Hematopoietic Stem and Progenitor Cell Maintenance and Multiple Lineage Differentiation Is an Integral Function of NFATc1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Flow Cytometry
2.3. Cell Isolation and Sorting
2.4. Immunofluorescence Staining
2.5. Adoptive Cell Transfer
2.6. Semiquantitative RT-PCR
2.7. Photographs
2.8. Statistics
3. Results
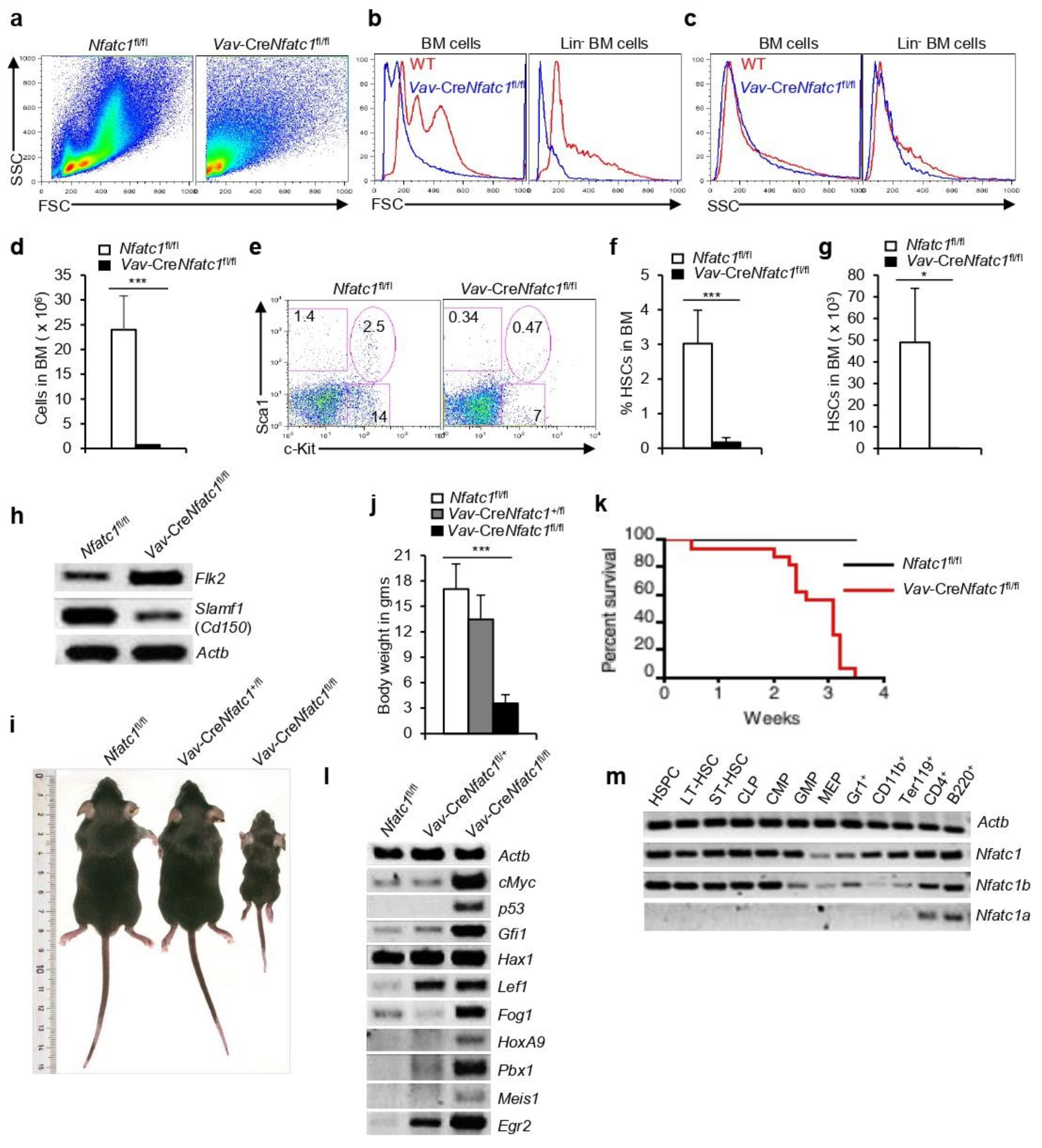
3.1. NFAT Expression in HSPC Population
3.2. NFATc1 Critically Regulates HSPC Development
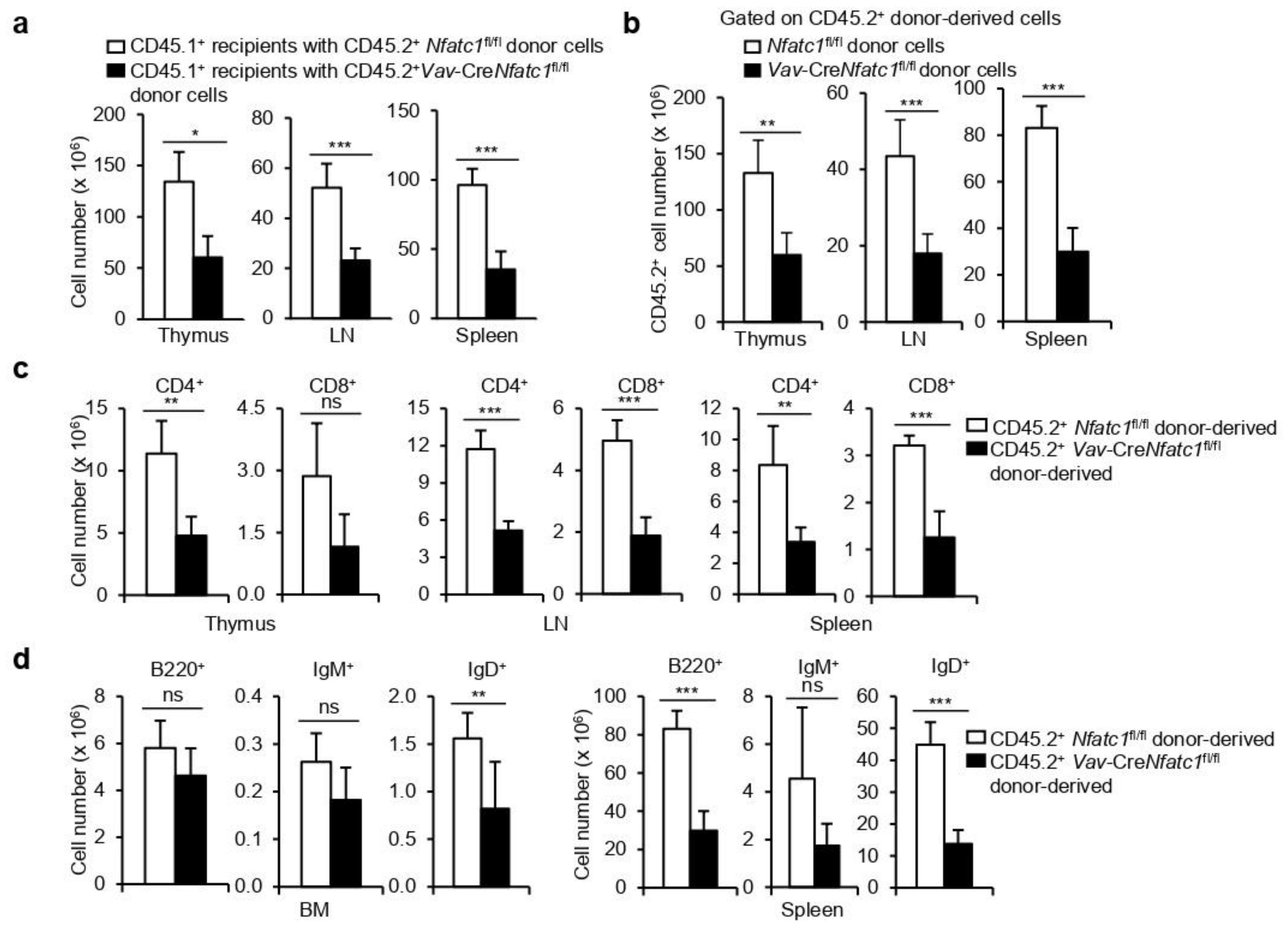
3.3. HSPC-Specific Defects Result in Severe Lymphopenia in Vav-CreNfatc1fl/fl Mice
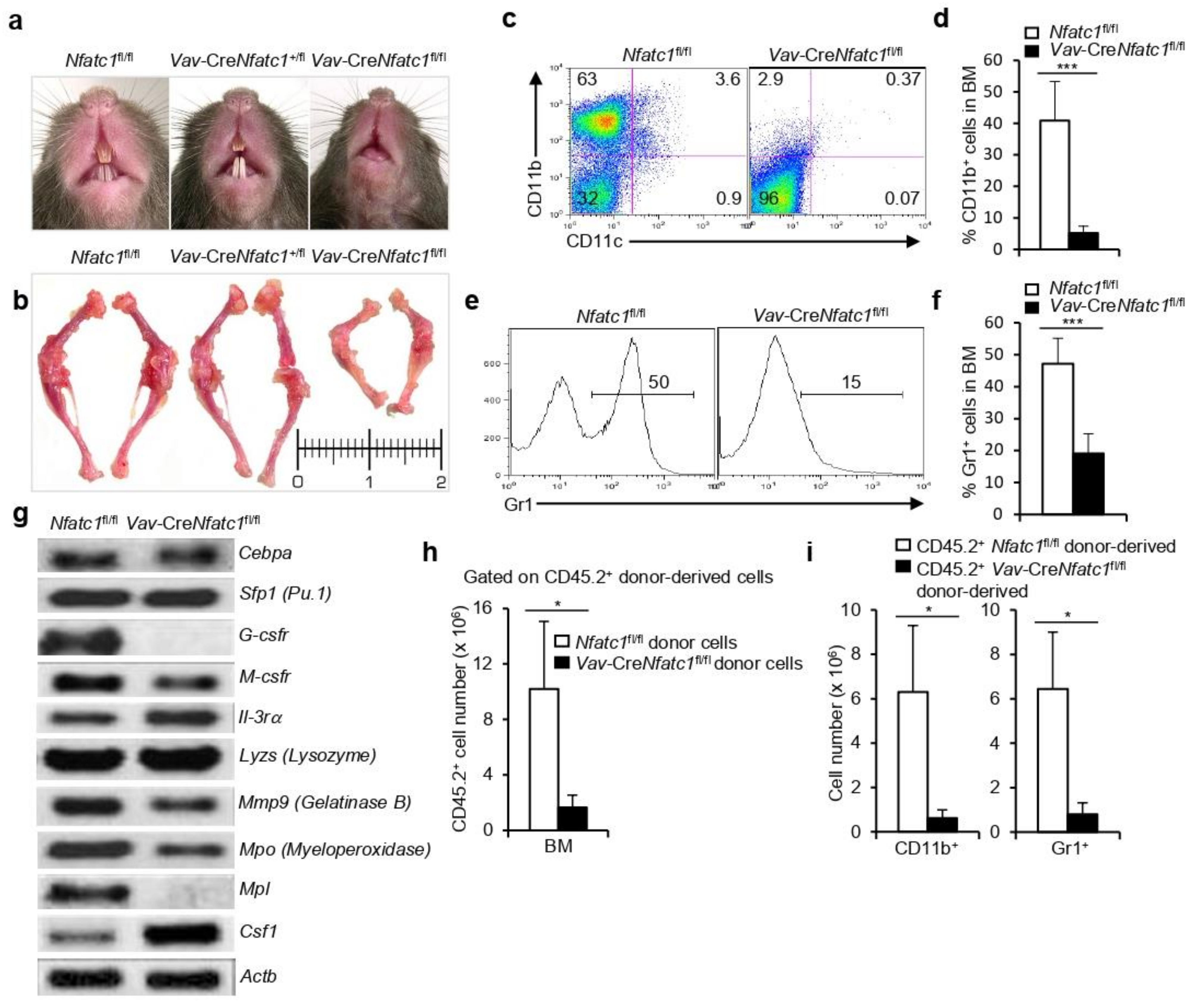
3.4. NFATc1 Deficiency Severely Impairs Myelopoiesis
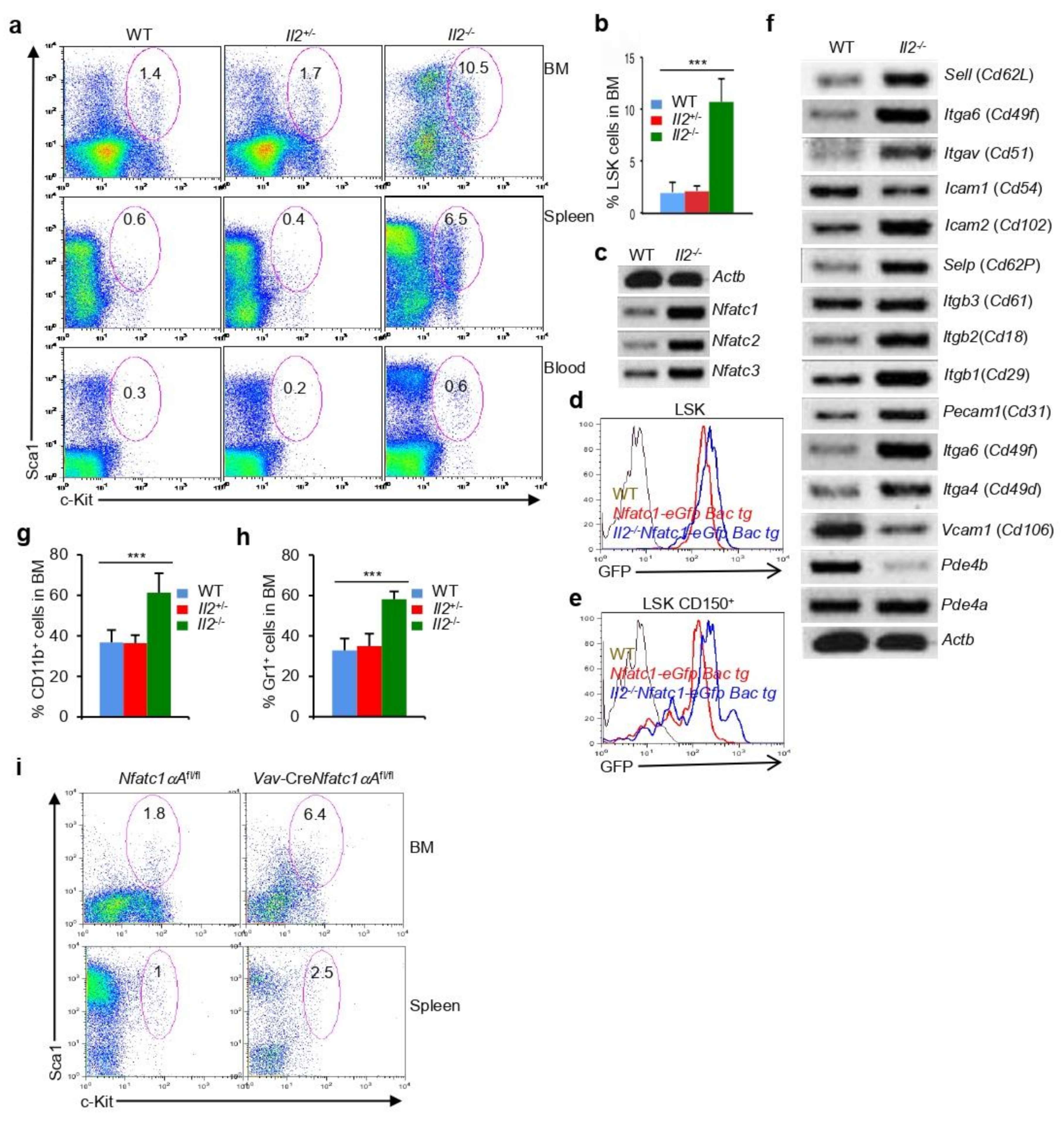
3.5. NFATc1 Activity Is Indispensable for Normal Hematopoiesis
3.6. Increase in NFATc1 Activity Impairs Hematopoiesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Rieger, M.A.; Schroeder, T. Hematopoiesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008250. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan-Bogdan, M.; Zon, L.I. Hematopoiesis. Development 2013, 140, 2463–2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Pinho, S.; Frenette, P.S. Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. 2019, 20, 303–320. [Google Scholar] [CrossRef]
- Boulais, P.E.; Frenette, P.S. Making sense of hematopoietic stem cell niches. Blood 2015, 125, 2621–2629. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, A.C.; Gottgens, B. Transcriptional regulation of haematopoietic stem cells. Adv. Exp. Med. Biol. 2013, 786, 187–212. [Google Scholar]
- Zhu, J.; Emerson, S.G. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene 2002, 21, 3295–3313. [Google Scholar] [CrossRef] [Green Version]
