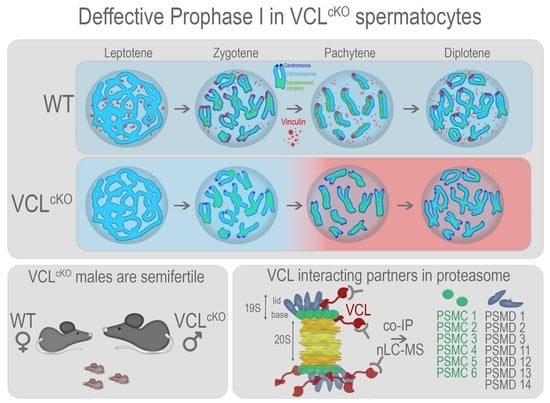
Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
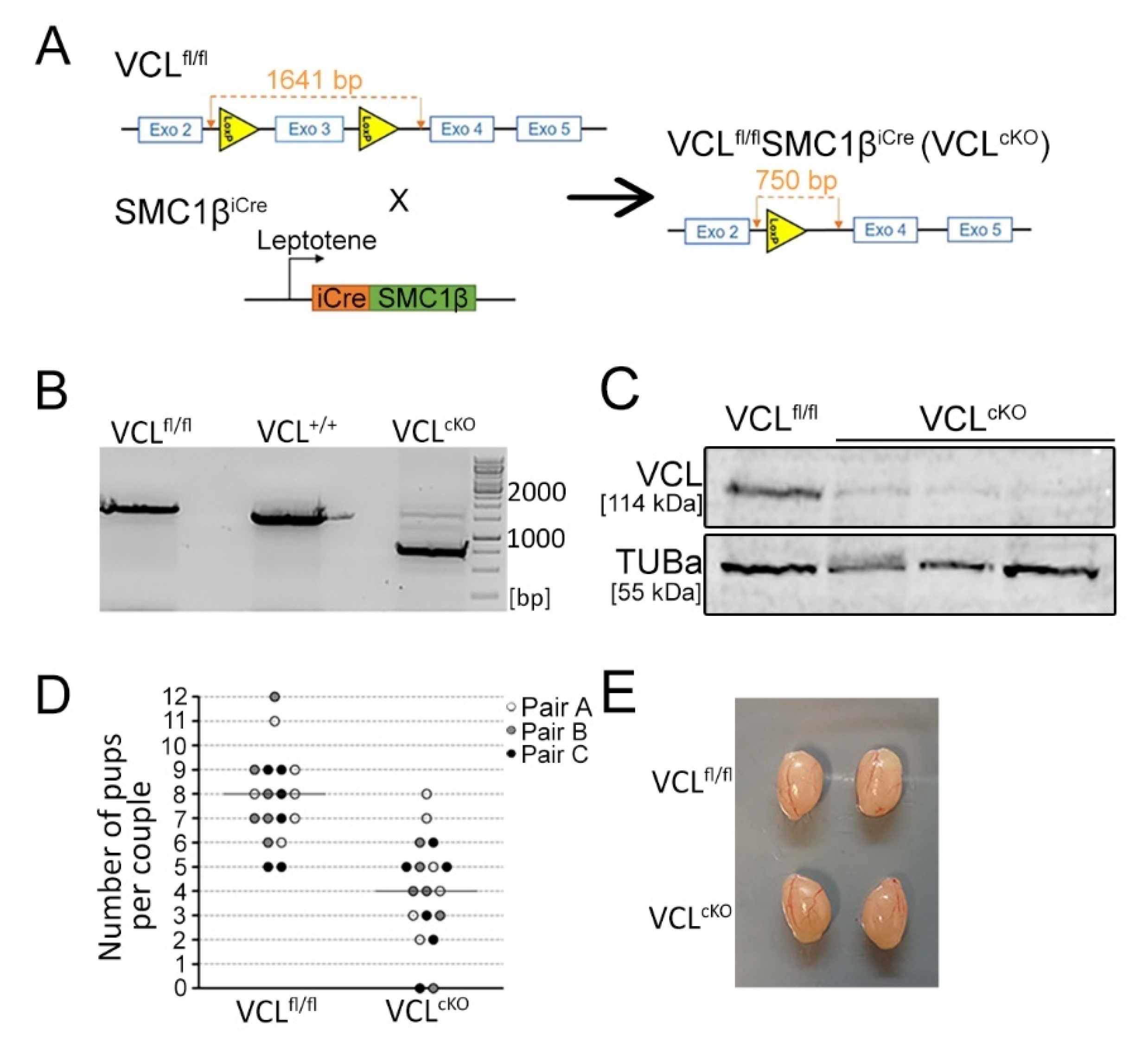
2.2. Generation of Vinculin Conditional Knock-out Mouse (VCL cKO) and Genotyping
2.3. Testes Cryo-Sections and Spermatocytes Cell Spreads
2.4. Seminiferous Tubules Squash
2.5. Immunofluorescence and Microscopy
2.6. TUNEL Assay
2.7. Short-Term Culture of Spermatocytes and Okadaic Acid Treatment
2.8. Testicular Single-Cell Suspension, Sperm Count, and Sperm Head Evaluation
2.9. FACS of Tubular Cells and RT-qPCR
2.10. Colchicine Treatment and Chromosome Segregation Analysis
2.11. Western Blot Protein Analysis
2.12. Preparation of Cell and Nuclear Protein Extracts
2.13. Co-Immunoprecipitation and Protein Digestion
2.14. nLC-MS 2 Analysis and Data Analysis
2.15. Computational and Statistical Analysis
3. Results
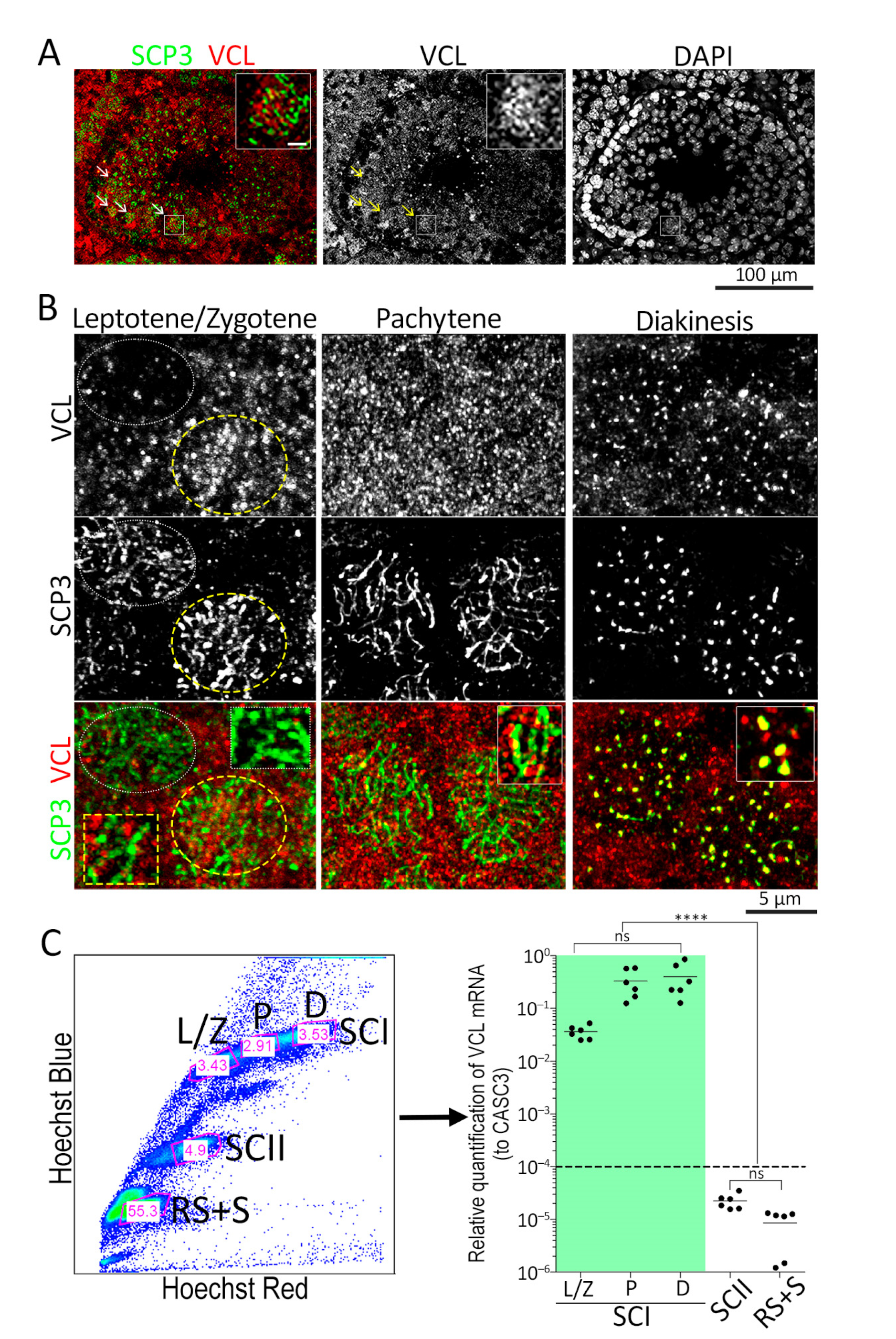
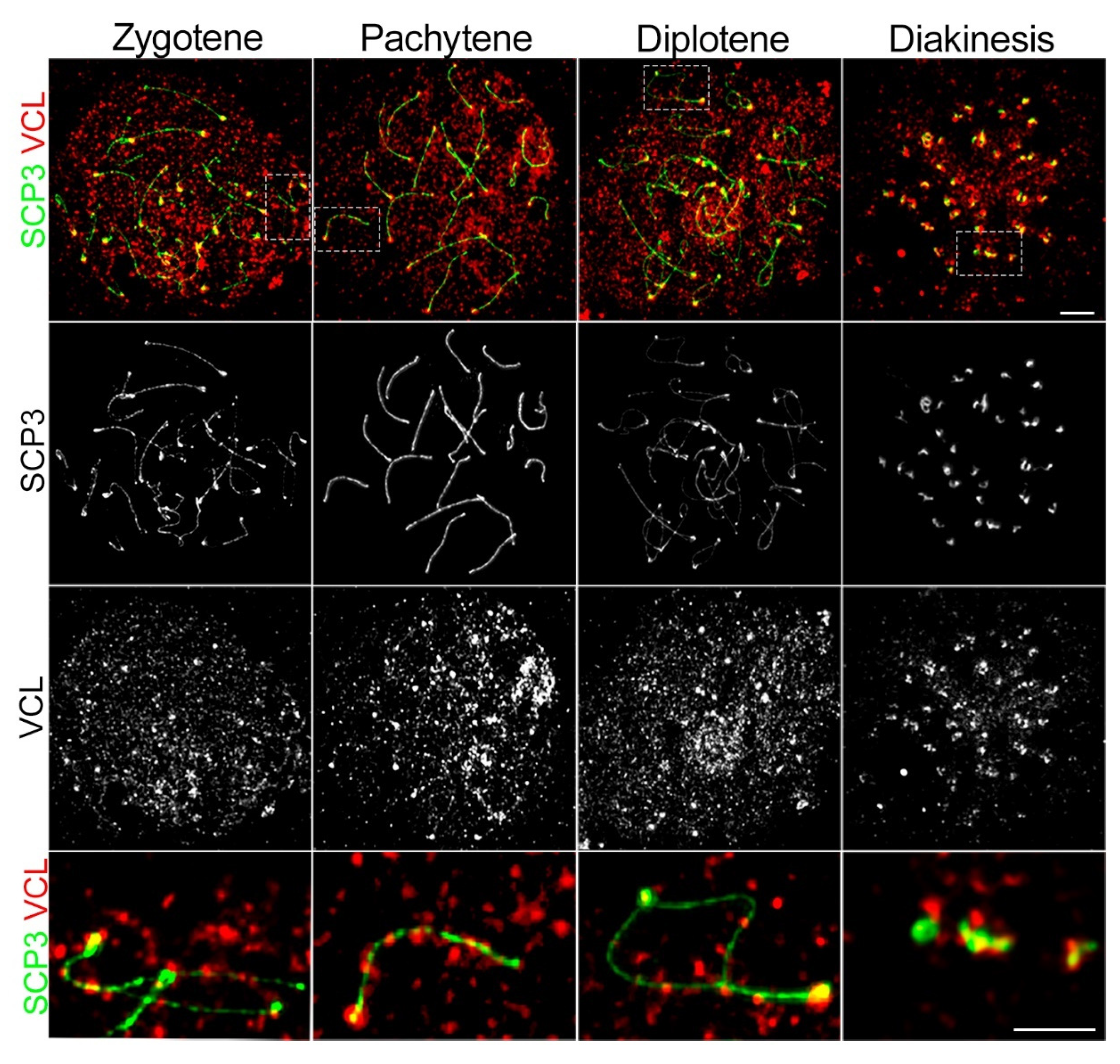
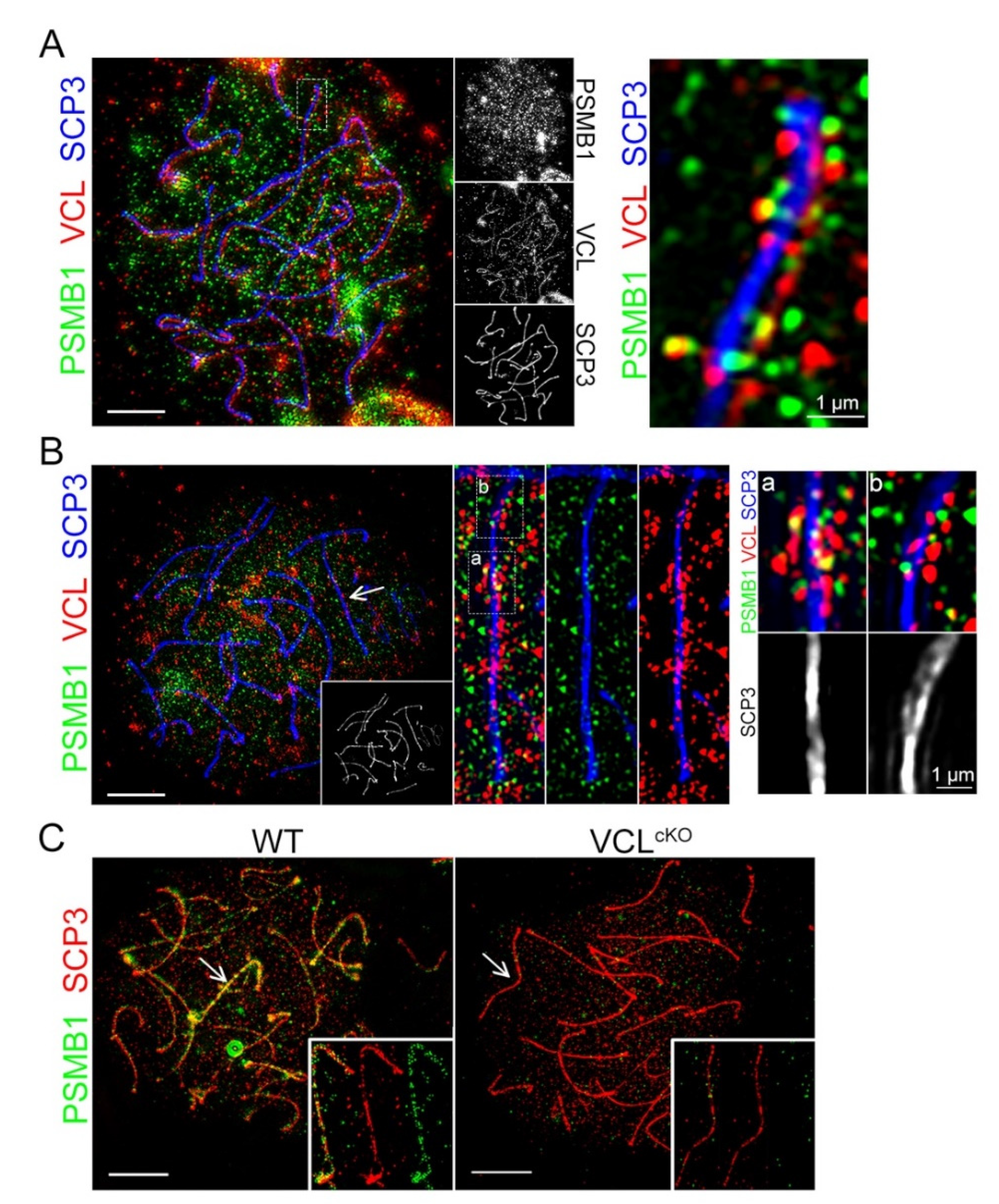
3.1. VCL localizes to Meiotic Nuclei during Prophase I
3.2. Depletion of VCL in Primary Spermatocytes Causes Decreased Fertility in Males
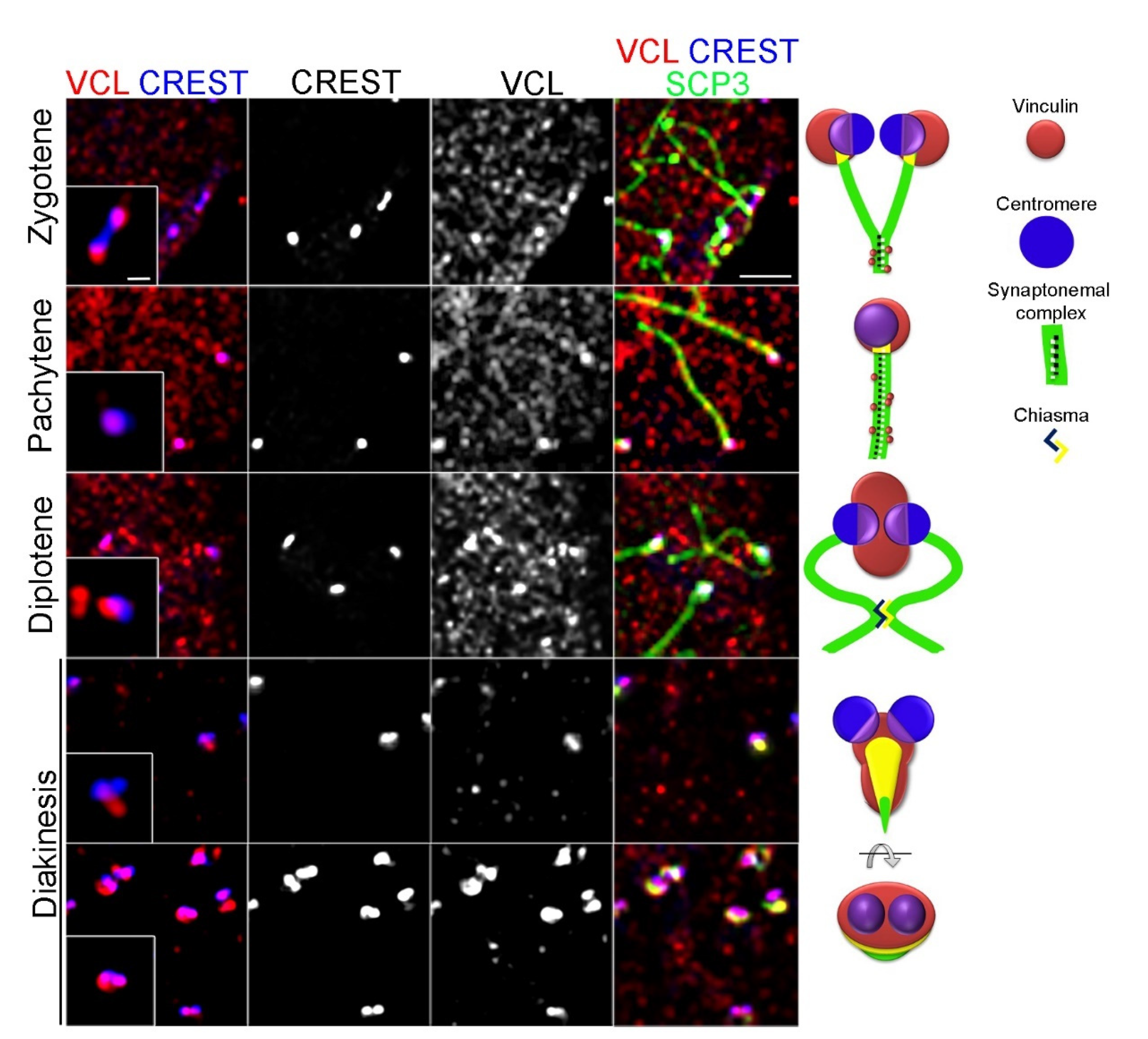
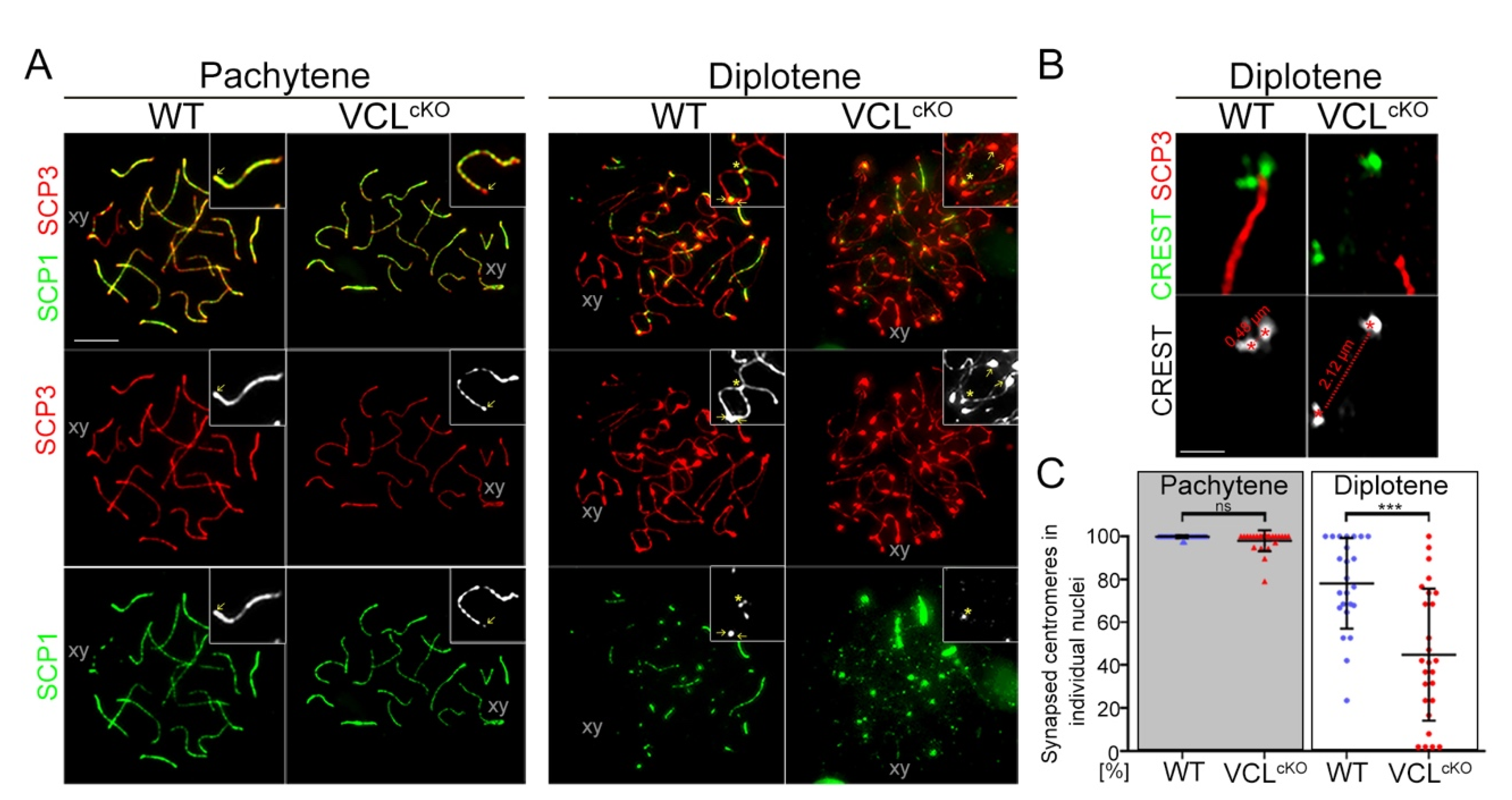
3.3. Depletion of VCL Causes Unwanted Centromeric De-Synapsis in Diplotene
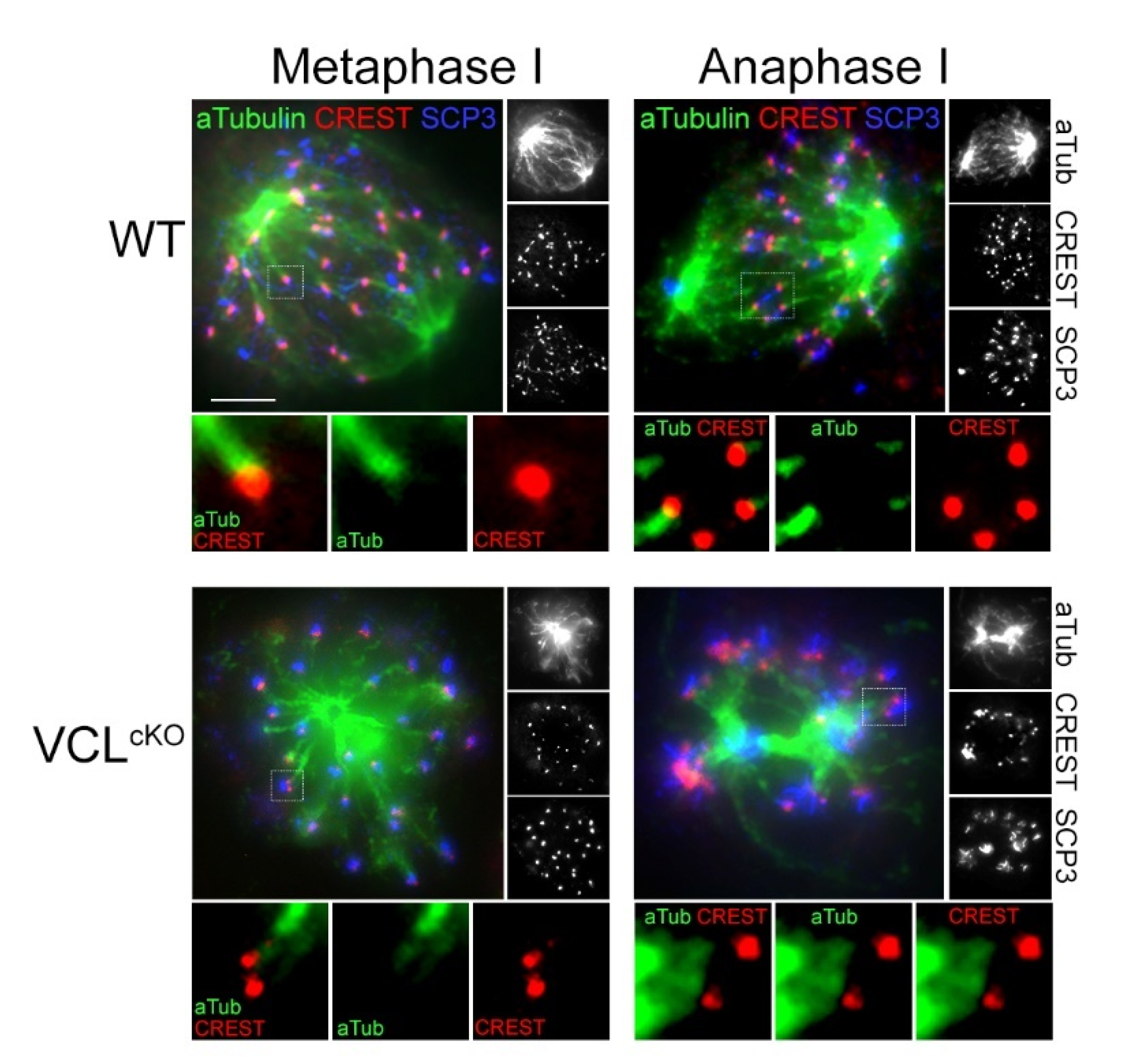
3.4. VCL Is Required for Meiotic Progression
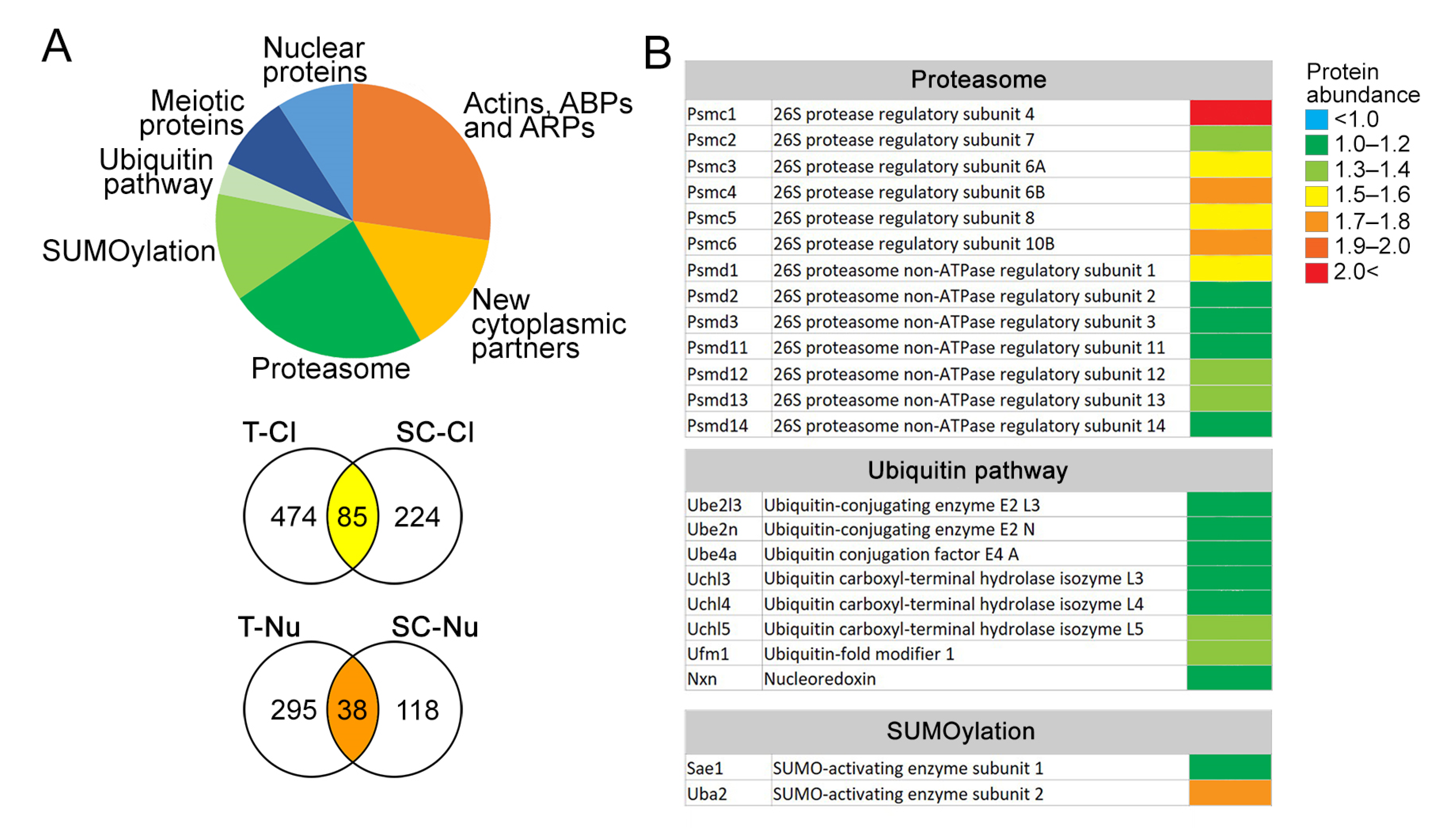
3.5. VCL Associates with Proteasome Subunits in Pachytene Nuclei
4. Discussion
4.1. VCL Might Be a New Kinetochore Component
4.2. Vinculin Associates with UPS and Probably Plays a Dual Role
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kleckner, N. Meiosis: How could it work? Proc. Natl. Acad. Sci. USA 1996, 93, 8167–8174. [Google Scholar] [PubMed]
- Nasmyth, K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001, 35, 673–745. [Google Scholar] [CrossRef] [PubMed]
- Petronczki, M.; Siomos, M.F.; Nasmyth, K. Un menage a quatre: The molecular biology of chromosome segregation in meiosis. Cell 2003, 112, 423–440. [Google Scholar] [CrossRef]
