Venetoclax Decreases the Expression of the Spike Protein through Amino Acids Q493 and S494 in SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
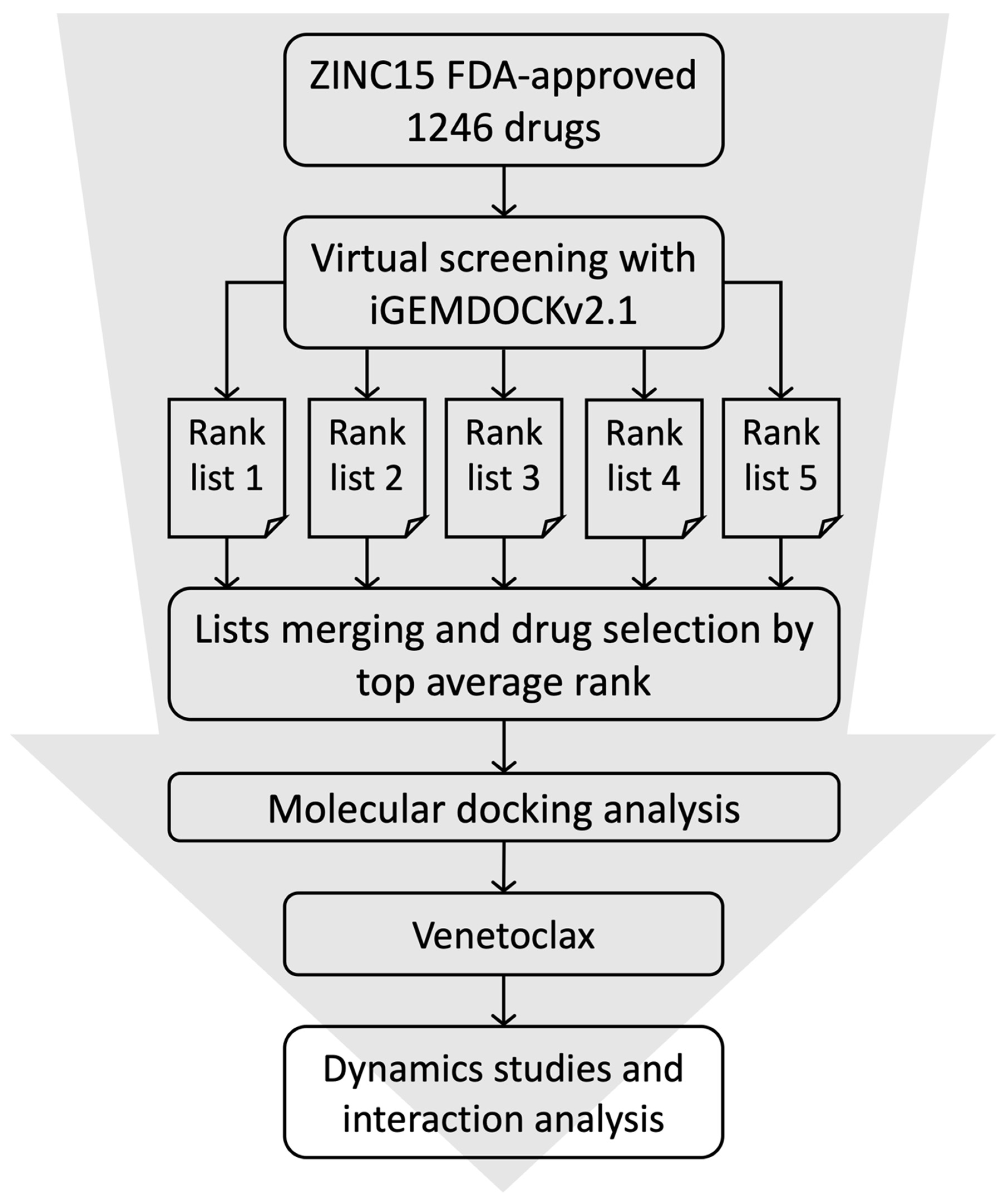
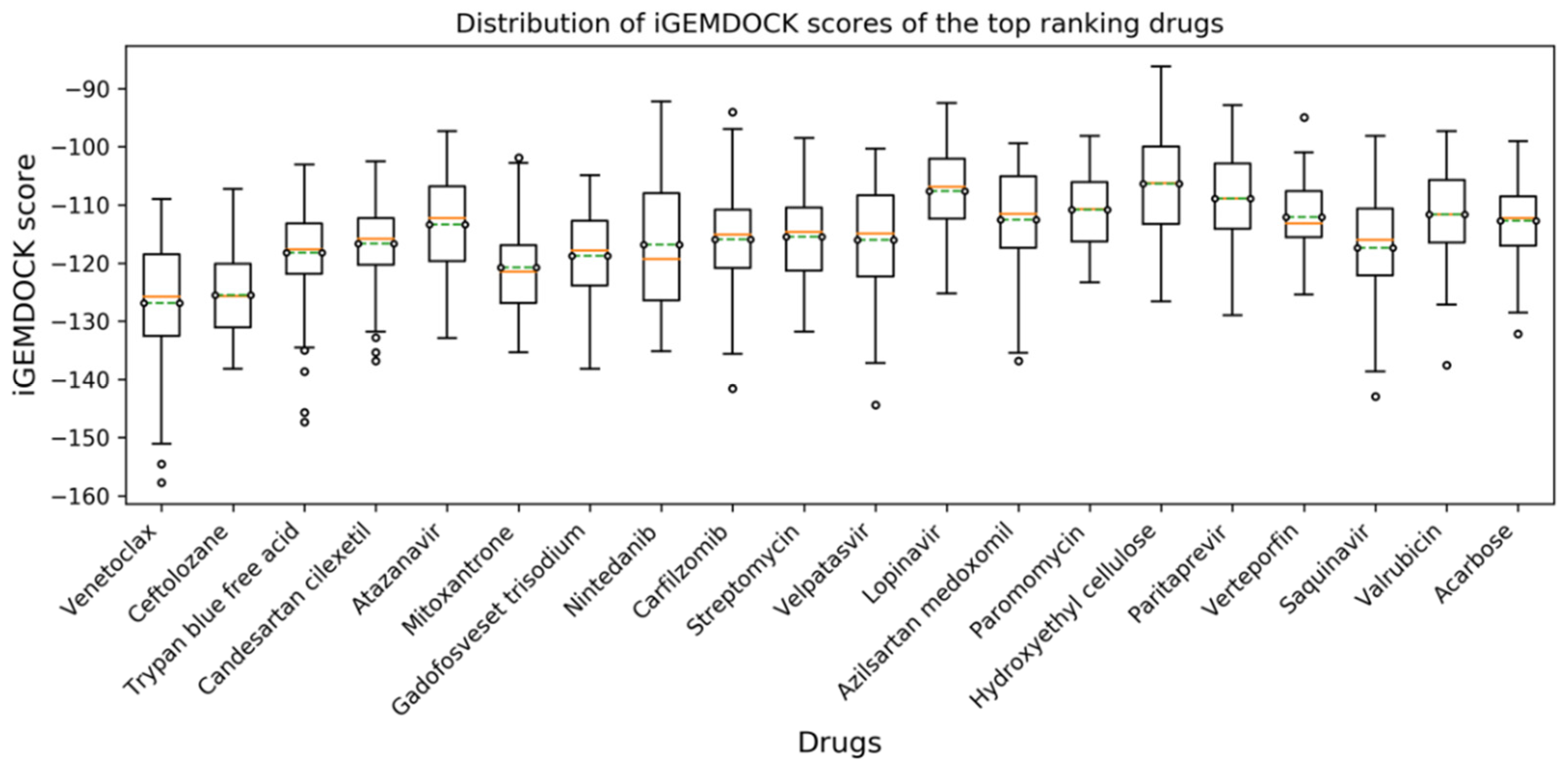
2.1. High-Throughput Virtual Screening and Molecular Docking
2.2. Molecular Dynamics Simulations
2.3. Binding Free Energy Calculation
2.4. Cell Lines and Drug
2.5. Transfection and Plasmid
2.6. Cell Growth
2.7. Inhibitor Screening Assay
2.8. Western Blotting
2.9. Immunofluorescence Staining
3. Results
3.1. Virtual Screening and Molecular Dynamics Simulation of S1-RBD:Venetoclax
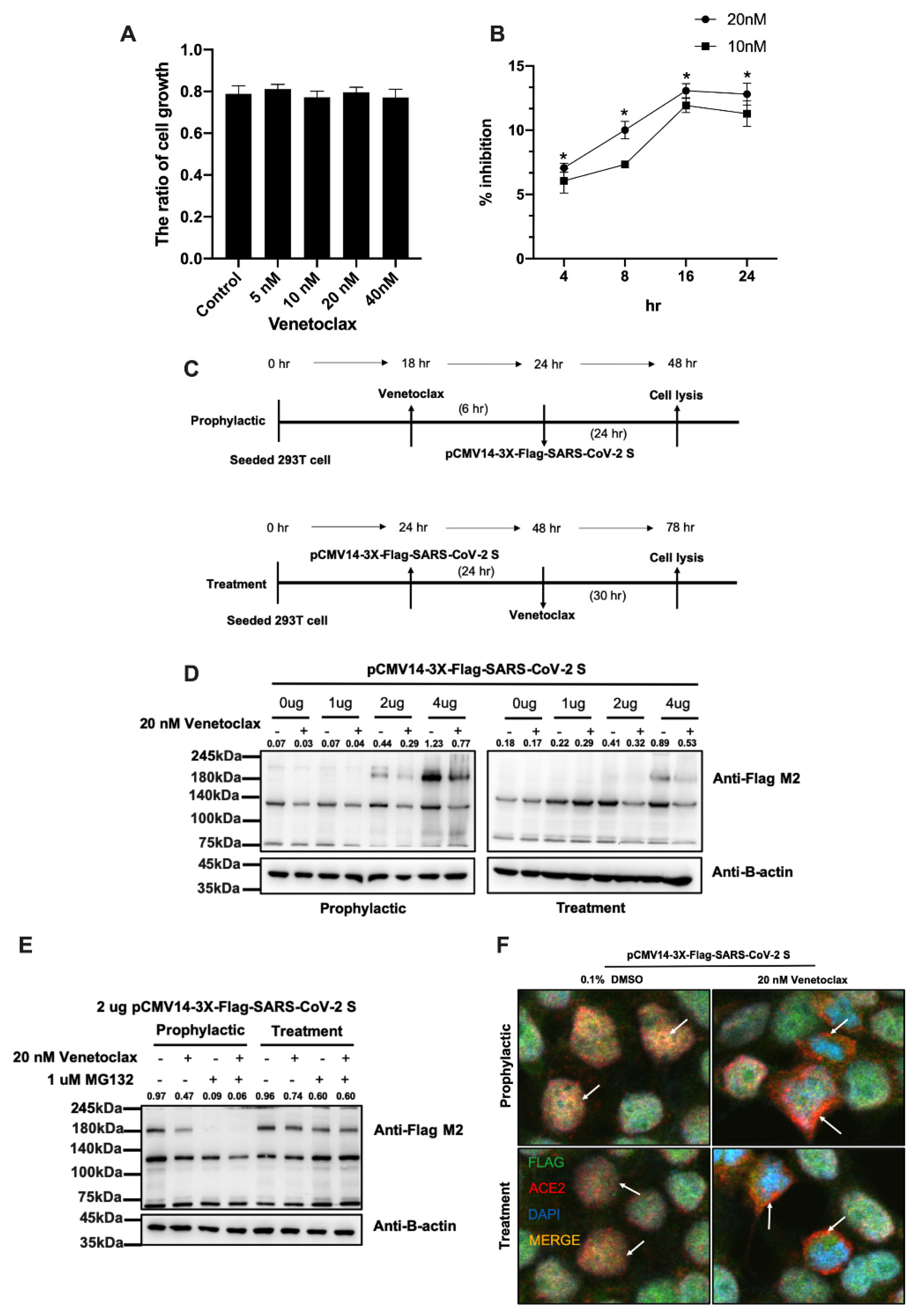
3.2. Venetoclax Blocked the Interaction between the SARS-CoV-2 Spike Protein and the ACE2 Receptor through Spike Protein Expression Degradation
3.3. Analysis of the S1-RBD:Venetoclax Complex Structures and Their Binding Free Energy between WT and MT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.F.; Yuan, S.; Kok, K.H.; To, K.K.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.; Poon, R.W.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes. Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe COVID-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Graham, F. Daily briefing: Inside Merck’s COVID drug, molnupiravir. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.X.; et al. Compassionate Use of Remdesivir for Patients with Severe COVID-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchholz, U.J.; Bukreyev, A.; Yang, L.; Lamirande, E.W.; Murphy, B.R.; Subbarao, K.; Collins, P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA 2004, 101, 9804–9809. