Distinct Motifs in ATAD5 C-Terminal Domain Modulate PCNA Unloading Process
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Purification
2.2. DNA Substrate for Loading/Unloading Reaction
2.3. PCNA-Loading and Unloading Reaction
2.4. Antibodies
2.5. Plasmids (or) DNA Constructs and siRNAs
2.6. Cell Culture
2.7. Transfections and RNA Interference
2.8. Chromatin Fractionation
2.9. Immunoprecipitation and Western Blot
3. Results
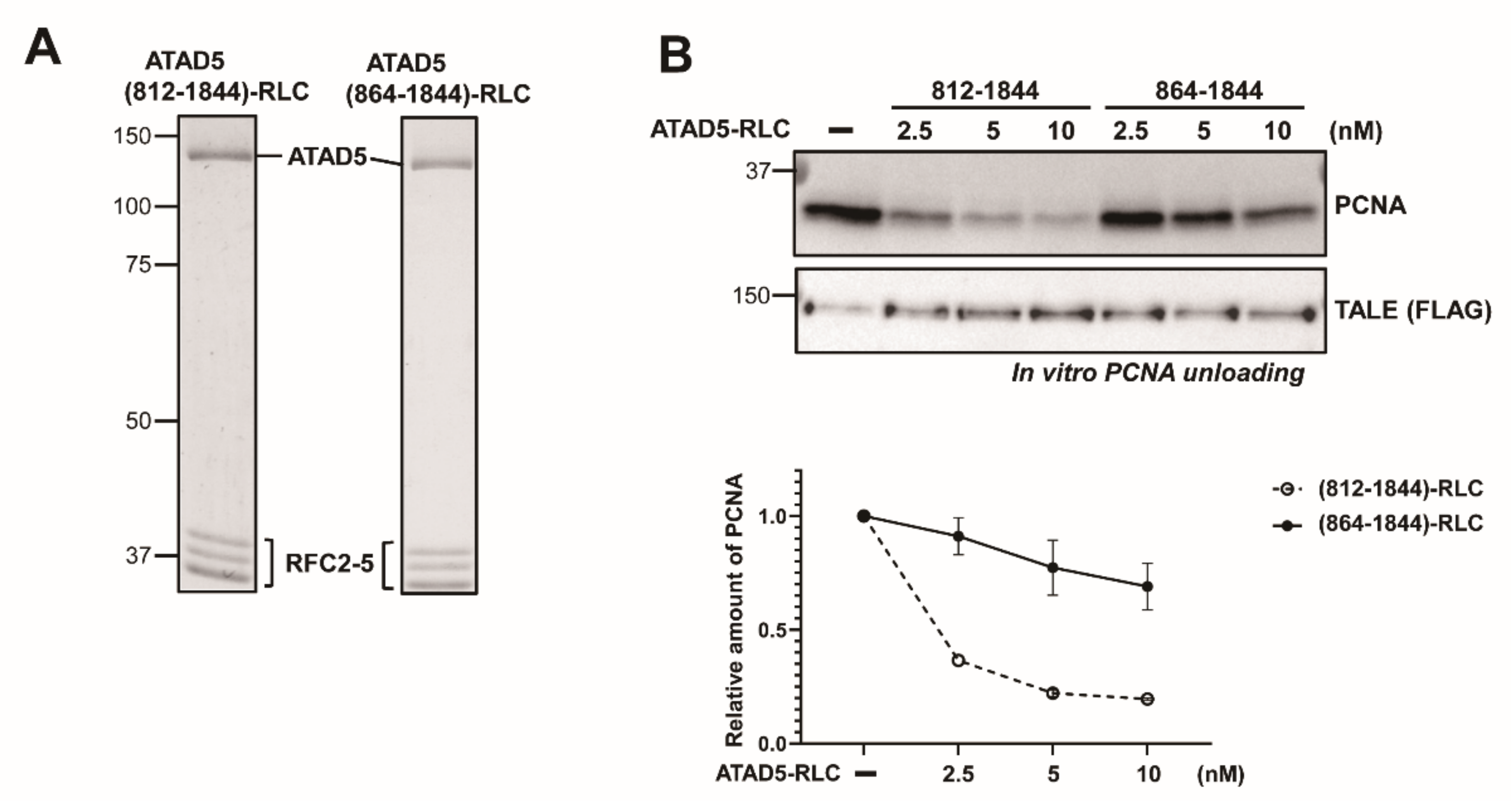
3.1. ATAD5 (812–1844) Is a Minimal PCNA-Unloading Domain
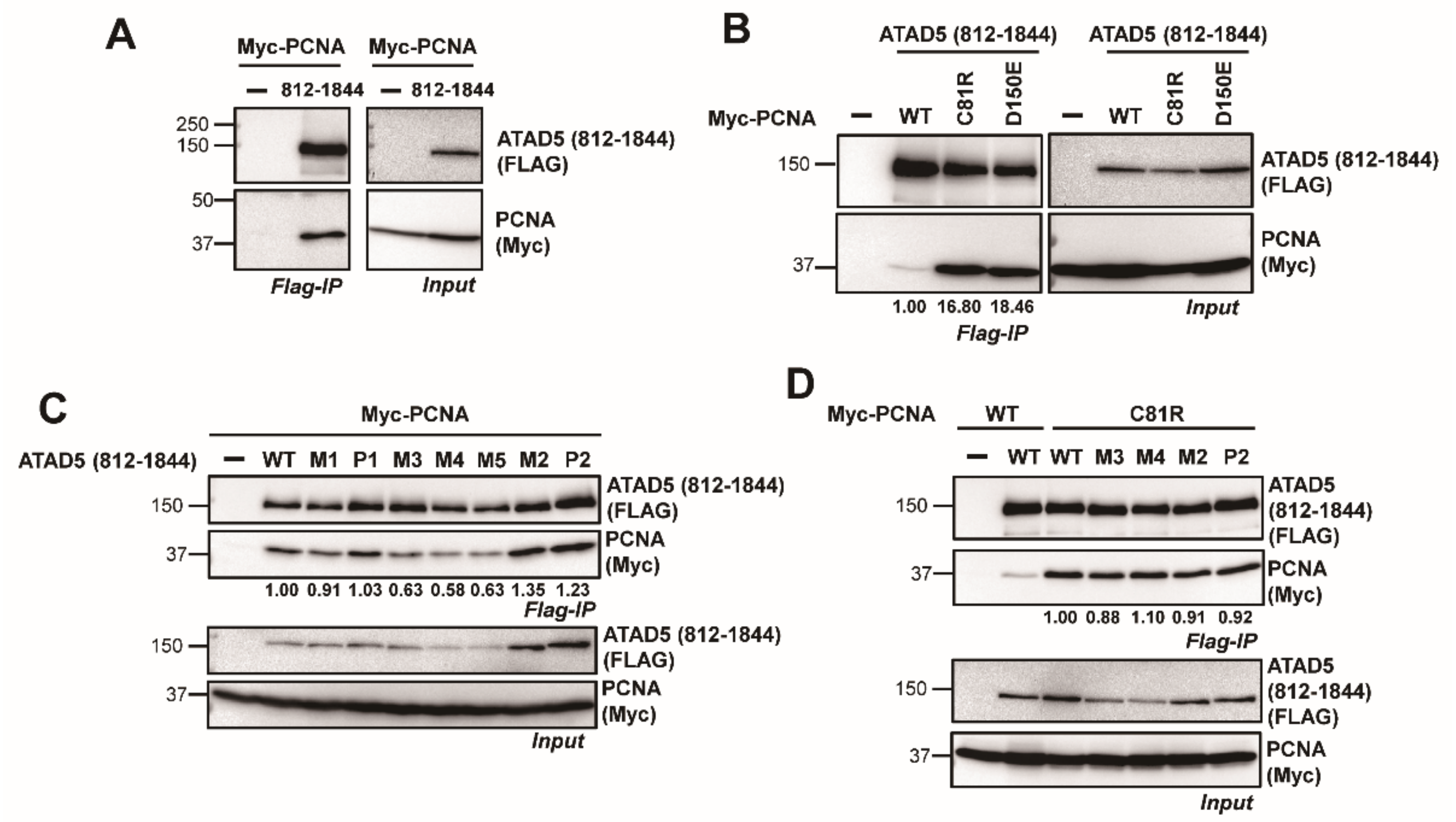
3.2. ATAD5 URM Is Required for PCNA Ring Opening
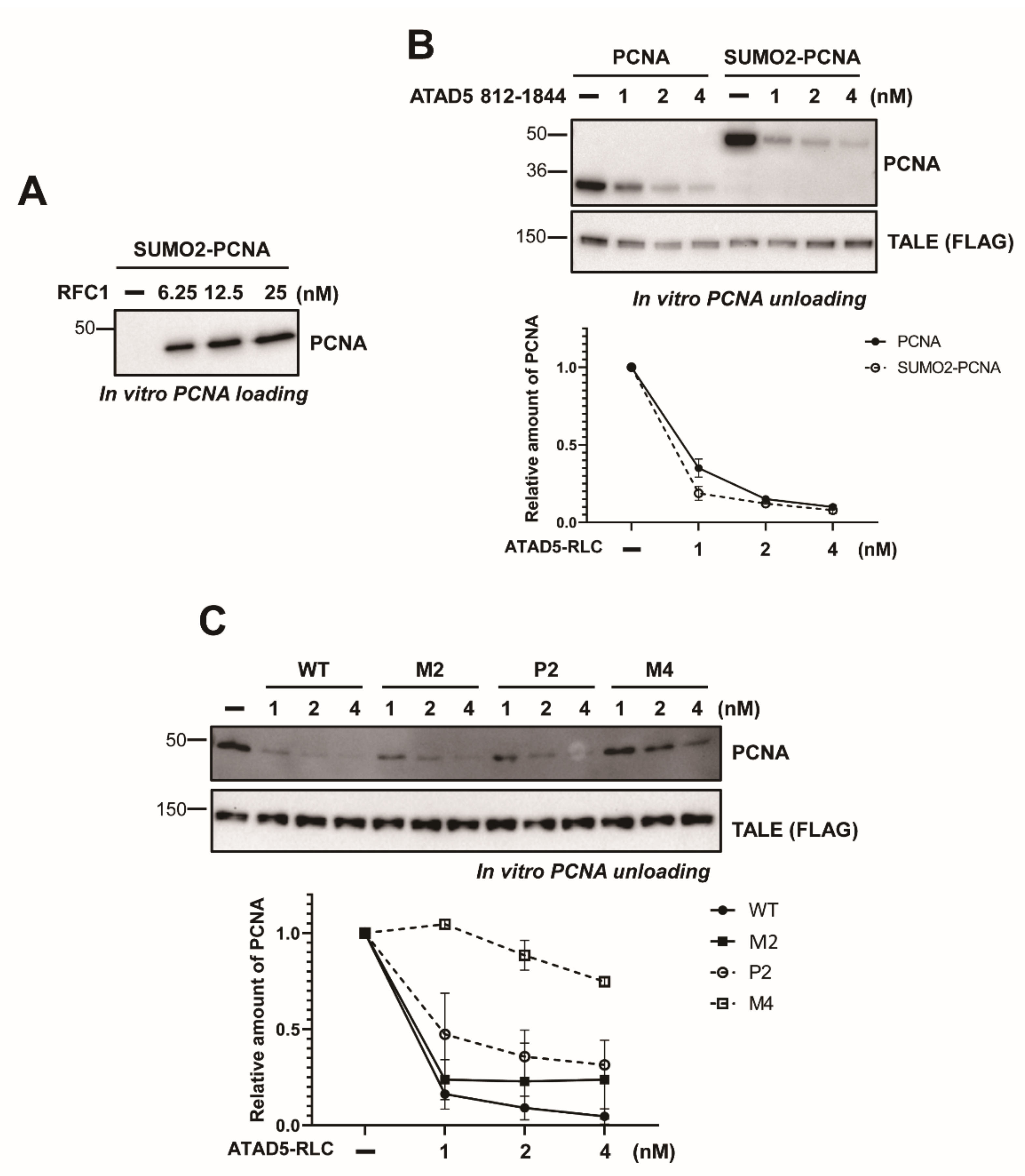
3.3. ATAD5 URM Contributes to the Opening of PCNA Ring
3.4. SUMO2 Fusion to PCNA Does not Enhance Unloading
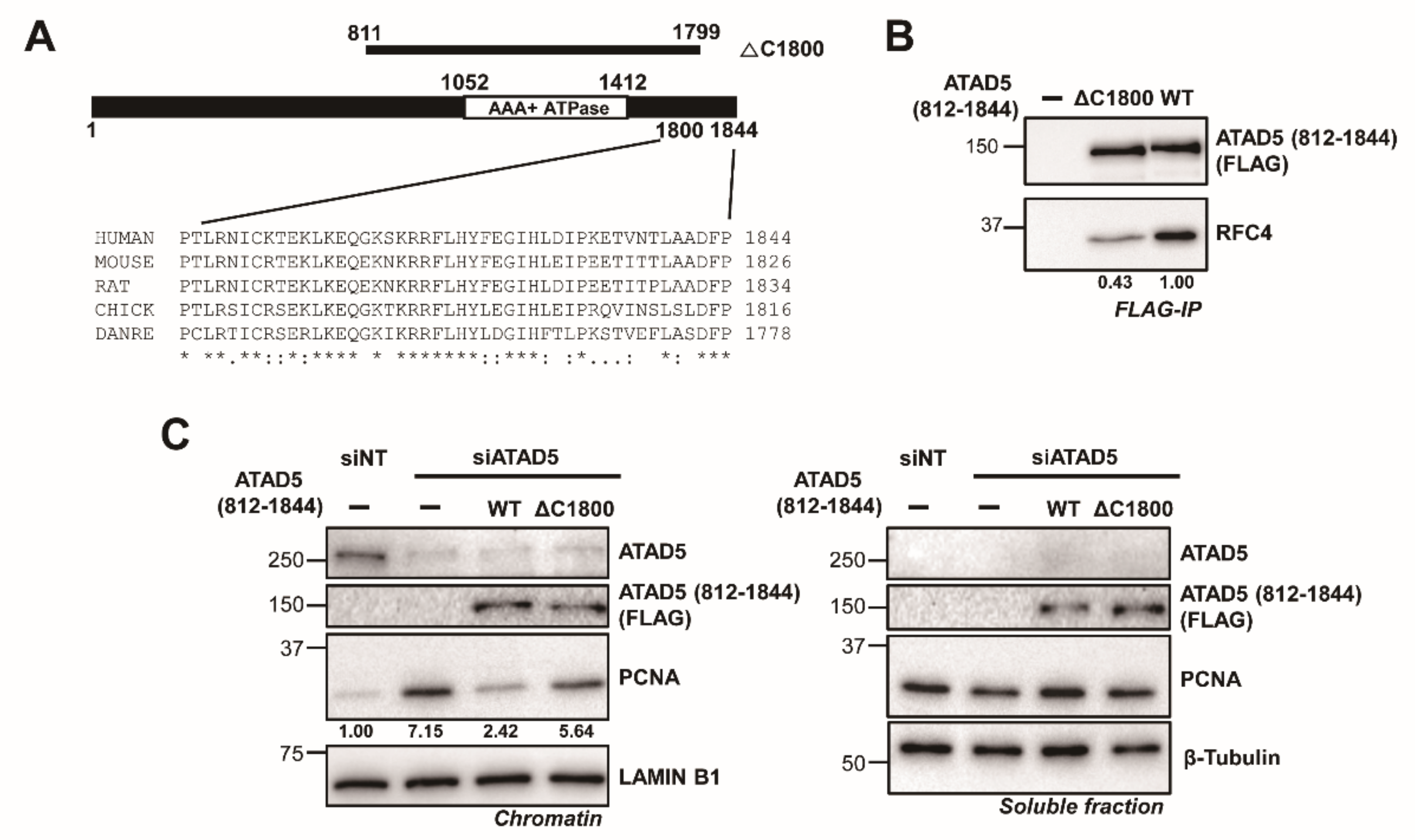
3.5. The ATAD5 C-Terminus Is Important for Stable RFC2-5 Binding
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, S.; Kang, M.-S.; Ryu, E.; Myung, K. Eukaryotic DNA replication: Orchestrated action of multi-subunit protein complexes. Mutat. Res. 2018, 809, 58–69. [Google Scholar] [CrossRef]