- Gerton, J.L.; Hawley, R.S. Homologous chromosome interactions in meiosis: Diversity amidst conservation. Nat. Rev. Genet. 2005, 6, 477–487. [Google Scholar] [CrossRef]
- Zickler, D. From early homologue recognition to synaptonemal complex formation. Chromosoma 2006, 115, 158–174. [Google Scholar] [CrossRef]
- Bhalla, N.; Dernburg, A.F. Prelude to a division. Annu. Rev. Cell Dev. Biol. 2008, 24, 397–424. [Google Scholar] [CrossRef]
- Hassold, T.J.; Jacobs, P.A. Trisomy in man. Annu. Rev. Genet. 1984, 18, 69–97. [Google Scholar] [CrossRef]
- Von Wettstein, D.; Rasmussen, S.W.; Holm, P.B. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 1984, 18, 331–413. [Google Scholar] [CrossRef]
- Page, S.L.; Hawley, R.S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004, 20, 525–558. [Google Scholar] [CrossRef]
- Yang, F.; Wang, P.J. The Mammalian synaptonemal complex: A scaffold and beyond. Genome Dyn. 2009, 5, 69–80. [Google Scholar] [CrossRef]
- Stewart, M.N.; Dawson, D.S. Changing partners: Moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet. TIG 2008, 24, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Chen, J.K.; Reynolds, A.; Hoog, C.; Paddy, M.; Hunter, N. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 2012, 8, e1002790. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, D.W.; Mao, Y.; Sullivan, K.F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 2003, 112, 407–421. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Edelmaier, C.; Lamson, A.R.; Gergely, Z.R.; Ansari, S.; Blackwell, R.; McIntosh, J.R.; Glaser, M.A.; Betterton, M.D. Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling. Elife 2020, 9, e48787. [Google Scholar] [CrossRef]
- Sun, F.; Handel, M.A. Regulation of the meiotic prophase I to metaphase I transition in mouse spermatocytes. Chromosoma 2008, 117, 471–485. [Google Scholar] [CrossRef]
- Rao, H.B.; Qiao, H.; Bhatt, S.K.; Bailey, L.R.; Tran, H.D.; Bourne, S.L.; Qiu, W.; Deshpande, A.; Sharma, A.N.; Beebout, C.J.; et al. A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 2017, 355, 403–407. [Google Scholar] [CrossRef]
- Brown, P.W.; Hwang, K.; Schlegel, P.N.; Morris, P.L. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum. Reprod. 2008, 23, 2850–2857. [Google Scholar] [CrossRef]
