S.; Rohwedder, I.; Weber, J.; Pick, R.; Mittmann, L.; Min-Weißenhorn, S.J.; Reichel, C.A.; Scheiermann, C.; Sperandio, M.; et al. Binding of Rap1 and Riam to Talin1 Fine-Tune Β2 Integrin Activity During Leukocyte Trafficking. Front. Immunol. 2021, 12, 702345. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Lim, C.J.; Puzon-McLaughlin, W.; Shattil, S.J.; Ginsberg, M.H. RIAM Activates Integrins by Linking Talin to Ras GTPase Membrane-Targeting Sequences. J. Biol. Chem. 2009, 284, 5119–5127. [Google Scholar] [CrossRef] [Green Version]
- Bos, J.L.; de Rooij, J.; Reedquist, K.A. Rap1 Signalling: Adhering to New Models. Nat. Rev. Mol. Cell Biol. 2001, 2, 369–377. [Google Scholar] [CrossRef]
- Bos, J.L.; de Bruyn, K.; Enserink, J.; Kuiperij, B.; Rangarajan, S.; Rehmann, H.; Riedl, J.; de Rooij, J.; van Mansfeld, F.; Zwartkruis, F. The Role of Rap1 in Integrin-Mediated Cell Adhesion. Biochem. Soc. Trans. 2003, 31, 83–86. [Google Scholar] [CrossRef]
- Bromberger, T.; Klapproth, S.; Rohwedder, I.; Zhu, L.; Mittmann, L.; Reichel, C.A.; Sperandio, M.; Qin, J.; Moser, M. Direct Rap1/Talin1 Interaction Regulates Platelet and Neutrophil Integrin Activity in Mice. Blood 2018, 132, 2754–2762. [Google Scholar] [CrossRef] [Green Version]
- Klapproth, S.; Moretti, F.A.; Zeiler, M.; Ruppert, R.; Breithaupt, U.; Mueller, S.; Haas, R.; Mann, M.; Sperandio, M.; Fässler, R.; et al. Minimal Amounts of Kindlin-3 Suffice for Basal Platelet and Leukocyte Functions in Mice. Blood 2015, 126, 2592–2600. [Google Scholar] [CrossRef] [Green Version]
- Ghiran, I.; Klickstein, L.B.; Nicholson-Weller, A. Calreticulin Is at the Surface of Circulating Neutrophils and Uses CD59 as an Adaptor Molecule. J. Biol. Chem. 2003, 278, 21024–21031. [Google Scholar] [CrossRef] [Green Version]
- Ohkuro, M.; Kim, J.-D.; Kuboi, Y.; Hayashi, Y.; Mizukami, H.; Kobayashi-Kuramochi, H.; Muramoto, K.; Shirato, M.; Michikawa-Tanaka, F.; Moriya, J.; et al. Calreticulin and Integrin Alpha Dissociation Induces Anti-Inflammatory Programming in Animal Models of Inflammatory Bowel Disease. Nat. Commun. 2018, 9, 1982. [Google Scholar] [CrossRef] [PubMed]
- Kinashi, T.; Katagiri, K. Regulation of Immune Cell Adhesion and Migration by Regulator of Adhesion and Cell Polarization Enriched in Lymphoid Tissues. Immunology 2005, 116, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Hyduk, S.J.; Oh, J.; Xiao, H.; Chen, M.; Cybulsky, M.I. Paxillin Selectively Associates with Constitutive and Chemoattractant-Induced High-Affinity Alpha4beta1 Integrins: Implications for Integrin Signaling. Blood 2004, 104, 2818–2824. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Bao, Y.; Ge, S.; Sun, P.; Sun, J.; Liu, J.; Chen, F.; Han, L.; Cao, Z.; Qin, J.; et al. Sharpin Suppresses Β1-Integrin Activation by Complexing with the Β1 Tail and Kindlin-1. Cell Commun. Signal. 2019, 17, 101. [Google Scholar] [CrossRef] [Green Version]
- Roberts, G.C.K.; Critchley, D.R. Structural and Biophysical Properties of the Integrin-Associated Cytoskeletal Protein Talin. Biophys. Rev. 2009, 1, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Monkley, S.J.; Pritchard, C.A.; Critchley, D.R. Analysis of the Mammalian Talin2 Gene TLN2. Biochem. Biophys. Res. Commun. 2001, 286, 880–885. [Google Scholar] [CrossRef]
- Senetar, M.A.; Moncman, C.L.; McCann, R.O. Talin2 Is Induced during Striated Muscle Differentiation and Is Targeted to Stable Adhesion Complexes in Mature Muscle. Cell Motil. Cytoskelet. 2007, 64, 157–173. [Google Scholar] [CrossRef]
- Lefort, C.T.; Rossaint, J.; Moser, M.; Petrich, B.G.; Zarbock, A.; Monkley, S.J.; Critchley, D.R.; Ginsberg, M.H.; Fässler, R.; Ley, K. Distinct Roles for Talin-1 and Kindlin-3 in LFA-1 Extension and Affinity Regulation. Blood 2012, 119, 4275–4282. [Google Scholar] [CrossRef]
- Lim, J.; Wiedemann, A.; Tzircotis, G.; Monkley, S.J.; Critchley, D.R.; Caron, E. An Essential Role for Talin during AMβ2-Mediated Phagocytosis. Mol. Biol. Cell 2007, 18, 976–985. [Google Scholar] [CrossRef] [Green Version]
- Calderwood, D.A.; Campbell, I.D.; Critchley, D.R. Talins and Kindlins: Partners in Integrin-Mediated Adhesion. Nat. Rev. Mol. Cell Biol. 2013, 14, 503–517. [Google Scholar] [CrossRef] [Green Version]
- García-Alvarez, B.; de Pereda, J.M.; Calderwood, D.A.; Ulmer, T.S.; Critchley, D.; Campbell, I.D.; Ginsberg, M.H.; Liddington, R.C. Structural Determinants of Integrin Recognition by Talin. Mol. Cell 2003, 11, 49–58. [Google Scholar] [CrossRef]