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, H.; Hattermann, K.; Marzi, A.; Gramberg, T.; Geier, M.; Krumbiegel, M.; Kuate, S.; Uberla, K.; Niedrig, M.; Pohlmann, S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004, 78, 6134–6142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, Y.Y.; Chou, C.J.; Lo, H.L.; Yang, P.M. Repositioning of a cyclin-dependent kinase inhibitor GW8510 as a ribonucleotide reductase M2 inhibitor to treat human colorectal cancer. Cell Death Discov. 2016, 2, 16027. [Google Scholar] [CrossRef] [Green Version]
- Malcomson, B.; Wilson, H.; Veglia, E.; Thillaiyampalam, G.; Barsden, R.; Donegan, S.; el Banna, A.; Elborn, J.S.; Ennis, M.; Kelly, C.; et al. Connectivity mapping (ssCMap) to predict A20-inducing drugs and their antiinflammatory action in cystic fibrosis. P Natl. Acad. Sci. USA 2016, 113, E3725–E3734. [Google Scholar] [CrossRef] [Green Version]
- Mirza, N.; Sills, G.J.; Pirmohamed, M.; Marson, A.G. Identifying new antiepileptic drugs through genomics-based drug repurposing. Hum. Mol. Genet. 2017, 26, 527–537. [Google Scholar] [CrossRef]
- Shin, E.; Lee, Y.C.; Kim, S.R.; Kim, S.H.; Park, J. Drug Signature-based Finding of Additional Clinical Use of LC28-0126 for Neutrophilic Bronchial Asthma. Sci. Rep.-UK 2015, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Smith, M.; Smith, J.C. Repurposing Therapeutics for COVID-19: Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Senathilake, K.; Samarakoon, S.; Tennekoon, K. Virtual Screening of Inhibitors Against Spike Glycoprotein of SARS-CoV-2: A Drug Repurposing Approach. Preprints 2020, 2020030042. [Google Scholar] [CrossRef]
- Calligari, P.; Bobone, S.; Ricci, G.; Bocedi, A. Molecular Investigation of SARS-CoV-2 Proteins and Their Interactions with Antiviral Drugs. Viruses 2020, 12, 445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trezza, A.; Iovinelli, D.; Santucci, A.; Prischi, F.; Spiga, O. An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors. Sci. Rep. 2020, 10, 13866. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Saeed, M.; Greten, H.J.; Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Comput. Biol. Med. 2021, 133, 104359. [Google Scholar] [CrossRef] [PubMed]