- Krishna, T.S.; Kong, X.-P.; Gary, S.; Burgers, P.M.; Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994, 79, 1233–1243. [Google Scholar] [CrossRef]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, N.Y.; O’Donnell, M. The RFC clamp loader: Structure and function. Subcell. Biochem. 2012, 62, 259–279. [Google Scholar] [PubMed] [Green Version]
- Bowman, G.D.; O’Donnell, M.; Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 2004, 429, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Suzuki, H.; Oyama, T.; Mayanagi, K.; Ishino, Y.; Morikawa, K. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc. Natl. Acad. Sci. USA 2005, 102, 13795–13800. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Z.; Yoder, B.L.; Burgers, P.M.J.; Benkovic, S.J. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc. Natl. Acad. Sci. USA 2006, 103, 2546–2551. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.-Y.; Park, S.H. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp. Mol. Med. 2020, 52, 1948–1958. [Google Scholar] [CrossRef]
- Ohashi, E.; Tsurimoto, T. Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication. Adv. Exp. Med. Biol. 2017, 1042, 135–162. [Google Scholar]
- Fujisawa, R.; Ohashi, E.; Hirota, K.; Tsurimoto, T. Human CTF18-RFC clamp-loader complexed with non-synthesising DNA polymerase epsilon efficiently loads the PCNA sliding clamp. Nucleic Acids Res. 2017, 45, 4550–4563. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.S.; Ryu, E.; Lee, S.-W.; Park, J.; Ha, N.Y.; Ra, J.S.; Kim, Y.J.; Kim, J.; Abdel-Rahman, M.; Park, S.H.; et al. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat. Commun. 2019, 10, 2420. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Fu, H.; Aladjem, M.I.; Myung, K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J. Cell. Biol. 2013, 200, 31–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.; Gali, V.K.; Takahashi, T.S.; Kubota, T. PCNA Retention on DNA into G2/M Phase Causes Genome Instability in Cells Lacking Elg1. Cell Rep. 2016, 16, 684–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kang, N.; Park, S.H.; Wells, J.; Hwang, T.; Ryu, E.; Kim, B.-G.; Hwang, S.; Kim, S.-J.; Kang, S.; et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020, 48, 7218–7238. [Google Scholar] [CrossRef]