- Hemmings, L.; Rees, D.J.; Ohanian, V.; Bolton, S.J.; Gilmore, A.P.; Patel, B.; Priddle, H.; Trevithick, J.E.; Hynes, R.O.; Critchley, D.R. Talin Contains Three Actin-Binding Sites Each of Which Is Adjacent to a Vinculin-Binding Site. J. Cell Sci. 1996, 109, 2715–2726. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Bidirectional, Allosteric Signaling Machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Ye, F.; Ginsberg, M.H. Regulation of Integrin Activation. Annu. Rev. Cell Dev. Biol. 2011, 27, 321–345. [Google Scholar] [CrossRef]
- Shattil, S.J.; Kim, C.; Ginsberg, M.H. The Final Steps of Integrin Activation: The End Game. Nat. Rev. Mol. Cell Biol. 2010, 11, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Wegener, K.L.; Partridge, A.W.; Han, J.; Pickford, A.R.; Liddington, R.C.; Ginsberg, M.H.; Campbell, I.D. Structural Basis of Integrin Activation by Talin. Cell 2007, 128, 171–182. [Google Scholar] [CrossRef]
- Yago, T.; Petrich, B.G.; Zhang, N.; Liu, Z.; Shao, B.; Ginsberg, M.H.; McEver, R.P. Blocking Neutrophil Integrin Activation Prevents Ischemia–Reperfusion Injury. J. Exp. Med. 2015, 212, 1267–1281. [Google Scholar] [CrossRef]
- Lagarrigue, F.; Kim, C.; Ginsberg, M.H. The Rap1-RIAM-Talin Axis of Integrin Activation and Blood Cell Function. Blood 2016, 128, 479–487. [Google Scholar] [CrossRef] [Green Version]
- Lagarrigue, F.; Gingras, A.R.; Paul, D.S.; Valadez, A.J.; Cuevas, M.N.; Sun, H.; Lopez-Ramirez, M.A.; Goult, B.T.; Shattil, S.J.; Bergmeier, W.; et al. Rap1 Binding to the Talin 1 F0 Domain Makes a Minimal Contribution to Murine Platelet GPIIb-IIIa Activation. Blood Adv. 2018, 2, 2358–2368. [Google Scholar] [CrossRef] [Green Version]
- Lagarrigue, F.; Paul, D.S.; Gingras, A.R.; Valadez, A.J.; Sun, H.; Lin, J.; Cuevas, M.N.; Ablack, J.N.; Lopez-Ramirez, M.A.; Bergmeier, W.; et al. Talin-1 Is the Principal Platelet Rap1 Effector of Integrin Activation. Blood 2020, 136, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Lagarrigue, F.; Ginsberg, M.H. The Connection Between Rap1 and Talin1 in the Activation of Integrins in Blood Cells. Front. Cell Dev. Biol. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Moser, M.; Legate, K.R.; Zent, R.; Fässler, R. The Tail of Integrins, Talin, and Kindlins. Science 2009, 324, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.T.; Nygren, P.; Jo, H.; Boesze-Battaglia, K.; Bennett, J.S.; DeGrado, W.F. Affinity of Talin-1 for the Β3-Integrin Cytosolic Domain Is Modulated by Its Phospholipid Bilayer Environment. Proc. Natl. Acad. Sci. USA 2012, 109, 793–798. [Google Scholar] [CrossRef] [Green Version]
- Gingras, A.R.; Ziegler, W.H.; Bobkov, A.A.; Joyce, M.G.; Fasci, D.; Himmel, M.; Rothemund, S.; Ritter, A.; Grossmann, J.G.; Patel, B.; et al. Structural Determinants of Integrin Binding to the Talin Rod*. J. Biol. Chem. 2009, 284, 8866–8876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moes, M.; Rodius, S.; Coleman, S.J.; Monkley, S.J.; Goormaghtigh, E.; Tremuth, L.; Kox, C.; van der Holst, P.P.G.; Critchley, D.R.; Kieffer, N. The Integrin Binding Site 2 (IBS2) in the Talin Rod Domain Is Essential for Linking Integrin β Subunits to the Cytoskeleton*. J. Biol. Chem. 2007, 282, 17280–17288. [Google Scholar] [CrossRef] [Green Version]
- Rodius, S.; Chaloin, O.; Moes, M.; Schaffner-Reckinger, E.; Landrieu, I.; Lippens, G.; Lin, M.; Zhang, J.; Kieffer, N. The Talin Rod IBS2 α-Helix Interacts with the Β3 Integrin Cytoplasmic Tail Membrane-Proximal Helix by Establishing Charge Complementary Salt Bridges*. J. Biol. Chem. 2008, 283, 24212–24223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haage, A.; Goodwin, K.; Whitewood, A.; Camp, D.; Bogutz, A.; Turner, C.T.; Granville, D.J.; Lefebvre, L.; Plotnikov, S.; Goult, B.T.; et al. Talin Autoinhibition Regulates Cell-ECM Adhesion Dynamics and Wound Healing In Vivo. Cell Rep. 2018, 25, 2401–2416.e5. [Google Scholar] [CrossRef] [Green Version]
- Goult, B.T.; Xu, X.-P.; Gingras, A.R.; Swift, M.; Patel, B.; Bate, N.; Kopp, P.M.; Barsukov, I.L.; Critchley, D.R.; Volkmann, N.; et al. Structural Studies on Full-Length Talin1 Reveal a Compact Auto-Inhibited Dimer: Implications for Talin Activation. J. Struct. Biol. 2013, 184, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Saltel, F.; Mortier, E.; Hytönen, V.P.; Jacquier, M.-C.; Zimmermann, P.; Vogel, V.; Liu, W.; Wehrle-Haller, B. New PI(4,5)P2- and Membrane Proximal Integrin-Binding Motifs in the Talin Head Control Beta3-Integrin Clustering. J. Cell Biol. 2009, 187, 715–731. [Google Scholar] [CrossRef] [Green Version]