- Culletta, G.; Gulotta, M.R.; Perricone, U.; Zappalà, M.; Almerico, A.M.; Tutone, M. Exploring the SARS-CoV-2 Proteome in the Search of Potential Inhibitors via Structure-Based Pharmacophore Modeling/Docking Approach. Computation 2020, 8, 77. [Google Scholar] [CrossRef]
- Hsu, K.C.; Chen, Y.F.; Lin, S.R.; Yang, J.M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011, 12 (Suppl. S1), S33. [Google Scholar] [CrossRef] [Green Version]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Gapsys, V.; Michielssens, S.; Seeliger, D.; de Groot, B.L. pmx: Automated protein structure and topology generation for alchemical perturbations. J. Comput. Chem. 2015, 36, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gapsys, V.; de Groot, B.L. pmx Webserver: A User Friendly Interface for Alchemistry. J. Chem. Inf. Model 2017, 57, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, M.J.; Tirado-Rives, J.; Jorgensen, W.L. Performance of Protein-Ligand Force Fields for the Flavodoxin-Flavin Mononucleotide System. J. Phys. Chem. Lett. 2016, 7, 3032–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, C.H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976, 22, 245–268. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, L.J.; Davies, F.E.; Monohan, G.P.; Kovacsovics, T.; Burwick, N.; Jakubowiak, A.; Kaufman, J.L.; Hong, W.J.; Dail, M.; Salem, A.H.; et al. Phase 2 study of venetoclax plus carfilzomib and dexamethasone in patients with relapsed/refractory multiple myeloma. Blood Adv. 2021, 5, 3748–3759. [Google Scholar] [CrossRef] [PubMed]