- Bhagwat, N.R.; Owens, S.N.; Ito, M.; Boinapalli, J.V.; Poa, P.; Ditzel, A.; Kopparapu, S.; Mahalawat, M.; Davies, O.R.; Collins, S.R.; et al. SUMO is a pervasive regulator of meiosis. Elife 2021, 10, e57720. [Google Scholar] [CrossRef]
- Savulescu, A.F.; Glickman, M.H. Proteasome activator 200: The heat is on. Mol. Cell. Proteom. 2011, 10, R110 006890. [Google Scholar] [CrossRef]
- Ahuja, J.S.; Sandhu, R.; Mainpal, R.; Lawson, C.; Henley, H.; Hunt, P.A.; Yanowitz, J.L.; Borner, G.V. Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 2017, 355, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Dixit, V.M. Cross talk between ubiquitination and demethylation. Mol. Cell. Biol. 2011, 31, 3682–3683. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hebert, A.S.; Richards, A.L.; Bailey, D.J.; Ulbrich, A.; Coughlin, E.E.; Westphall, M.S.; Coon, J.J. The one hour yeast proteome. Mol. Cell. Proteom. 2014, 13, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.C.; Yang, W.X. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Mol. Biol. Rep. 2013, 40, 3213–3230. [Google Scholar] [CrossRef]
- Thievessen, I.; Thompson, P.M.; Berlemont, S.; Plevock, K.M.; Plotnikov, S.V.; Zemljic-Harpf, A.; Ross, R.S.; Davidson, M.W.; Danuser, G.; Campbell, S.L.; et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 2013, 202, 163–177. [Google Scholar] [CrossRef]
- Saunders, R.M.; Holt, M.R.; Jennings, L.; Sutton, D.H.; Barsukov, I.L.; Bobkov, A.; Liddington, R.C.; Adamson, E.A.; Dunn, G.A.; Critchley, D.R. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 2006, 85, 487–500. [Google Scholar] [CrossRef]
- Philimonenko, V.V.; Zhao, J.; Iben, S.; Dingova, H.; Kysela, K.; Kahle, M.; Zentgraf, H.; Hofmann, W.A.; de Lanerolle, P.; Hozak, P.; et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 2004, 6, 1165–1172. [Google Scholar] [CrossRef]
- Visa, N.; Percipalle, P. Nuclear functions of actin. Cold Spring Harb. Perspect. Biol. 2010, 2, a000620. [Google Scholar] [CrossRef]