- Fong, K.P.; Molnar, K.S.; Agard, N.; Litvinov, R.I.; Kim, O.V.; Wells, J.A.; Weisel, J.W.; DeGrado, W.F.; Bennett, J.S. Cleavage of Talin by Calpain Promotes Platelet-Mediated Fibrin Clot Contraction. Blood Adv. 2021, 5, 4901–4909. [Google Scholar] [CrossRef]
- Huang, C.; Rajfur, Z.; Yousefi, N.; Chen, Z.; Jacobson, K.; Ginsberg, M.H. Talin Phosphorylation by Cdk5 Regulates Smurf1-Mediated Talin Head Ubiquitylation and Cell Migration. Nat. Cell Biol. 2009, 11, 624–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouaouina, M.; Lad, Y.; Calderwood, D.A. The N-Terminal Domains of Talin Cooperate with the Phosphotyrosine Binding-like Domain to Activate Beta1 and Beta3 Integrins. J. Biol. Chem. 2008, 283, 6118–6125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goult, B.T.; Bouaouina, M.; Elliott, P.R.; Bate, N.; Patel, B.; Gingras, A.R.; Grossmann, J.G.; Roberts, G.C.K.; Calderwood, D.A.; Critchley, D.R.; et al. Structure of a Double Ubiquitin-like Domain in the Talin Head: A Role in Integrin Activation. EMBO J. 2010, 29, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-C.; Su, W.; Cho, E.-A.; Zhang, H.; Huang, Q.; Philips, M.R.; Wu, J. Molecular Basis for Autoinhibition of RIAM Regulated by FAK in Integrin Activation. Proc. Natl. Acad. Sci. USA 2019, 116, 3524–3529. [Google Scholar] [CrossRef] [Green Version]
- Cho, E.-A.; Zhang, P.; Kumar, V.; Kavalchuk, M.; Zhang, H.; Huang, Q.; Duncan, J.S.; Wu, J. Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM. Structure 2021, 29, 320–329.e4. [Google Scholar] [CrossRef]
- Goult, B.T.; Zacharchenko, T.; Bate, N.; Tsang, R.; Hey, F.; Gingras, A.R.; Elliott, P.R.; Roberts, G.C.K.; Ballestrem, C.; Critchley, D.R.; et al. RIAM and Vinculin Binding to Talin Are Mutually Exclusive and Regulate Adhesion Assembly and Turnover. J. Biol. Chem. 2013, 288, 8238–8249. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-S.; Anekal, P.; Lim, C.J.; Liu, C.-C.; Ginsberg, M.H. Two Modes of Integrin Activation Form a Binary Molecular Switch in Adhesion Maturation. Mol. Biol. Cell 2013, 24, 1354–1362. [Google Scholar] [CrossRef]
- Watanabe, N.; Bodin, L.; Pandey, M.; Krause, M.; Coughlin, S.; Boussiotis, V.A.; Ginsberg, M.H.; Shattil, S.J. Mechanisms and Consequences of Agonist-Induced Talin Recruitment to Platelet Integrin AlphaIIbbeta3. J. Cell Biol. 2008, 181, 1211–1222. [Google Scholar] [CrossRef] [Green Version]
- Stritt, S.; Wolf, K.; Lorenz, V.; Vögtle, T.; Gupta, S.; Bösl, M.R.; Nieswandt, B. Rap1-GTP–Interacting Adaptor Molecule (RIAM) Is Dispensable for Platelet Integrin Activation and Function in Mice. Blood 2015, 125, 219–222. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Wynne, J.; Pinheiro, E.M.; Strazza, M.; Mor, A.; Montenont, E.; Berger, J.; Paul, D.S.; Bergmeier, W.; Gertler, F.B.; et al. Rap1 and Its Effector RIAM Are Required for Lymphocyte Trafficking. Blood 2015, 126, 2695–2703. [Google Scholar] [CrossRef] [Green Version]
- Ménasché, G.; Kliche, S.; Chen, E.J.H.; Stradal, T.E.B.; Schraven, B.; Koretzky, G. RIAM Links the ADAP/SKAP-55 Signaling Module to Rap1, Facilitating T-Cell-Receptor-Mediated Integrin Activation. Mol. Cell Biol. 2007, 27, 4070–4081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, J.L. Ras-like GTPases. Biochim. Biophys. Acta 1997, 1333, M19–M31. [Google Scholar] [CrossRef]
- Stork, P.J.S.; Dillon, T.J. Multiple Roles of Rap1 in Hematopoietic Cells: Complementary versus Antagonistic Functions. Blood 2005, 106, 2952–2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polakis, P.G.; Rubinfeld, B.; Evans, T.; McCormick, F. Purification of a Plasma Membrane-Associated GTPase-Activating Protein Specific for Rap1/Krev-1 from HL60 Cells. Proc. Natl. Acad. Sci. USA 1991, 88, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurachi, H.; Wada, Y.; Tsukamoto, N.; Maeda, M.; Kubota, H.; Hattori, M.; Iwai, K.; Minato, N. Human SPA-1 Gene Product Selectively Expressed in Lymphoid Tissues Is a Specific GTPase-Activating Protein for Rap1 and Rap2. Segregate Expression Profiles from a Rap1GAP Gene Product. J. Biol. Chem. 1997, 272, 28081–28088. [Google Scholar] [CrossRef] [Green Version]