- Cang, S.; Iragavarapu, C.; Savooji, J.; Song, Y.; Liu, D. ABT-199 (venetoclax) and BCL-2 inhibitors in clinical development. J. Hematol. Oncol. 2015, 8, 129. [Google Scholar] [CrossRef]
- Punnoose, E.A.; Leverson, J.D.; Peale, F.; Boghaert, E.R.; Belmont, L.D.; Tan, N.; Young, A.; Mitten, M.; Ingalla, E.; Darbonne, W.C.; et al. Expression Profile of BCL-2, BCL-XL, and MCL-1 Predicts Pharmacological Response to the BCL-2 Selective Antagonist Venetoclax in Multiple Myeloma Models. Mol. Cancer Ther. 2016, 15, 1132–1144. [Google Scholar] [CrossRef] [Green Version]
- Salem, A.H.; Agarwal, S.K.; Dunbar, M.; Enschede, S.L.; Humerickhouse, R.A.; Wong, S.L. Pharmacokinetics of Venetoclax, a Novel BCL-2 Inhibitor, in Patients With Relapsed or Refractory Chronic Lymphocytic Leukemia or Non-Hodgkin Lymphoma. J. Clin. Pharmacol. 2017, 57, 484–492. [Google Scholar] [CrossRef]
- Gentile, M.; Petrungaro, A.; Uccello, G.; Vigna, E.; Recchia, A.G.; Caruso, N.; Bossio, S.; de Stefano, L.; Palummo, A.; Storino, F.; et al. Venetoclax for the treatment of chronic lymphocytic leukemia. Expert Opin. Investig. Drugs 2017, 26, 1307–1316. [Google Scholar] [CrossRef]
- Chen, Y.W.; Yiu, C.B.; Wong, K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research 2020, 9, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Bhattacherjee, D.; Satpati, P.; Bhabak, K.P. Venetoclax: A promising repurposed drug against SARS-CoV-2 main protease. J. Biomol. Struct. Dyn. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Inde, Z.; Croker, B.A.; Yapp, C.; Joshi, G.N.; Spetz, J.; Fraser, C.; Qin, X.; Xu, L.; Deskin, B.; Ghelfi, E.; et al. Age-dependent regulation of SARS-CoV-2 cell entry genes and cell death programs correlates with COVID-19 severity. Sci. Adv. 2021, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Donalisio, M.; Bonotto, R.M.; Schneider, E.; Arduino, I.; Boni, F.; Lembo, D.; Marcello, A.; Mastrangelo, E. Combined in silico and in vitro approaches identified the antipsychotic drug lurasidone and the antiviral drug elbasvir as SARS-CoV2 and HCoV-OC43 inhibitors. Antivir. Res. 2021, 189, 105055. [Google Scholar] [CrossRef]