- Marasek, P.; Dzijak, R.; Studenyak, I.; Fiserova, J.; Ulicna, L.; Novak, P.; Hozak, P. Paxillin-dependent regulation of IGF2 and H19 gene cluster expression. J. Cell Sci. 2015, 128, 3106–3116. [Google Scholar] [CrossRef]
- Baarlink, C.; Plessner, M.; Sherrard, A.; Morita, K.; Misu, S.; Virant, D.; Kleinschnitz, E.M.; Harniman, R.; Alibhai, D.; Baumeister, S.; et al. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 2017, 19, 1389–1399. [Google Scholar] [CrossRef]
- Oda, H.; Shirai, N.; Ura, N.; Ohsumi, K.; Iwabuchi, M. Chromatin tethering to the nuclear envelope by nuclear actin filaments: A novel role of the actin cytoskeleton in the Xenopus blastula. Genes Cells: Devoted Mol. Cell. Mech. 2017, 22, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, J.A.; Vartiainen, M.K. Diverse functions for different forms of nuclear actin. Curr. Opin. Cell Biol. 2017, 46, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Riveline, D.; Zamir, E.; Balaban, N.Q.; Schwarz, U.S.; Ishizaki, T.; Narumiya, S.; Kam, Z.; Geiger, B.; Bershadsky, A.D. Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 2001, 153, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Atherton, P.; Stutchbury, B.; Wang, D.Y.; Jethwa, D.; Tsang, R.; Meiler-Rodriguez, E.; Wang, P.; Bate, N.; Zent, R.; Barsukov, I.L.; et al. Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 2015, 6, 10038. [Google Scholar] [CrossRef]
- Thompson, P.M.; Ramachandran, S.; Case, L.B.; Tolbert, C.E.; Tandon, A.; Pershad, M.; Dokholyan, N.V.; Waterman, C.M.; Campbell, S.L. A Structural Model for Vinculin Insertion into PIP2-Containing Membranes and the Effect of Insertion on Vinculin Activation and Localization. Structure 2017, 25, 264–275. [Google Scholar] [CrossRef]
- Balaban, C.; Sztacho, M.; Blazikova, M.; Hozak, P. The F-Actin-Binding MPRIP Forms Phase-Separated Condensates and Associates with PI(4,5)P2 and Active RNA Polymerase II in the Cell Nucleus. Cells 2021, 10, 848. [Google Scholar] [CrossRef]
- Hoboth, P.; Sztacho, M.; Sebesta, O.; Schatz, M.; Castano, E.; Hozak, P. Nanoscale mapping of nuclear phosphatidylinositol phosphate landscape by dual-color dSTORM. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158890. [Google Scholar] [CrossRef]