- Frische, E.W.; Zwartkruis, F.J.T. Rap1, a Mercenary among the Ras-like GTPases. Dev. Biol. 2010, 340, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Jaśkiewicz, A.; Pająk, B.; Orzechowski, A. The Many Faces of Rap1 GTPase. Int. J. Mol. Sci. 2018, 19, 2848. [Google Scholar] [CrossRef] [Green Version]
- Schwamborn, J.C.; Püschel, A.W. The Sequential Activity of the GTPases Rap1B and Cdc42 Determines Neuronal Polarity. Nat. Neurosci. 2004, 7, 923–929. [Google Scholar] [CrossRef]
- Bivona, T.G.; Wiener, H.H.; Ahearn, I.M.; Silletti, J.; Chiu, V.K.; Philips, M.R. Rap1 Up-Regulation and Activation on Plasma Membrane Regulates T Cell Adhesion. J. Cell Biol. 2004, 164, 461–470. [Google Scholar] [CrossRef] [Green Version]
- Bromberger, T.; Zhu, L.; Klapproth, S.; Qin, J.; Moser, M. Rap1 and Membrane Lipids Cooperatively Recruit Talin to Trigger Integrin Activation. J. Cell Sci. 2019, 132, jcs235531. [Google Scholar] [CrossRef]
- Hogan, C.; Serpente, N.; Cogram, P.; Hosking, C.R.; Bialucha, C.U.; Feller, S.M.; Braga, V.M.M.; Birchmeier, W.; Fujita, Y. Rap1 Regulates the Formation of E-Cadherin-Based Cell-Cell Contacts. Mol. Cell. Biol. 2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawant, K.; Chen, Y.; Kotian, N.; Preuss, K.M.; McDonald, J.A. Rap1 GTPase Promotes Coordinated Collective Cell Migration in Vivo. Mol. Biol. Cell 2018, 29, 2656–2673. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-C.; Wu, J.-W.; Hsieh, Y.-C.; Huang, T.-H.; Liao, Z.-M.; Huang, Y.-S.; Mondo, J.A.; Montell, D.; Jang, A.C.-C. Rap1 Negatively Regulates the Hippo Pathway to Polarize Directional Protrusions in Collective Cell Migration. Cell Rep. 2018, 22, 2160–2175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Liu, Z.; Liu, X.; Wang, S.; Zhang, Y.; He, X.; Sun, S.; Ma, S.; Shyh-Chang, N.; Liu, F.; et al. Telomere-Dependent and Telomere-Independent Roles of RAP1 in Regulating Human Stem Cell Homeostasis. Protein Cell 2019, 10, 649–667. [Google Scholar] [CrossRef] [Green Version]
- Camp, D.; Haage, A.; Solianova, V.; Castle, W.M.; Xu, Q.A.; Lostchuck, E.; Goult, B.T.; Tanentzapf, G. Direct Binding of Talin to Rap1 Is Required for Cell-ECM Adhesion in Drosophila. J. Cell Sci. 2018, 131, jcs225144. [Google Scholar] [CrossRef] [Green Version]
- Gingras, A.R.; Lagarrigue, F.; Cuevas, M.N.; Valadez, A.J.; Zorovich, M.; McLaughlin, W.; Lopez-Ramirez, M.A.; Seban, N.; Ley, K.; Kiosses, W.B.; et al. Rap1 Binding and a Lipid-Dependent Helix in Talin F1 Domain Promote Integrin Activation in Tandem. J. Cell Biol. 2019, 218, 1799–1809. [Google Scholar] [CrossRef] [Green Version]
- Stefanini, L.; Lee, R.H.; Paul, D.S.; O’Shaughnessy, E.C.; Ghalloussi, D.; Jones, C.I.; Boulaftali, Y.; Poe, K.O.; Piatt, R.; Kechele, D.O.; et al. Functional Redundancy between RAP1 Isoforms in Murine Platelet Production and Function. Blood 2018, 132, 1951–1962. [Google Scholar] [CrossRef] [Green Version]
- Lagarrigue, F.; Tan, B.; Du, Q.; Fan, Z.; Lopez-Ramirez, M.A.; Gingras, A.R.; Wang, H.; Qi, W.; Sun, H. Direct Binding of Rap1 to Talin1 and to MRL Proteins Promotes Integrin Activation in CD4+ T Cells. J. Immunol. 2022, 208, 1378–1388. [Google Scholar] [CrossRef]
- Lozano, M.L.; Cook, A.; Bastida, J.M.; Paul, D.S.; Iruin, G.; Cid, A.R.; Adan-Pedroso, R.; Ramón González-Porras, J.; Hernández-Rivas, J.M.; Fletcher, S.J.; et al. Novel Mutations in RASGRP2, Which Encodes CalDAG-GEFI, Abrogate Rap1 Activation, Causing Platelet Dysfunction. Blood 2016, 128, 1282–1289. [Google Scholar] [CrossRef] [Green Version]
- Bergmeier, W.; Goerge, T.; Wang, H.-W.; Crittenden, J.R.; Baldwin, A.C.W.; Cifuni, S.M.; Housman, D.E.; Graybiel, A.M.; Wagner, D.D. Mice Lacking the Signaling Molecule CalDAG-GEFI Represent a Model for Leukocyte Adhesion Deficiency Type III. J. Clin. Investig. 2007, 117, 1699–1707. [Google Scholar] [CrossRef] [Green Version]