- Li, X.; Lidsky, P.V.; Xiao, Y.; Wu, C.T.; Garcia-Knight, M.; Yang, J.; Nakayama, T.; Nayak, J.V.; Jackson, P.K.; Andino, R.; et al. Ethacridine inhibits SARS-CoV-2 by inactivating viral particles. PLoS Pathog. 2021, 17, e1009898. [Google Scholar] [CrossRef]
- Chen, Y.; Lear, T.B.; Evankovich, J.W.; Larsen, M.B.; Lin, B.; Alfaras, I.; Kennerdell, J.R.; Salminen, L.; Camarco, D.P.; Lockwood, K.C.; et al. A high-throughput screen for TMPRSS2 expression identifies FDA-approved compounds that can limit SARS-CoV-2 entry. Nat. Commun. 2021, 12, 3907. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef]
- Malik, S.A.; Orhon, I.; Morselli, E.; Criollo, A.; Shen, S.; Marino, G.; BenYounes, A.; Benit, P.; Rustin, P.; Maiuri, M.C.; et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene 2011, 30, 3918–3929. [Google Scholar] [CrossRef] [Green Version]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mosbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 5770. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Gotte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, R.; Samanta, P.; Sharma, P.; Zhang, F.; Mishra, S.K.; Kucheryavy, P.; Kim, S.B.; Aderibigbe, A.O.; Linhardt, R.J.; Tandon, R.; et al. Structural and kinetic analyses of holothurian sulfated glycans suggest potential treatment for SARS-CoV-2 infection. J. Biol. Chem. 2021, 297, 101207. [Google Scholar] [CrossRef] [PubMed]
