A Unique Spectrum of Spontaneous Tumors in Dino Knockout Mice Identifies Tissue-Specific Requirements for Tumor Suppression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Pathology
2.3. Total Body Irradiation
2.4. Survival Analysis
2.5. RNA Isolation and Quantitative Reverse Transcription-PCR
2.6. Statistics
3. Results
3.1. Spontaneous Tumorigenesis in Dino−/− Mice Reveals a Predisposition for Sarcoma
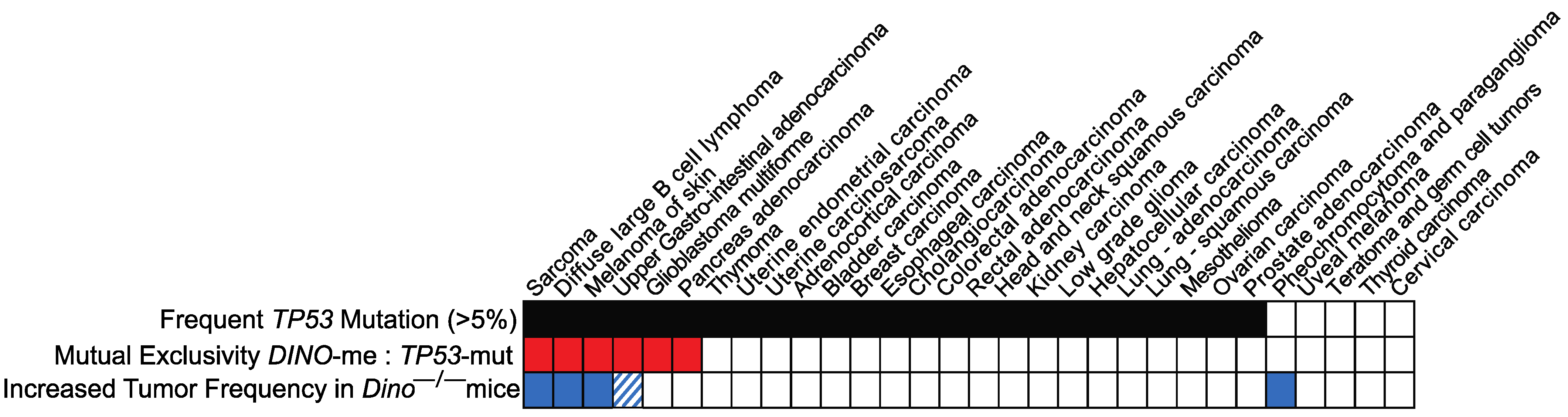
3.2. The Spectrum of Spontaneous Tumors in Dino−/− Mice Is Similar to Types of Human Cancers in Which Epigenetic Silencing of DINO Is Mutually Exclusive with TP53 Mutations
3.3. Loss of Dino Is Insufficient for Tumorigenesis in Selected Tissues That Depend on p53 for Tumor Suppression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.E.; The ENCODE Project Consortium; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Andergassen, D.; Rinn, J.L. From genotype to phenotype: Genetics of mammalian long non-coding RNAs in vivo. Nat. Rev. Genet. 2022, 23, 229–243. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology. Cold Spring Harb. Perspect. Med. 2017, 7, a026492. [Google Scholar] [CrossRef] [Green Version]
- Winkler, L.; Dimitrova, N. A mechanistic view of long noncoding RNAs in cancer. Wiley Interdiscip. Rev. RNA 2021, 13, e1699. [Google Scholar] [CrossRef]
- Lane, D.; Levine, A. p53 Research: The Past Thirty Years and the Next Thirty Years. Cold Spring Harb. Perspect. Biol. 2010, 2, a000893. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Thorne, R.F.; Zhang, X.D.; Wu, M.; Liu, L. Non-coding RNAs, guardians of the p53 galaxy. Semin. Cancer Biol. 2021, 75, 72–83. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Kusafuka, T.; Fukuzawa, M.; Oue, T.; Komoto, Y.; Yoneda, A.; Okada, A. Mutation analysis of p53 gene in childhood malignant solid tumors. J. Pediatr. Surg. 1997, 32, 1175–1180. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valente, L.J.; Gray, D.H.D.; Michalak, E.M.; Pinon-Hofbauer, J.; Egle, A.; Scott, C.L.; Janic, A.; Strasser, A. p53 Efficiently Suppresses Tumor Development in the Complete Absence of Its Cell-Cycle Inhibitory and Proapoptotic Effectors p21, Puma, and Noxa. Cell Rep. 2013, 3, 1339–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Kon, N.; Jiang, L.; Tan, M.; Ludwig, T.; Zhao, Y.; Baer, R.; Gu, W. Tumor Suppression in the Absence of p53-Mediated Cell-Cycle Arrest, Apoptosis, and Senescence. Cell 2012, 149, 1269–1283. [Google Scholar] [CrossRef] [Green Version]
- Grossi, E.; Sánchez, Y.; Huarte, M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim. Biophys. Acta 2016, 1859, 200–208. [Google Scholar] [CrossRef]
- Schmitt, A.; Garcia, J.T.; Hung, T.; Flynn, R.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-Da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef] [Green Version]
- Marney, C.B.; Anderson, E.S.; Adnan, M.; Peng, K.-L.; Hu, Y.; Weinhold, N.; Schmitt, A.M. p53-intact cancers escape tumor suppression through loss of long noncoding RNA Dino. Cell Rep. 2021, 35, 109329. [Google Scholar] [CrossRef]
- Jacks, T.; Remington, L.; Williams, B.O.; Schmitt, E.M.; Halachmi, S.; Bronson, R.T.; Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994, 4, 1–7. [Google Scholar] [CrossRef]