- Kubota, T.; Katou, Y.; Nakato, R.; Shirahige, K.; Donaldson, A.D. Replication-Coupled PCNA Unloading by the Elg1 Complex Occurs Genome-wide and Requires Okazaki Fragment Ligation. Cell Rep. 2015, 12, 774–787. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Yang, K.; Cohn, M.A.; Sikdar, N.; D’Andrea, A.D.; Myung, K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 2010, 285, 10362–10369. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Kang, N.; Song, E.; Wie, M.; Lee, E.A.; Hwang, S.; Lee, D.; Ra, J.S.; Park, I.B.; Park, J.; et al. ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress. Nat. Commun. 2019, 10, 5718. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Kim, Y.; Ra, J.S.; Wie, M.W.; Kang, M.-S.; Kang, S.; Myung, K.; Lee, K.-Y. Timely termination of repair DNA synthesis by ATAD5 is important in oxidative DNA damage-induced single-strand break repair. Nucleic Acids Res. 2021, 49, 11746–11764. [Google Scholar] [CrossRef]
- Kang, M.S.; Kim, J.; Ryu, E.; Ha, N.Y.; Hwang, S.; Kim, B.-G.; Ra, J.S.; Kim, Y.J.; Hwang, J.M.; Myung, K.; et al. PCNA Unloading Is Negatively Regulated by BET Proteins. Cell Rep. 2019, 29, 4632–4645.e5. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.; D’Souza, R.C.J.; Tyanova, S.; Schaab, C.; Wiśniewski, J.R.; Cox, J.; Mann, M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef] [Green Version]