- Côte, M.; Fos, C.; Canonigo-Balancio, A.J.; Ley, K.; Bécart, S.; Altman, A. SLAT Promotes TCR-Mediated, Rap1-Dependent LFA-1 Activation and Adhesion through Interaction of Its PH Domain with Rap1. J. Cell Sci. 2015, 128, 4341–4352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, L.; Marshall, A.J.; Liu, L. Suppressive Role of Bam32/DAPP1 in Chemokine-Induced Neutrophil Recruitment. Int. J. Mol. Sci. 2021, 22, 1825. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Sun, H.; Lagarrigue, F.; Fox, J.W.; Sherman, N.E.; Gingras, A.R.; Ginsberg, M.H. Phostensin Enables Lymphocyte Integrin Activation and Population of Peripheral Lymphoid Organs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Huang, K.-Y.; Wang, T.-F.; Huang, H.; Yu, H.-C.; Yen, J.; Hung, S.; Liu, S.-Q.; Lai, N.-S.; Huang, H. Immunolocalization of Phostensin in Lymphatic Cells and Tissues. J. Histochem. Cytochem. 2011, 59, 741–749. [Google Scholar] [CrossRef] [Green Version]
- Ussar, S.; Wang, H.-V.; Linder, S.; Fässler, R.; Moser, M. The Kindlins: Subcellular Localization and Expression during Murine Development. Exp. Cell Res. 2006, 312, 3142–3151. [Google Scholar] [CrossRef]
- Bialkowska, K.; Ma, Y.-Q.; Bledzka, K.; Sossey-Alaoui, K.; Izem, L.; Zhang, X.; Malinin, N.; Qin, J.; Byzova, T.; Plow, E.F. The Integrin Co-Activator Kindlin-3 Is Expressed and Functional in a Non-Hematopoietic Cell, the Endothelial Cell. J. Biol. Chem. 2010, 285, 18640–18649. [Google Scholar] [CrossRef] [Green Version]
- Rognoni, E.; Ruppert, R.; Fässler, R. The Kindlin Family: Functions, Signaling Properties and Implications for Human Disease. J. Cell Sci. 2016, 129, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Moser, M.; Bauer, M.; Schmid, S.; Ruppert, R.; Schmidt, S.; Sixt, M.; Wang, H.-V.; Sperandio, M.; Fässler, R. Kindlin-3 Is Required for Beta2 Integrin-Mediated Leukocyte Adhesion to Endothelial Cells. Nat. Med. 2009, 15, 300–305. [Google Scholar] [CrossRef]
- Svensson, L.; Howarth, K.; McDowall, A.; Patzak, I.; Evans, R.; Ussar, S.; Moser, M.; Metin, A.; Fried, M.; Tomlinson, I.; et al. Leukocyte Adhesion Deficiency-III Is Caused by Mutations in KINDLIN3 Affecting Integrin Activation. Nat. Med. 2009, 15, 306–312. [Google Scholar] [CrossRef] [Green Version]
- Malinin, N.L.; Zhang, L.; Choi, J.; Ciocea, A.; Razorenova, O.; Ma, Y.-Q.; Podrez, E.A.; Tosi, M.; Lennon, D.P.; Caplan, A.I.; et al. A Point Mutation in KINDLIN3 Ablates Activation of Three Integrin Subfamilies in Humans. Nat. Med. 2009, 15, 313–318. [Google Scholar] [CrossRef] [Green Version]
- Dixit, N.; Kim, M.-H.; Rossaint, J.; Yamayoshi, I.; Zarbock, A.; Simon, S.I. Leukocyte Function Antigen-1, Kindlin-3, and Calcium Flux Orchestrate Neutrophil Recruitment during Inflammation. J. Immunol. 2012, 189, 5954–5964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerholm, S.C.; Lek, H.S.; Morrison, V.L. Kindlin-3 in the Immune System. Am. J. Clin. Exp. Immunol. 2014, 3, 37–42. [Google Scholar] [PubMed]
- Karaköse, E.; Schiller, H.B.; Fässler, R. The Kindlins at a Glance. J. Cell Sci. 2010, 123, 2353–2356. [Google Scholar] [CrossRef] [Green Version]
- Yates, L.A.; Füzéry, A.K.; Bonet, R.; Campbell, I.D.; Gilbert, R.J.C. Biophysical Analysis of Kindlin-3 Reveals an Elongated Conformation and Maps Integrin Binding to the Membrane-Distal β-Subunit NPXY Motif. J. Biol. Chem. 2012, 287, 37715–37731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, V.L.; MacPherson, M.; Savinko, T.; San Lek, H.; Prescott, A.; Fagerholm, S.C. The Β2 Integrin–Kindlin-3 Interaction Is Essential for T-Cell Homing but Dispensable. for T-Cell Activation in Vivo. Blood 2013, 122, 1428–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Chen, X.; Zhi, H.; Gao, J.; Bialkowska, K.; Byzova, T.V.; Pluskota, E.; White, G.C.; Liu, J.; Plow, E.F.; et al. Direct Interaction of Kindlin-3 With Integrin AIIbβ3 in Platelets Is Required for Supporting Arterial Thrombosis in Mice. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1961–1967. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Cai, J.; Gao, J.; White, G.C.; Chen, F.; Ma, Y.-Q. Interaction of Kindlin-3 and Β2-Integrins Differentially Regulates Neutrophil Recruitment and NET Release in Mice. Blood 2015, 126, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Ni, B.; Cao, Z.; Zielonka, J.; Gao, J.; Chen, F.; Kalyanaraman, B.; White, G.C.; Ma, Y.-Q. Kindlin-3 Negatively Regulates the Release of Neutrophil Extracellular Traps. J. Leukoc. Biol. 2018, 104, 597–602. [Google Scholar] [CrossRef]
- Klapproth, S.; Bromberger, T.; Türk, C.; Krüger, M.; Moser, M. A Kindlin-3–Leupaxin–Paxillin Signaling Pathway Regulates Podosome Stability. J. Cell Biol. 2019, 218, 3436–3454. [Google Scholar] [CrossRef] [Green Version]