- Taurino, D.; Frigeni, M.; Grassi, A.; Cavallaro, G.; Salmoiraghi, S.; Spinelli, O.; Rambaldi, A.; Lussana, F. Concurrent Diagnosis of Acute Myeloid Leukemia and Symptomatic COVID-19 Infection: A Case Report Successfully Treated with Azacitidine-Venetoclax Combination. Mediterr. J. Hematol. Infect. Dis. 2021, 13, e2021057. [Google Scholar] [CrossRef] [PubMed]
- Goetz, L.; Yang, J.; Greene, W.; Zhu, Y. A COVID-19 Patient with Repeatedly Undetectable SARS-CoV-2 Antibodies. J. Appl. Lab. Med. 2020, 5, 1401–1405. [Google Scholar] [CrossRef]
- Furstenau, M.; Langerbeins, P.; de Silva, N.; Fink, A.M.; Robrecht, S.; von Tresckow, J.; Simon, F.; Hohloch, K.; Droogendijk, J.; van der Klift, M.; et al. COVID-19 among fit patients with CLL treated with venetoclax-based combinations. Leukemia 2020, 34, 2225–2229. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-C.; Zhuang, Z.-J.; Wu, C.-W.; Tan, Y.-L.; Huang, C.-H.; Hsu, C.-Y.; Tsai, E.-M.; Hsieh, T.-H. Venetoclax Decreases the Expression of the Spike Protein through Amino Acids Q493 and S494 in SARS-CoV-2. Cells 2022, 11, 1924. https://doi.org/10.3390/cells11121924
Chen C-C, Zhuang Z-J, Wu C-W, Tan Y-L, Huang C-H, Hsu C-Y, Tsai E-M, Hsieh T-H. Venetoclax Decreases the Expression of the Spike Protein through Amino Acids Q493 and S494 in SARS-CoV-2. Cells. 2022; 11(12):1924. https://doi.org/10.3390/cells11121924
Chicago/Turabian StyleChen, Chih-Chieh, Zhi-Jie Zhuang, Chia-Wei Wu, Yi-Ling Tan, Chen-Hsiu Huang, Chia-Yi Hsu, Eing-Mei Tsai, and Tsung-Hua Hsieh. 2022. "Venetoclax Decreases the Expression of the Spike Protein through Amino Acids Q493 and S494 in SARS-CoV-2" Cells 11, no. 12: 1924. https://doi.org/10.3390/cells11121924
APA StyleChen, C.-C., Zhuang, Z.-J., Wu, C.-W., Tan, Y.-L., Huang, C.-H., Hsu, C.-Y., Tsai, E.-M., & Hsieh, T.-H. (2022). Venetoclax Decreases the Expression of the Spike Protein through Amino Acids Q493 and S494 in SARS-CoV-2. Cells, 11(12), 1924. https://doi.org/10.3390/cells11121924