- Edginton-White, B.; Bonifer, C. The transcriptional regulation of normal and malignant blood cell development. FEBS J. 2021, 289, 1240–1255. [Google Scholar] [CrossRef]
- Kiel, M.J.; Yilmaz, O.H.; Iwashita, T.; Yilmaz, O.H.; Terhorst, C.; Morrison, S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 2005, 121, 1109–1121. [Google Scholar] [CrossRef] [Green Version]
- Christensen, J.L.; Weissman, I.L. Flk-2 is a marker in hematopoietic stem cell differentiation, a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA 2001, 98, 14541–14546. [Google Scholar] [CrossRef] [Green Version]
- Gazit, R.; Garrison, B.S.; Rao, T.N.; Shay, T.; Costello, J.; Ericson, J.; Kim, F.; Collins, J.J.; Regev, A.; Wagers, A.J.; et al. Transcriptome analysis identifies regulators of hematopoietic stem and progenitor cells. Stem Cell Rep. 2013, 1, 266–280. [Google Scholar] [CrossRef] [Green Version]
- Crabtree, G.R.; Olson, E.N. NFAT signaling, choreographing the social lives of cells. Cell 2002, 109 (Suppl. S6), 7–79. [Google Scholar]
- Hogan, P.G.; Chen, L.; Nardone, J.; Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003, 17, 2205–2232. [Google Scholar] [CrossRef] [Green Version]
- Patra, A.K.; Avots, A.; Zahedi, R.P.; Schuler, T.; Sickmann, A.; Bommhardt, U.; Serfling, E. An alternative NFAT-activation pathway mediated by IL-7 is critical for early thymocyte development. Nat. Immunol. 2013, 14, 127–135. [Google Scholar] [CrossRef]
- Macian, F. NFAT proteins, key regulators of T-cell development and function. Nat. Rev. Immunol. 2005, 5, 472–484. [Google Scholar] [CrossRef]
- Peng, S.L.; Gerth, A.J.; Ranger, A.M.; Glimcher, L.H. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 2001, 14, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Fric, J.; Zelante, T.; Wong, A.Y.; Mertes, A.; Yu, H.B.; Ricciardi-Castagnoli, P. NFAT control of innate immunity. Blood 2012, 120, 1380–1389. [Google Scholar] [CrossRef]