- Gaubitz, C.; Liu, X.; Pajak, J.; Stone, N.P.; A Hayes, J.; Demo, G.; A Kelch, B. Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader. eLife 2022, 11. [Google Scholar] [CrossRef]
- Devakumar, L.J.P.S.; Gaubitz, C.; Lundblad, V.; A Kelch, B.; Kubota, T. Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex. Nucleic Acids Res. 2019, 47, 6826–6841. [Google Scholar]
- Dieckman, L.M.; Boehm, E.M.; Hingorani, M.M.; Washington, M.T. Distinct structural alterations in proliferating cell nuclear antigen block DNA mismatch repair. Biochemistry 2013, 52, 5611–5619. [Google Scholar] [CrossRef] [Green Version]
- Arbel, M.; Bronstein, A.; Sau, S.; Liefshitz, B.; Kupiec, M. Access to PCNA by Srs2 and Elg1 Controls the Choice between Alternative Repair Pathways in Saccharomyces cerevisiae. mBio 2020, 11, e00705-20. [Google Scholar] [CrossRef]
- Shemesh, K.; Sebesta, M.; Pacesa, M.; Sau, S.; Bronstein, A.; Parnas, O.; Liefshitz, B.; Venclovas, C.; Krejci, L.; Kupiec, M. A structure-function analysis of the yeast Elg1 protein reveals the importance of PCNA unloading in genome stability maintenance. Nucleic Acids Res. 2017, 45, 3189–3203. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, A.A.; Mohideen, F.; Lima, C.D. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 2012, 483, 59–63. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, E.; Ha, N.Y.; Jung, W.; Yoo, J.; Myung, K.; Kang, S. Distinct Motifs in ATAD5 C-Terminal Domain Modulate PCNA Unloading Process. Cells 2022, 11, 1832. https://doi.org/10.3390/cells11111832
Ryu E, Ha NY, Jung W, Yoo J, Myung K, Kang S. Distinct Motifs in ATAD5 C-Terminal Domain Modulate PCNA Unloading Process. Cells. 2022; 11(11):1832. https://doi.org/10.3390/cells11111832
Chicago/Turabian StyleRyu, Eunjin, Na Young Ha, Woojae Jung, Juyeong Yoo, Kyungjae Myung, and Sukhyun Kang. 2022. "Distinct Motifs in ATAD5 C-Terminal Domain Modulate PCNA Unloading Process" Cells 11, no. 11: 1832. https://doi.org/10.3390/cells11111832
APA StyleRyu, E., Ha, N. Y., Jung, W., Yoo, J., Myung, K., & Kang, S. (2022). Distinct Motifs in ATAD5 C-Terminal Domain Modulate PCNA Unloading Process. Cells, 11(11), 1832. https://doi.org/10.3390/cells11111832





