Renal Ischemia Induces Epigenetic Changes in Apoptotic, Proteolytic, and Mitochondrial Genes in Swine Scattered Tubular-like Cells
Abstract
:1. Introduction
2. Materials and Methods
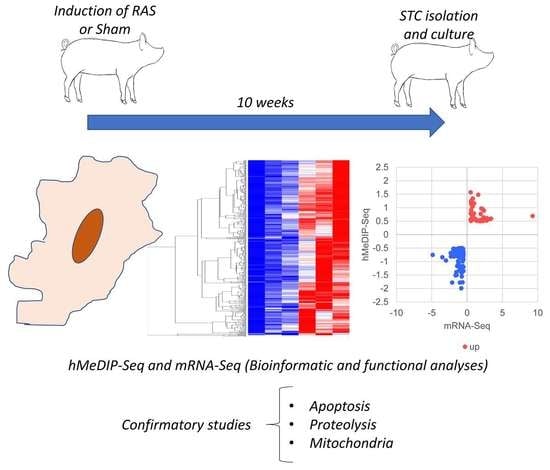
2.1. Experimental Design
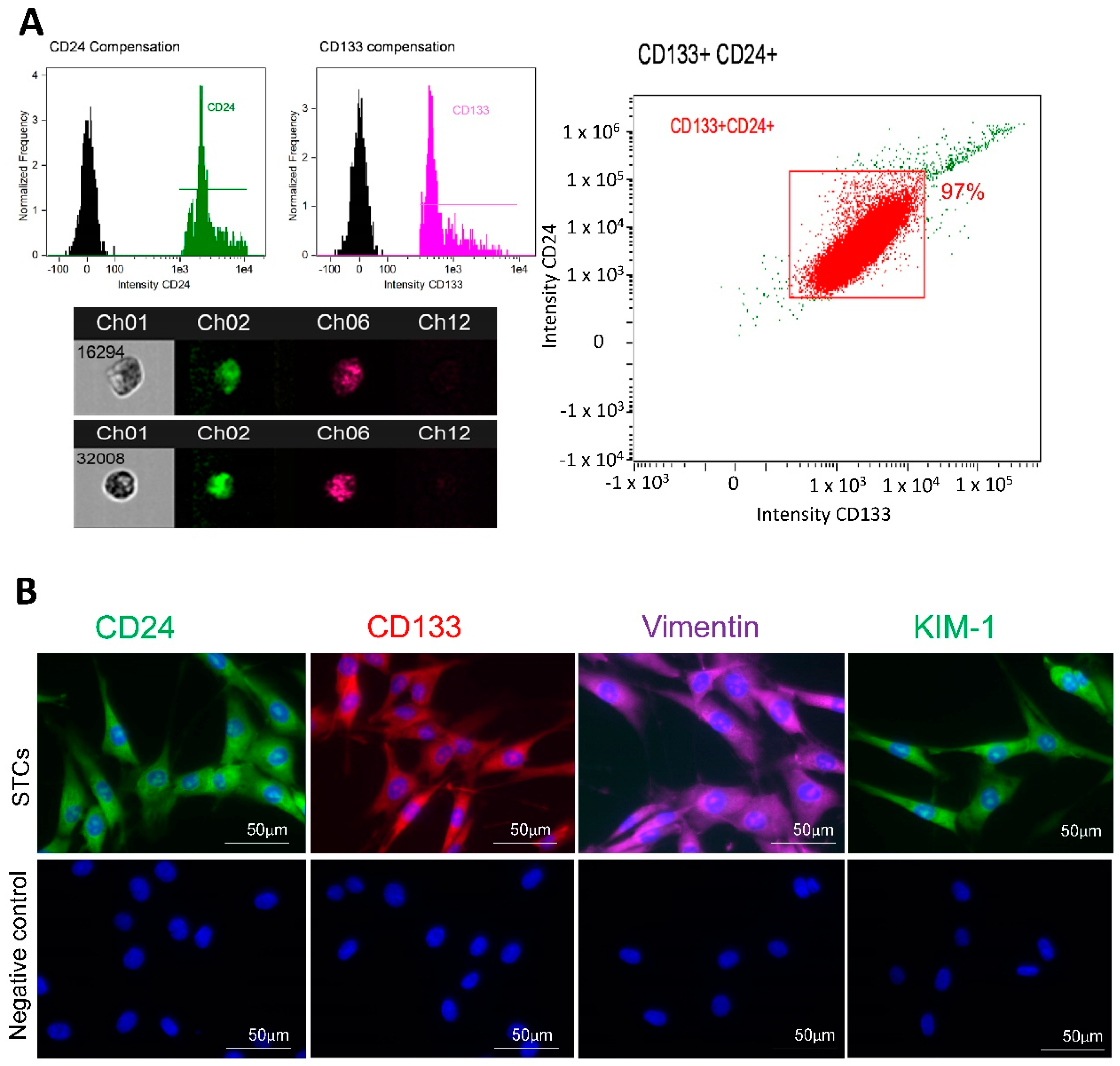
2.2. STC Isolation and Characterization
2.3. MeDIP- and hMeDIP-seq
2.4. mRNA-seq and Integrated (MeDIP- and hMeDIP-seq/mRNA-seq) Analysis
2.5. Validation of MeDIP- and hMeDIP-seq/mRNA-seq Analysis
2.6. Functional Studies