- Plow, E.F.; Qin, J. The Kindlin Family of Adapter Proteins: A Past, Present and Future Prospectus. Circ. Res. 2019, 124, 202–204. [Google Scholar] [CrossRef]
- Wen, L.; Marki, A.; Roy, P.; McArdle, S.; Sun, H.; Fan, Z.; Gingras, A.R.; Ginsberg, M.H.; Ley, K. Kindlin-3 Recruitment to the Plasma Membrane Precedes High-Affinity Β2-Integrin and Neutrophil Arrest from Rolling. Blood 2021, 137, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Li, Y.-F.; Yau, Y.-H.; Lee, H.-S.; Tang, X.-Y.; Xue, Z.-H.; Zhou, Y.-C.; Lim, W.-M.; Cornvik, T.C.; Ruedl, C.; et al. Kindlin-3 Mediates Integrin ALβ2 Outside-in Signaling, and It Interacts with Scaffold Protein Receptor for Activated-C Kinase 1 (RACK1). J. Biol. Chem. 2012, 287, 10714–10726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouaouina, M.; Goult, B.T.; Huet-Calderwood, C.; Bate, N.; Brahme, N.N.; Barsukov, I.L.; Critchley, D.R.; Calderwood, D.A. A Conserved Lipid-Binding Loop in the Kindlin FERM F1 Domain Is Required for Kindlin-Mediated AIIbβ3 Integrin Coactivation*. J. Biol. Chem. 2012, 287, 6979–6990. [Google Scholar] [CrossRef] [Green Version]
- Chua, G.-L.; Tan, S.-M.; Bhattacharjya, S. NMR Characterization and Membrane Interactions of the Loop Region of Kindlin-3 F1 Subdomain. PLoS ONE 2016, 11, e0153501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orré, T.; Joly, A.; Karatas, Z.; Kastberger, B.; Cabriel, C.; Böttcher, R.T.; Lévêque-Fort, S.; Sibarita, J.-B.; Fässler, R.; Wehrle-Haller, B.; et al. Molecular Motion and Tridimensional Nanoscale Localization of Kindlin Control Integrin Activation in Focal Adhesions. Nat. Commun. 2021, 12, 3104. [Google Scholar] [CrossRef]
- Margraf, A.; Germena, G.; Drexler, H.C.A.; Rossaint, J.; Ludwig, N.; Prystaj, B.; Mersmann, S.; Thomas, K.; Block, H.; Gottschlich, W.; et al. The Integrin-Linked Kinase Is Required for Chemokine-Triggered High-Affinity Conformation of the Neutrophil Β2-Integrin LFA-1. Blood 2020, 136, 2200–2205. [Google Scholar] [CrossRef]
- Shao, B.; Yago, T.; Coghill, P.A.; Klopocki, A.G.; Mehta-D’souza, P.; Schmidtke, D.W.; Rodgers, W.; McEver, R.P. Signal-Dependent Slow Leukocyte Rolling Does Not Require Cytoskeletal Anchorage of P-Selectin Glycoprotein Ligand-1 (PSGL-1) or Integrin ALβ2. J. Biol. Chem. 2012, 287, 19585–19598. [Google Scholar] [CrossRef] [Green Version]
- Critchley, D.R. Biochemical and Structural Properties of the Integrin-Associated Cytoskeletal Protein Talin. Annu. Rev. Biophys. 2009, 38, 235–254. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, J.; Bromberger, T.; Holly, A.; Lu, F.; Liu, H.; Sun, K.; Klapproth, S.; Hirbawi, J.; Byzova, T.V.; et al. Structure of Rap1b Bound to Talin Reveals a Pathway for Triggering Integrin Activation. Nat. Commun. 2017, 8, 1744. [Google Scholar] [CrossRef] [Green Version]
- Bledzka, K.; Bialkowska, K.; Sossey-Alaoui, K.; Vaynberg, J.; Pluskota, E.; Qin, J.; Plow, E.F. Kindlin-2 Directly Binds Actin and Regulates Integrin Outside-in Signaling. J. Cell Biol. 2016, 213, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Patsoukis, N.; Bardhan, K.; Weaver, J.D.; Sari, D.; Torres-Gomez, A.; Li, L.; Strauss, L.; Lafuente, E.M.; Boussiotis, V.A. The Adaptor Molecule RIAM Integrates Signaling Events Critical for Integrin-Mediated Control of Immune Function and Cancer Progression. Sci. Signal. 2017, 10, eaam8298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otey, C.A.; Pavalko, F.M.; Burridge, K. An Interaction between Alpha-Actinin and the Beta 1 Integrin Subunit in Vitro. J. Cell Biol. 1990, 111, 721–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpén, O.; Pallai, P.; Staunton, D.E.; Springer, T.A. Association of Intercellular Adhesion Molecule-1 (ICAM-1) with Actin-Containing Cytoskeleton and Alpha-Actinin. J. Cell Biol. 1992, 118, 1223–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roca-Cusachs, P.; del Rio, A.; Puklin-Faucher, E.; Gauthier, N.C.; Biais, N.; Sheetz, M.P. Integrin-Dependent Force Transmission to the Extracellular Matrix by α-Actinin Triggers Adhesion Maturation. Proc. Natl. Acad. Sci. USA 2013, 110, E1361–E1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arandjelovic, S.; Perry, J.S.A.; Lucas, C.D.; Penberthy, K.K.; Kim, T.-H.; Zhou, M.; Rosen, D.A.; Chuang, T.-Y.; Bettina, A.M.; Shankman, L.S.; et al. A Noncanonical Role for the Engulfment Gene ELMO1 in Neutrophils That Promotes Inflammatory Arthritis. Nat. Immunol. 