- Diez, G.; Auernheimer, V.; Fabry, B.; Goldmann, W.H. Head/tail interaction of vinculin influences cell mechanical behavior. Biochem. Biophys. Res. Commun. 2011, 406, 85–88. [Google Scholar] [CrossRef]
- Plotnikov, S.V.; Sabass, B.; Schwarz, U.S.; Waterman, C.M. High-resolution traction force microscopy. Methods Cell Biol. 2014, 123, 367–394. [Google Scholar] [CrossRef]
- Dumbauld, D.W.; Lee, T.T.; Singh, A.; Scrimgeour, J.; Gersbach, C.A.; Zamir, E.A.; Fu, J.; Chen, C.S.; Curtis, J.E.; Craig, S.W.; et al. How vinculin regulates force transmission. Proc. Natl. Acad. Sci. USA 2013, 110, 9788–9793. [Google Scholar] [CrossRef]
- Zemljic-Harpf, A.E.; Miller, J.C.; Henderson, S.A.; Wright, A.T.; Manso, A.M.; Elsherif, L.; Dalton, N.D.; Thor, A.K.; Perkins, G.A.; McCulloch, A.D.; et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol. Cell. Biol. 2007, 27, 7522–7537. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Baribault, H.; Adamson, E.D. Vinculin knockout results in heart and brain defects during embryonic development. Development 1998, 125, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Revenkova, E.; Eijpe, M.; Heyting, C.; Gross, B.; Jessberger, R. Novel meiosis-specific isoform of mammalian SMC1. Mol. Cell. Biol. 2001, 21, 6984–6998. [Google Scholar] [CrossRef] [PubMed]
- Revenkova, E.; Eijpe, M.; Heyting, C.; Hodges, C.A.; Hunt, P.A.; Liebe, B.; Scherthan, H.; Jessberger, R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 2004, 6, 555–562. [Google Scholar] [CrossRef]
- Anderson, L.K.; Reeves, A.; Webb, L.M.; Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 1999, 151, 1569–1579. [Google Scholar] [CrossRef]
- Page, J.; Suja, J.A.; Santos, J.L.; Rufas, J.S. Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 1998, 6, 639–642. [Google Scholar] [CrossRef]
- La Salle, S.; Sun, F.; Handel, M.A. Isolation and short-term culture of mouse spermatocytes for analysis of meiosis. Methods Mol. Biol. 2009, 558, 279–297. [Google Scholar] [CrossRef]
- Bastos, H.; Lassalle, B.; Chicheportiche, A.; Riou, L.; Testart, J.; Allemand, I.; Fouchet, P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytom. A 2005, 65, 40–49. [Google Scholar] [CrossRef]