- Klein-Hessling, S.; Rudolf, R.; Muhammad, K.; Knobeloch, K.P.; Maqbool, M.A.; Cauchy, P.; Andrau, J.C.; Avots, A.; Talora, C.; Ellenrieder, V.; et al. A threshold level of NFATc1 activity facilitates thymocyte differentiation and opposes notch-driven leukaemia development. Nat. Commun. 2016, 7, 11841. [Google Scholar] [CrossRef]
- Giampaolo, S.; Wojcik, G.; Klein-Hessling, S.; Serfling, E.; Patra, A.K. B cell development is critically dependent on NFATc1 activity. Cell Mol. Immunol. 2019, 16, 508–520. [Google Scholar] [CrossRef]
- Giampaolo, S.; Wojcik, G.; Klein-Hessling, S.; Serfling, E.; Patra, A.K. NFAT-mediated defects in erythropoiesis cause anemia in Il2(−/−) mice. Oncotarget 2018, 9, 9632–9644. [Google Scholar] [CrossRef] [Green Version]
- Patra, A.K.; Drewes, T.; Engelmann, S.; Chuvpilo, S.; Kishi, H.; Hunig, T.; Serfling, E.; Bommhardt, U.H. PKB rescues calcineurin/NFAT-induced arrest of Rag expression and pre-T cell differentiation. J. Immunol. 2006, 177, 4567–4576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Pompa, J.L.; Timmerman, L.A.; Takimoto, H.; Yoshida, H.; Elia, A.J.; Samper, E.; Potter, J.; Wakeham, A.; Marengere, L.; Langille, B.L.; et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 1998, 392, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Hock, M.; Vaeth, M.; Rudolf, R.; Patra, A.K.; Pham, D.A.; Muhammad, K.; Pusch, T.; Bopp, T.; Schmitt, E.; Rost, R.; et al. NFATc1 Induction in Peripheral T and B Lymphocytes. J. Immunol. 2013, 190, 2345–2353. [Google Scholar] [CrossRef] [PubMed]
- Chuvpilo, S.; Avots, A.; Berberich-Siebelt, F.; Glockner, J.; Fischer, C.; Kerstan, A.; Escher, C.; Inashkina, I.; Hlubek, F.; Jankevics, E.; et al. Multiple NF-ATc isoforms with individual transcriptional properties are synthesized in T lymphocytes. J. Immunol. 1999, 162, 7294–7301. [Google Scholar]