- Lowe, S.W.; Bodis, S.; McClatchey, A.; Remington, L.; Ruley, H.E.; Fisher, D.E.; Housman, D.E.; Jacks, T. p53 Status and the Efficacy of Cancer Therapy in Vivo. Science 1994, 266, 807–810. [Google Scholar] [CrossRef]
- Martín-Caballero, J.; Flores, J.M.; García-Palencia, P.; Serrano, M. Tumor Susceptibility of p21Waf1/Cip1-deficient Mice1. Cancer Res. 2001, 61, 6234–6238. [Google Scholar]
- Pettan-Brewer, C.; Treuting, P.M.M. Practical pathology of aging mice. Pathobiol. Aging Age-Relat. Dis. 2011, 1, 7202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacroix-Triki, M.; Lacoste-Collin, L.; Jozan, S.; Charlet, J.-P.; Caratero, C.; Courtade, M. Histiocytic Sarcoma in C57BL/6J Female Mice is Associated with Liver Hematopoiesis: Review of 41 Cases. Toxicol. Pathol. 2003, 31, 304–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BBlackwell, B.-N.; Bucci, T.J.; Hart, R.W.; Turturro, A. Longevity, Body Weight, and Neoplasia in Ad Libitum-Fed and Diet-Restricted C57BL6 Mice Fed NIH-31 Open Formula Diet. Toxicol. Pathol. 1995, 23, 570–582. [Google Scholar] [CrossRef]
- Robles, A.I.; Linke, S.P.; Harris, C.C. The p53 network in lung carcinogenesis. Oncogene 2002, 21, 6898–6907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciriello, G.; Cerami, E.; Sander, C.; Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012, 22, 398–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donehower, L.A.; Harvey, M.; Slagle, B.L.; McArthur, M.J.; Montgomery, C.A., Jr.; Butel, J.S.; Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992, 356, 215–221. [Google Scholar] [CrossRef]
- Liu, Q.; Xiao, Y.; Cai, P.; Li, J.; Li, D. Long noncoding RNA DINO (damage induced noncoding) represses the development of gastric cancer by modulating p21 and Bcl-2 Associated X Protein (Bax) expression. J. Cell. Biochem. 2019, 120, 11190–11195. [Google Scholar] [CrossRef]
- Kemp, C.J.; Wheldon, T.; Balmain, A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat. Genet. 1994, 8, 66–69. [Google Scholar] [CrossRef]
- Lowe, S.W.; Schmitt, E.M.; Smitht, S.W.; Osbornet, B.A.; Jacks, T. pS3 is required for radiation-induced apoptosis in mouse thymocytes. Nature 1993, 362, 847–849. [Google Scholar] [CrossRef]
- Clarke, A.R.; Purdie, C.A.; Harrison, D.J.; Morris, R.G.; Bird, C.C.; Hooper, M.L.; Wyllie, A.H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 1993, 362, 849–852. [Google Scholar] [CrossRef]
- Kaplan, H.S.; Brown, M.B. A quantitative dose-response study of lymphoid-tumor development in irradiated C 57 black mice. J. Natl. Cancer Inst. 1952, 13, 185–208. [Google Scholar] [PubMed]
- De La Cueva, E.; García-Cao, I.; Herranz, M.; López, P.; García-Palencia, P.; Flores, J.M.; Serrano, M.; Fernández-Piqueras, J.; Martín-Caballero, J. Tumorigenic activity of p21Waf1/Cip1 in thymic lymphoma. Oncogene 2006, 25, 4128–4132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Brady, C.A.; Johnson, T.M.; Lee, E.Y.; Park, E.J.; Scott, M.P.; Attardi, L.D. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc. Natl. Acad. Sci. USA 2011, 108, 17123–17128. [Google Scholar] [CrossRef] [Green Version]