- Gu, N.H.; Zhao, W.L.; Wang, G.S.; Sun, F. Comparative analysis of mammalian sperm ultrastructure reveals relationships between sperm morphology, mitochondrial functions and motility. Reprod. Biol. Endocrinol. 2019, 17, 66. [Google Scholar] [CrossRef]
- Reynolds, A.; Qiao, H.; Yang, Y.; Chen, J.K.; Jackson, N.; Biswas, K.; Holloway, J.K.; Baudat, F.; de Massy, B.; Wang, J.; et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 2013, 45, 269–278. [Google Scholar] [CrossRef]
- Vrooman, L.A.; Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Evidence for paternal age-related alterations in meiotic chromosome dynamics in the mouse. Genetics 2014, 196, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Matunis, M.J. Isolation and fractionation of rat liver nuclear envelopes and nuclear pore complexes. Methods 2006, 39, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Tomita, M.; Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008, 7, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Bartles, J.R.; Wierda, A.; Zheng, L. Identification and characterization of espin, an actin-binding protein localized to the F-actin-rich junctional plaques of Sertoli cell ectoplasmic specializations. J. Cell Sci. 1996, 109 (Pt 6), 1229–1239. [Google Scholar] [CrossRef]
- Young, J.S.; Vogl, A.W. Focal adhesion proteins Zyxin and Vinculin are co-distributed at tubulobulbar complexes. Spermatogenesis 2012, 2, 63–68. [Google Scholar] [CrossRef][Green Version]
- Young, J.S.; De Asis, M.; Guttman, J.; Vogl, A.W. Cortactin depletion results in short tubulobulbar complexes and spermiation failure in rat testes. Biol. Open 2012, 1, 1069–1077. [Google Scholar] [CrossRef][Green Version]
- Lie, P.P.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Epidermal growth factor receptor pathway substrate 8 (Eps8) is a novel regulator of cell adhesion and the blood-testis barrier integrity in the seminiferous epithelium. FASEB J. 2009, 23, 2555–2567. [Google Scholar] [CrossRef]