- Bhattacharyya, S.; Deb, J.; Patra, A.K.; Thuy Pham, D.A.; Chen, W.; Vaeth, M.; Berberich-Siebelt, F.; Klein-Hessling, S.; Lamperti, E.D.; Reifenberg, K.; et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin—NFAT signaling network. J. Exp. Med. 2011, 208, 823–839. [Google Scholar] [CrossRef]
- Winslow, M.M.; Pan, M.; Starbuck, M.; Gallo, E.M.; Deng, L.; Karsenty, G.; Crabtree, G.R. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev. Cell 2006, 10, 771–782. [Google Scholar] [CrossRef] [Green Version]
- Fromigue, O.; Hay, E.; Barbara, A.; Marie, P.J. Essential role of nuclear factor of activated T cells (NFAT)-mediated Wnt signaling in osteoblast differentiation induced by strontium ranelate. J. Biol. Chem. 2010, 285, 25251–25258. [Google Scholar] [CrossRef] [Green Version]
- Asagiri, M.; Sato, K.; Usami, T.; Ochi, S.; Nishina, H.; Yoshida, H.; Morita, I.; Wagner, E.F.; Mak, T.W.; Serfling, E.; et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 2005, 202, 1261–1269. [Google Scholar] [CrossRef] [Green Version]
- Asagiri, M.; Takayanagi, H. The molecular understanding of osteoclast differentiation. Bone 2007, 40, 251–264. [Google Scholar] [CrossRef]
- Giampaolo, S.; Wojcik, G.; Serfling, E.; Patra, A.K. Interleukin-2-regulatory T cell axis critically regulates maintenance of hematopoietic stem cells. Oncotarget 2017, 8, 29625–29642. [Google Scholar] [CrossRef] [Green Version]
- Carrera, A.C.; Rincon, M.; De Landazuri, M.O.; Lopez-Botet, M. CD2 is involved in regulating cyclic AMP levels in T cells. Eur. J. Immunol. 1988, 18, 961–964. [Google Scholar] [CrossRef]