- Ocasio, J.; Babcock, B.; Malawsky, D.; Weir, S.J.; Loo, L.; Simon, J.M.; Zylka, M.J.; Hwang, D.; Dismuke, T.; Sokolsky, M.; et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat. Commun. 2019, 10, 5829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schüller, U.; Heine, V.M.; Mao, J.; Kho, A.T.; Dillon, A.K.; Han, Y.-G.; Huillard, E.; Sun, T.; Ligon, A.H.; Qian, Y.; et al. Acquisition of Granule Neuron Precursor Identity Is a Critical Determinant of Progenitor Cell Competence to Form Shh-Induced Medulloblastoma. Cancer Cell 2008, 14, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.-J.; Ellis, T.; Markant, S.L.; Read, T.-A.; Kessler, J.D.; Bourboulas, M.; Schüller, U.; Machold, R.; Fishell, G.; Rowitch, D.H.; et al. Medulloblastoma Can Be Initiated by Deletion of Patched in Lineage-Restricted Progenitors or Stem Cells. Cancer Cell 2008, 14, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Zhu, C.; Jin, Y. The Oncogenic and Tumor Suppressive Functions of the Long Noncoding RNA MALAT1: An Emerging Controversy. Front. Genet. 2020, 11, 93. Available online: https://www.frontiersin.org/article/10.3389/fgene.2020.00093 (accessed on 4 April 2022). [CrossRef] [Green Version]
- Eißmann, M.; Gutschner, T.; Hämmerle, M.; Günther, S.; Caudron-Herger, M.; Groß, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zörnig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Rep. 2012, 2, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Hewitson, J.P.; West, K.A.; James, K.R.; Rani, G.F.; Dey, N.; Romano, A.; Brown, N.; Teichmann, S.A.; Kaye, P.M.; Lagos, D. Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells. J. Immunol. 2020, 204, 2949–2960. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, Z.; Liu, L.; Yang, Z.; Liu, S.; Ma, Z.; Liu, Y.; Ma, Y.; Zhang, L.; Zhang, X.; et al. LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity. Proc. Natl. Acad. Sci. USA 2020, 117, 23695–23706. [Google Scholar] [CrossRef] [PubMed]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sae, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.B.; Kobayashi, N.; Todo, Y.; Watari, H. Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front. Genet. 2018, 9, 471. Available online: https://www.frontiersin.org/article/10.3389/fgene.2018.00471 (accessed on 31 March 2022). [CrossRef] [PubMed] [Green Version]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Sharpless, N.E.; Ramsey, M.R.; Balasubramanian, P.; Castrillon, D.H.; DePinho, R.A. The differential impact of p16INK4A or p19ARF deficiency on cell growth and tumorigenesis. Oncogene 2004, 23, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Genik, P.C.; Bielefeldt-Ohmann, H.; Liu, X.; Story, M.D.; Ding, L.; Bush, J.M.; Fallgren, C.M.; Weil, M.M. Strain Background Determines Lymphoma Incidence in Atm Knockout Mice. Neoplasia 2014, 16, 129–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.N.; Hancock, A.R.; Vogel, H.; Donehower, L.A.; Bradley, A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 15608–15612. [Google Scholar] [CrossRef] [Green Version]
- Jeffers, J.R.; Parganas, E.; Lee, Y.; Yang, C.; Wang, J.; Brennan, J.; MacLean, K.H.; Han, J.; Chittenden, T.; Ihle, J.N.; et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 2003, 4, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Carson-Walter, E.B.; Kuan, S.F.; Zhang, L.; Yu, J. PUMA Suppresses Intestinal Tumorigenesis in Mice. Cancer Res. 2009, 69, 4999–5006. [Google Scholar] [CrossRef] [Green Version]