2019, 20, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Tang, W.; Bruscia, E.M.; Zhang, P.-X.; Lin, A.; Gaines, P.; Wu, D.; Halene, S. SRF Is Required for Neutrophil Migration in Response to Inflammation. Blood 2014, 123, 3027–3036. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, G.; Langen, H.; Naito, M.; Pieters, J. A Coat Protein on Phagosomes Involved in the Intracellular Survival of Mycobacteria. Cell 1999, 97, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Pick, R.; Begandt, D.; Stocker, T.J.; Salvermoser, M.; Thome, S.; Böttcher, R.T.; Montanez, E.; Harrison, U.; Forné, I.; Khandoga, A.G.; et al. Coronin 1A, a Novel Player in Integrin Biology, Controls Neutrophil Trafficking in Innate Immunity. Blood 2017, 130, 847–858. [Google Scholar] [CrossRef]
- Riley, D.R.J.; Khalil, J.S.; Pieters, J.; Naseem, K.M.; Rivero, F. Coronin 1 Is Required for Integrin Β2 Translocation in Platelets. Int. J. Mol. Sci. 2020, 21, 356. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Yang, H.; Zhang, R.; Sun, H.; Zhao, B.; Gao, C.; Zhu, F.; Jiao, J. The Upregulation of TRPC6 Contributes to Ca2+ Signaling and Actin Assembly in Human Mesangial Cells after Chronic Hypoxia. Biochem. Biophys. Res. Commun. 2012, 421, 750–756. [Google Scholar] [CrossRef]
- Lindemann, O.; Rossaint, J.; Najder, K.; Schimmelpfennig, S.; Hofschröer, V.; Wälte, M.; Fels, B.; Oberleithner, H.; Zarbock, A.; Schwab, A. Intravascular Adhesion and Recruitment of Neutrophils in Response to CXCL1 Depends on Their TRPC6 Channels. J. Mol. Med. 2020, 98, 349–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazaki, Y.; Takada, S.; Nio-Kobayashi, J.; Maekawa, S.; Higashi, T.; Onodera, Y.; Sabe, H. Mitofusin 2 Is Involved in Chemotaxis of Neutrophil-like Differentiated HL-60 cells. Biochem. Biophys. Res. Commun. 2019, 513, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hsu, A.Y.; Wang, Y.; Syahirah, R.; Wang, T.; Jeffries, J.; Wang, X.; Mohammad, H.; Seleem, M.N.; Umulis, D.; et al. Mitofusin 2 Regulates Neutrophil Adhesive Migration and the Actin Cytoskeleton. J. Cell Sci. 2020, 133, jcs248880. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Hsu, A.Y.; Wang, Y.; Lin, T.; Sun, H.; Pachter, J.S.; Groisman, A.; Imperioli, M.; Yungher, F.W.; Hu, L.; et al. Mitofusin-2 Regulates Leukocyte Adhesion and Β2 Integrin Activation. J. Leukoc. Biol. 2021. [Google Scholar] [CrossRef]
- Fan, Z.; Kiosses, W.B.; Sun, H.; Orecchioni, M.; Ghosheh, Y.; Zajonc, D.M.; Arnaout, M.A.; Gutierrez, E.; Groisman, A.; Ginsberg, M.H.; et al. High-Affinity Bent Β2-Integrin Molecules in Arresting Neutrophils Face Each Other through Binding to ICAMs In Cis. Cell Rep. 2019, 26, 119–130.e5. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zhi, K.; Hu, L.; Fan, Z. The Activation and Regulation of Β2 Integrins in Phagocytes and Phagocytosis. Front. Immunol. 2021, 12, 978. [Google Scholar] [CrossRef]
- Yao, C.-H.; Wang, R.; Wang, Y.; Kung, C.-P.; Weber, J.D.; Patti, G.J. Mitochondrial Fusion Supports Increased Oxidative Phosphorylation during Cell Proliferation. eLife 2019, 8, e41351. [Google Scholar] [CrossRef]
- de Brito, O.M.; Scorrano, L. Mitofusin 2 Tethers Endoplasmic Reticulum to Mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Kuo, I.Y.; Brill, A.L.; Lemos, F.O.; Jiang, J.Y.; Falcone, J.L.; Kimmerling, E.P.; Cai, Y.; Dong, K.; Kaplan, D.L.; Wallace, D.P.; et al. Polycystin 2 Regulates Mitochondrial Ca2+ Signaling, Bioenergetics, and Dynamics through Mitofusin 2. Sci. Signal. 2019, 12, eaat7397. [Google Scholar] [CrossRef]
- Khodzhaeva, V.; Schreiber, Y.; Geisslinger, G.; Brandes, R.P.; Brüne, B.; Namgaladze, D. Mitofusin 2 Deficiency Causes Pro-Inflammatory Effects in Human Primary Macrophages. Front. Immunol. 2021, 12, 723683. [Google Scholar] [CrossRef]
- Lloberas, J.; Muñoz, J.P.; Hernández-Álvarez, M.I.; Cardona, P.-J.; Zorzano, A.; Celada, A. Macrophage Mitochondrial MFN2 (Mitofusin 2) Links Immune Stress and Immune Response through Reactive Oxygen Species (ROS) Production. Autophagy 2020, 16, 2307–2309. [Google Scholar] [CrossRef]
- Tur, J.; Pereira-Lopes, S.; Vico, T.; Marín, E.A.; Muñoz, J.P.; Hernández-Alvarez, M.; Cardona, P.-J.; Zorzano, A.; Lloberas, J.; Celada, A. Mitofusin 2 in Macrophages Links Mitochondrial ROS Production, Cytokine Release, Phagocytosis, Autophagy, and Bactericidal Activity. Cell Rep. 2020, 32, 108079. [Google Scholar] [CrossRef] [PubMed]