- Hahn, W.C.; Rosenstein, Y.; Burakoff, S.J.; Bierer, B.E. Interaction of CD2 with its ligand lymphocyte function-associated antigen-3 induces adenosine 3′,5′-cyclic monophosphate production in T lymphocytes. J. Immunol. 1991, 147, 14–21. [Google Scholar]
- Meyer, C.J.; Alenghat, F.J.; Rim, P.; Fong, J.H.; Fabry, B.; Ingber, D.E. Mechanical control of cyclic AMP signalling and gene transcription through integrins. Nat. Cell Biol. 2000, 2, 666–668. [Google Scholar] [CrossRef]
- Rao, A.; Luo, C.; Hogan, P.G. Transcription factors of the NFAT family, regulation and function. Annu. Rev. Immunol. 1997, 15, 707–747. [Google Scholar] [CrossRef]
- Serfling, E.; Klein-Hessling, S.; Palmetshofer, A.; Bopp, T.; Stassen, M.; Schmitt, E. NFAT transcription factors in control of peripheral T cell tolerance. Eur. J. Immunol. 2006, 36, 2837–2843. [Google Scholar] [CrossRef]
- Vaeth, M.; Feske, S. NFAT Control of Immune Function, New Frontiers for an Abiding Trooper. F1000Research 2018, 7, 260. [Google Scholar] [CrossRef] [Green Version]
- Sugimura, R.; He, X.C.; Venkatraman, A.; Arai, F.; Box, A.; Semerad, C.; Haug, J.S.; Peng, L.; Zhong, X.B.; Suda, T.; et al. Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell 2012, 150, 351–365. [Google Scholar] [CrossRef] [Green Version]
- Luchsinger, L.L.; de Almeida, M.J.; Corrigan, D.J.; Mumau, M.; Snoeck, H.W. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature 2016, 529, 528–531. [Google Scholar] [CrossRef] [Green Version]
- Horsley, V.; Aliprantis, A.O.; Polak, L.; Glimcher, L.H.; Fuchs, E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 2008, 132, 299–310. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Grzywacz, B.; Sukovich, D.; McCullar, V.; Cao, Q.; Lee, A.B.; Blazar, B.R.; Cornfield, D.N.; Miller, J.S.; Verneris, M.R. The unexpected effect of cyclosporin A on CD56+CD16− and CD56+CD16+ natural killer cell subpopulations. Blood 2007, 110, 1530–1539. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.J.; Kim, N.; Kang, H.J.; Kim, E.O.; Kim, S.T.; Ahn, H.S.; Bluestone, J.A.; Lee, K.M. FK506 causes cellular and functional defects in human natural killer cells. J. Leukoc. Biol. 2010, 88, 1089–1097. [Google Scholar] [CrossRef]
- Zanoni, I.; Ostuni, R.; Capuano, G.; Collini, M.; Caccia, M.; Ronchi, A.E.; Rocchetti, M.; Mingozzi, F.; Foti, M.; Chirico, G.; et al. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature 2009, 460, 264–268. [Google Scholar] [CrossRef]
- Akimzhanov, A.; Krenacs, L.; Schlegel, T.; Klein-Hessling, S.; Bagdi, E.; Stelkovics, E.; Kondo, E.; Chuvpilo, S.; Wilke, P.; Avots, A.; et al. Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling. Am. J. Pathol. 2008, 172, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Solovey, M.; Wang, Y.; Michel, C.; Metzeler, K.H.; Herold, T.; Gothert, J.R.; Ellenrieder, V.; Hessmann, E.; Gattenlöhner, S.; Neubauer, A.; et al. Nuclear factor of activated T-cells, NFATC1, governs FLT3(ITD)-driven hematopoietic stem cell transformation and a poor prognosis in AML. J. Hematol. Oncol. 2019, 12, 72. [Google Scholar] [CrossRef]
- Hayashi, H.; Nakahama, K.; Sato, T.; Tuchiya, T.; Asakawa, Y.; Maemura, T.; Tanaka, M.; Morita, M.; Morita, I. The role of Mac-1 (CD11b/CD18) in osteoclast differentiation induced by receptor activator of nuclear factor-kappaB ligand. FEBS Lett. 2008, 582, 3243–3248. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Chen, X.; Yan, Z.; Zhu, Q.; Yang, C. CD11b promotes the differentiation of osteoclasts induced by RANKL through the spleen tyrosine kinase signalling pathway. J. Cell Mol. Med. 2017, 21, 3445–3452. [Google Scholar] [CrossRef]