- MacPherson, D.; Kim, J.; Kim, T.; Rhee, B.K.; Van Oostrom, C.T.M.; DiTullio, R.A.; Venere, M.; Halazonetis, T.D.; Bronson, R.; De Vries, A.; et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004, 23, 3689–3699. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Parant, J.M.; Lang, G.; Chau, P.; Chavez-Reyes, A.; El-Naggar, A.K.; Multani, A.S.; Chang, S.; Lozano, G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat. Genet. 2003, 36, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Landuzzi, L.; Ianzano, M.L.; Nicoletti, G.; Palladini, A.; Grosso, V.; Ranieri, D.; Ora, M.D.; Raschi, E.; Laranga, R.; Gambarotti, M.; et al. Genetic prevention of lymphoma in p53 knockout mice allows the early development of p53-related sarcomas. Oncotarget 2014, 5, 11924–11938. [Google Scholar] [CrossRef] [PubMed]
- Christophorou, M.A.; Ringshausen, I.; Finch, A.J.; Swigart, L.B.; Evan, G.I. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 2006, 443, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Hinkal, G.; Parikh, N.; Donehower, L.A. Timed Somatic Deletion of p53 in Mice Reveals Age-Associated Differences in Tumor Progression. PLoS ONE 2009, 4, e6654. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-L.; Castle, K.D.; Moding, E.J.; Blum, J.M.; Williams, N.; Luo, L.; Ma, Y.; Borst, L.; Kim, Y.; Kirsch, D.G. Acute DNA damage activates the tumour suppressor p53 to promote radiation-induced lymphoma. Nat. Commun. 2015, 6, 8477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramaswamy, V.; Nör, C.; Taylor, M.D. p53 and Meduloblastoma. Cold Spring Harb. Perspect. Med. 2016, 6, a026278. [Google Scholar] [CrossRef] [Green Version]


| Tumor | Dino+/+ (%) | Dino−/− (%) |
|---|---|---|
| Histiocytic sarcoma | 4 (21.1) | 9 (32.1) |
| Sarcoma (bone and soft tissue) | 0 (0) | 6 (21.4) 1 |
| B and non-T cell lymphoma | 3 (16.7) | 8 (28.6) |
| T cell lymphoma | 0 (0) | 0 (0) |
| Lung adenocarcinoma | 2 (10.5) | 2 (7.1) |
| Melanoma | 0 (0) | 1 (3.6) |
| Gastrointestinal adenocarcinoma | 0 (0) | 1 (3.6) |
| Pheochromocytoma | 0 (0) | 1 (3.6) |
| Total Mice with Tumors | 9 (47.4) | 22 (78.6) 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marney, C.B.; Anderson, E.S.; Baum, R.; Schmitt, A.M. A Unique Spectrum of Spontaneous Tumors in Dino Knockout Mice Identifies Tissue-Specific Requirements for Tumor Suppression. Cells 2022, 11, 1818. https://doi.org/10.3390/cells11111818
Marney CB, Anderson ES, Baum R, Schmitt AM. A Unique Spectrum of Spontaneous Tumors in Dino Knockout Mice Identifies Tissue-Specific Requirements for Tumor Suppression. Cells. 2022; 11(11):1818. https://doi.org/10.3390/cells11111818
Chicago/Turabian StyleMarney, Christina B., Erik S. Anderson, Rachel Baum, and Adam M. Schmitt. 2022. "A Unique Spectrum of Spontaneous Tumors in Dino Knockout Mice Identifies Tissue-Specific Requirements for Tumor Suppression" Cells 11, no. 11: 1818. https://doi.org/10.3390/cells11111818
APA StyleMarney, C. B., Anderson, E. S., Baum, R., & Schmitt, A. M. (2022). A Unique Spectrum of Spontaneous Tumors in Dino Knockout Mice Identifies Tissue-Specific Requirements for Tumor Suppression. Cells, 11(11), 1818. https://doi.org/10.3390/cells11111818