- Lie, P.P.; Chan, A.Y.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Restricted Arp3 expression in the testis prevents blood-testis barrier disruption during junction restructuring at spermatogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 11411–11416. [Google Scholar] [CrossRef] [PubMed]
- Gaysinskaya, V.; Soh, I.Y.; van der Heijden, G.W.; Bortvin, A. Optimized flow cytometry isolation of murine spermatocytes. Cytom. A 2014, 85, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.T.; Viera, A.; Gomez, R.; Page, J.; Benavente, R.; Santos, J.L.; Rufas, J.S.; Suja, J.A. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J. Cell Sci. 2004, 117, 1221–1234. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, V.V.; Hochwagen, A. The meiotic checkpoint network: Step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 2014, 6, a016675. [Google Scholar] [CrossRef]
- Khawar, M.B.; Gao, H.; Li, W. Mechanism of Acrosome Biogenesis in Mammals. Front. Cell Dev. Biol. 2019, 7, 195. [Google Scholar] [CrossRef]
- Gordon, S.G.; Kursel, L.E.; Xu, K.; Rog, O. Synaptonemal Complex dimerization regulates chromosome alignment and crossover patterning in meiosis. PLoS Genet. 2021, 17, e1009205. [Google Scholar] [CrossRef]
- Bisig, C.G.; Guiraldelli, M.F.; Kouznetsova, A.; Scherthan, H.; Hoog, C.; Dawson, D.S.; Pezza, R.J. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet. 2012, 8, e1002701. [Google Scholar] [CrossRef]
- De Pol, A.; Marzona, L.; Vaccina, F.; Negro, R.; Sena, P.; Forabosco, A. Apoptosis in different stages of human oogenesis. Anticancer Res. 1998, 18, 3457–3461. [Google Scholar]
- Walter, A.O.; Seghezzi, W.; Korver, W.; Sheung, J.; Lees, E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 2000, 19, 4906–4916. [Google Scholar] [CrossRef]
- Lee, J.Y.; Orr-Weaver, T.L. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 2001, 17, 753–777. [Google Scholar] [CrossRef]