- Lee, P.Y.; Wang, J.X.; Parisini, E.; Dascher, C.C.; Nigrovic, P.A. Ly6 family proteins in neutrophil biology. J. Leukoc. Biol. 2013, 94, 585–594. [Google Scholar] [CrossRef] [Green Version]
- Berdnikovs, S. The twilight zone, plasticity and mixed ontogeny of neutrophil and eosinophil granulocyte subsets. Semin. Immunopathol. 2021, 43, 337–346. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Simmons, R.M.; Underhill, D.M. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J. Immunol. 2007, 178, 3107–3115. [Google Scholar] [CrossRef] [Green Version]
- Chopra, M.; Langenhorst, D.; Beilhack, A.; Serfling, E.; Patra, A.K. Interleukin-2 critically regulates bone marrow erythropoiesis and prevents anemia development. Eur. J. Immunol. 2015, 45, 3362–3374. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barahona de Brito, C.; Klein-Hessling, S.; Serfling, E.; Patra, A.K. Hematopoietic Stem and Progenitor Cell Maintenance and Multiple Lineage Differentiation Is an Integral Function of NFATc1. Cells 2022, 11, 2012. https://doi.org/10.3390/cells11132012
Barahona de Brito C, Klein-Hessling S, Serfling E, Patra AK. Hematopoietic Stem and Progenitor Cell Maintenance and Multiple Lineage Differentiation Is an Integral Function of NFATc1. Cells. 2022; 11(13):2012. https://doi.org/10.3390/cells11132012
Chicago/Turabian StyleBarahona de Brito, Carlotta, Stefan Klein-Hessling, Edgar Serfling, and Amiya Kumar Patra. 2022. "Hematopoietic Stem and Progenitor Cell Maintenance and Multiple Lineage Differentiation Is an Integral Function of NFATc1" Cells 11, no. 13: 2012. https://doi.org/10.3390/cells11132012
APA StyleBarahona de Brito, C., Klein-Hessling, S., Serfling, E., & Patra, A. K. (2022). Hematopoietic Stem and Progenitor Cell Maintenance and Multiple Lineage Differentiation Is an Integral Function of NFATc1. Cells, 11(13), 2012. https://doi.org/10.3390/cells11132012