- Fagerholm, S.C.; Hilden, T.J.; Nurmi, S.M.; Gahmberg, C.G. Specific Integrin Alpha and Beta Chain Phosphorylations Regulate LFA-1 Activation through Affinity-Dependent and -Independent Mechanisms. J. Cell Biol. 2005, 171, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerholm, S.C.; Varis, M.; Stefanidakis, M.; Hilden, T.J.; Gahmberg, C.G. Alpha-Chain Phosphorylation of the Human Leukocyte CD11b/CD18 (Mac-1) Integrin Is Pivotal for Integrin Activation to Bind ICAMs and Leukocyte Extravasation. Blood 2006, 108, 3379–3386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uotila, L.M.; Aatonen, M.; Gahmberg, C.G. Integrin CD11c/CD18 α-Chain Phosphorylation Is Functionally Important. J. Biol. Chem. 2013, 288, 33494–33499. [Google Scholar] [CrossRef] [Green Version]
- Nurmi, S.M.; Autero, M.; Raunio, A.K.; Gahmberg, C.G.; Fagerholm, S.C. Phosphorylation of the LFA-1 Integrin Beta2-Chain on Thr-758 Leads to Adhesion, Rac-1/Cdc42 Activation, and Stimulation of CD69 Expression in Human T Cells. J. Biol. Chem. 2007, 282, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, T.; Bundo, K.; Hino, A.; Honda, K.; Inoue, A.; Shirakata, M.; Osawa, M.; Tamura, T.; Nariuchi, H.; Oda, H.; et al. Dok-1 and Dok-2 Are Negative Regulators of T Cell Receptor Signaling. Int. Immunol. 2007, 19, 487–495. [Google Scholar] [CrossRef]
- Mashima, R.; Hishida, Y.; Tezuka, T.; Yamanashi, Y. The Roles of Dok Family Adapters in Immunoreceptor Signaling. Immunol. Rev. 2009, 232, 273–285. [Google Scholar] [CrossRef]
- Lim, J.; Hotchin, N.A.; Caron, E. Ser756 of Β2 Integrin Controls Rap1 Activity during Inside-out Activation of AMβ2. Biochem. J. 2011, 437, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Sala, C.; Yoon, J.; Park, S.; Kuroda, S.; Sheng, M.; Kim, E. Sharpin, a Novel Postsynaptic Density Protein That Directly Interacts with the Shank Family of Proteins. Mol. Cell Neurosci. 2001, 17, 385–397. [Google Scholar] [CrossRef]
- Ikeda, F.; Deribe, Y.L.; Skånland, S.S.; Stieglitz, B.; Grabbe, C.; Franz-Wachtel, M.; van Wijk, S.J.L.; Goswami, P.; Nagy, V.; Terzic, J.; et al. SHARPIN Forms a Linear Ubiquitin Ligase Complex Regulating NF-ΚB Activity and Apoptosis. Nature 2011, 471, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, F.; Nakagawa, T.; Nakahara, M.; Saeki, Y.; Taniguchi, M.; Sakata, S.; Tanaka, K.; Nakano, H.; Iwai, K. SHARPIN Is a Component of the NF-ΚB-Activating Linear Ubiquitin Chain Assembly Complex. Nature 2011, 471, 633–636. [Google Scholar] [CrossRef] [PubMed]
- IWAI, K. LUBAC-Mediated Linear Ubiquitination: A Crucial Regulator of Immune Signaling. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021, 97, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Kirisako, T.; Kamei, K.; Murata, S.; Kato, M.; Fukumoto, H.; Kanie, M.; Sano, S.; Tokunaga, F.; Tanaka, K.; Iwai, K. A Ubiquitin Ligase Complex Assembles Linear Polyubiquitin Chains. EMBO J. 2006, 25, 4877–4887. [Google Scholar] [CrossRef] [PubMed]
- Rantala, J.K.; Pouwels, J.; Pellinen, T.; Veltel, S.; Laasola, P.; Potter, C.S.; Duffy, T.; Sundberg, J.P.; Kallioniemi, O.; Askari, J.A.; et al. SHARPIN Is an Endogenous Inhibitor of Beta1-Integrin Activation. Nat. Cell Biol. 2011, 13, 1315–1324. [Google Scholar] [CrossRef] [Green Version]
- Tokunaga, F.; Iwai, K. LUBAC, a Novel Ubiquitin Ligase for Linear Ubiquitination, Is Crucial for Inflammation and Immune Responses. Microbes Infect. 2012, 14, 563–572. [Google Scholar] [CrossRef]
- Gurung, P.; Sharma, B.R.; Kanneganti, T.-D. Distinct Role of IL-1β in Instigating Disease in Sharpincpdm Mice. Sci. Rep. 2016, 6, 36634. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pulikkot, S.; Hu, L.; Chen, Y.; Sun, H.; Fan, Z. Integrin Regulators in Neutrophils. Cells 2022, 11, 2025. https://doi.org/10.3390/cells11132025
Pulikkot S, Hu L, Chen Y, Sun H, Fan Z. Integrin Regulators in Neutrophils. Cells. 2022; 11(13):2025. https://doi.org/10.3390/cells11132025
Chicago/Turabian StylePulikkot, Sunitha, Liang Hu, Yunfeng Chen, Hao Sun, and Zhichao Fan. 2022. "Integrin Regulators in Neutrophils" Cells 11, no. 13: 2025. https://doi.org/10.3390/cells11132025
APA StylePulikkot, S., Hu, L., Chen, Y., Sun, H., & Fan, Z. (2022). Integrin Regulators in Neutrophils. Cells, 11(13), 2025. https://doi.org/10.3390/cells11132025