- Handel, M.A.; Hunt, P.A. Sex-chromosome pairing and activity during mammalian meiosis. Bioessays 1992, 14, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Tsubouchi, T.; Roeder, G.S. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 2005, 308, 870–873. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dokholyan, N.V. Insights into allosteric control of vinculin function from its large scale conformational dynamics. J. Biol. Chem. 2006, 281, 29148–29154. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kitajima, T.S.; Tanno, Y.; Yoshida, K.; Morita, T.; Miyano, T.; Miyake, M.; Watanabe, Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2008, 10, 42–52. [Google Scholar] [CrossRef]
- Llano, E.; Gomez, R.; Gutierrez-Caballero, C.; Herran, Y.; Sanchez-Martin, M.; Vazquez-Quinones, L.; Hernandez, T.; de Alava, E.; Cuadrado, A.; Barbero, J.L.; et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008, 22, 2400–2413. [Google Scholar] [CrossRef] [PubMed]
- Zierhut, C.; Funabiki, H. Nucleosome functions in spindle assembly and nuclear envelope formation. Bioessays 2015, 37, 1074–1085. [Google Scholar] [CrossRef]
- Jiao, X.; Chang, S.; Yang, L.; An, M.; Chen, W. Vinculin motion modes analysis with elastic network model. Int. J. Mol. Sci. 2012, 13, 208–220. [Google Scholar] [CrossRef]
- Stec, D.L.; Stec, B. Complete Model of Vinculin Suggests the Mechanism of Activation by Helical Super-Bundle Unfurling. Protein J. 2022, 41, 55–70. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrusová, J.; Havalda, R.; Flachs, P.; Venit, T.; Darášová, A.; Hůlková, L.; Sztacho, M.; Hozák, P. Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis. Cells 2022, 11, 2013. https://doi.org/10.3390/cells11132013
Petrusová J, Havalda R, Flachs P, Venit T, Darášová A, Hůlková L, Sztacho M, Hozák P. Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis. Cells. 2022; 11(13):2013. https://doi.org/10.3390/cells11132013
Chicago/Turabian StylePetrusová, Jana, Robert Havalda, Petr Flachs, Tomáš Venit, Alžběta Darášová, Lenka Hůlková, Martin Sztacho, and Pavel Hozák. 2022. "Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis" Cells 11, no. 13: 2013. https://doi.org/10.3390/cells11132013
APA StylePetrusová, J., Havalda, R., Flachs, P., Venit, T., Darášová, A., Hůlková, L., Sztacho, M., & Hozák, P. (2022). Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis. Cells, 11(13), 2013. https://doi.org/10.3390/cells11132013