2.7. Statistical Analysis
3. Results
3.1. STC Characterization
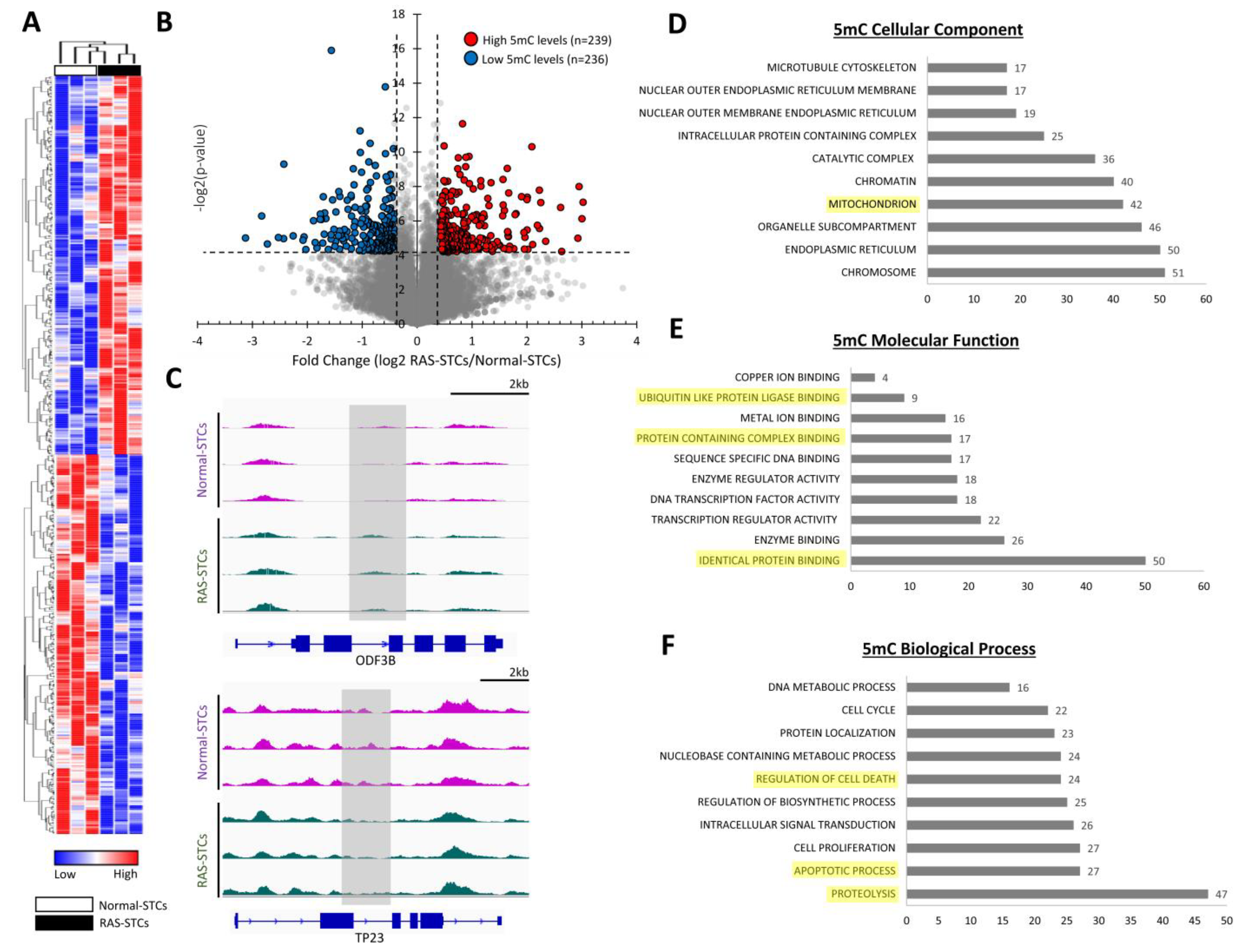
3.2. RAS Alters 5mC and 5hmC Levels in Swine STCs
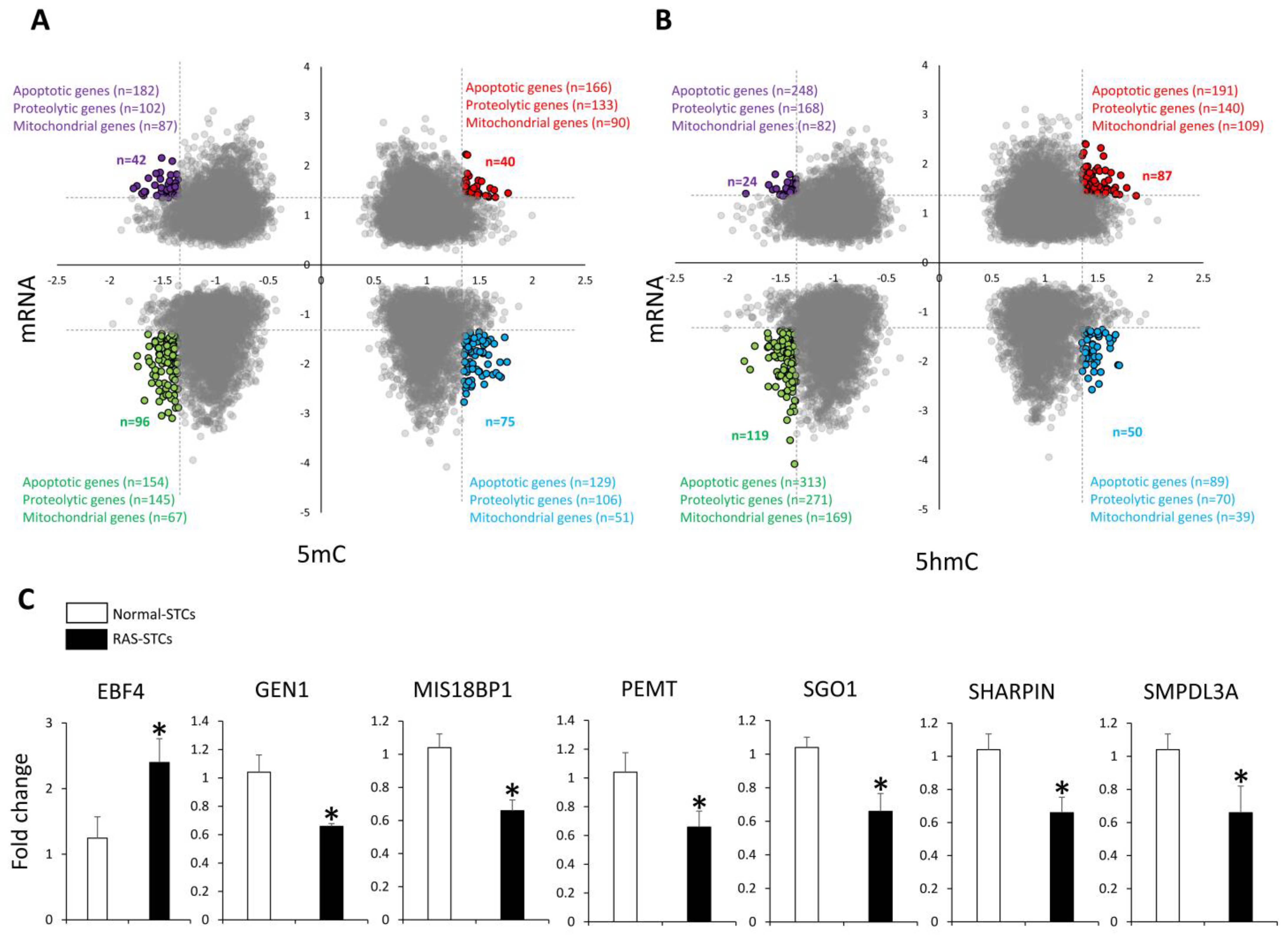
3.3. RAS Induces Changes in Gene Expression in Swine STCs
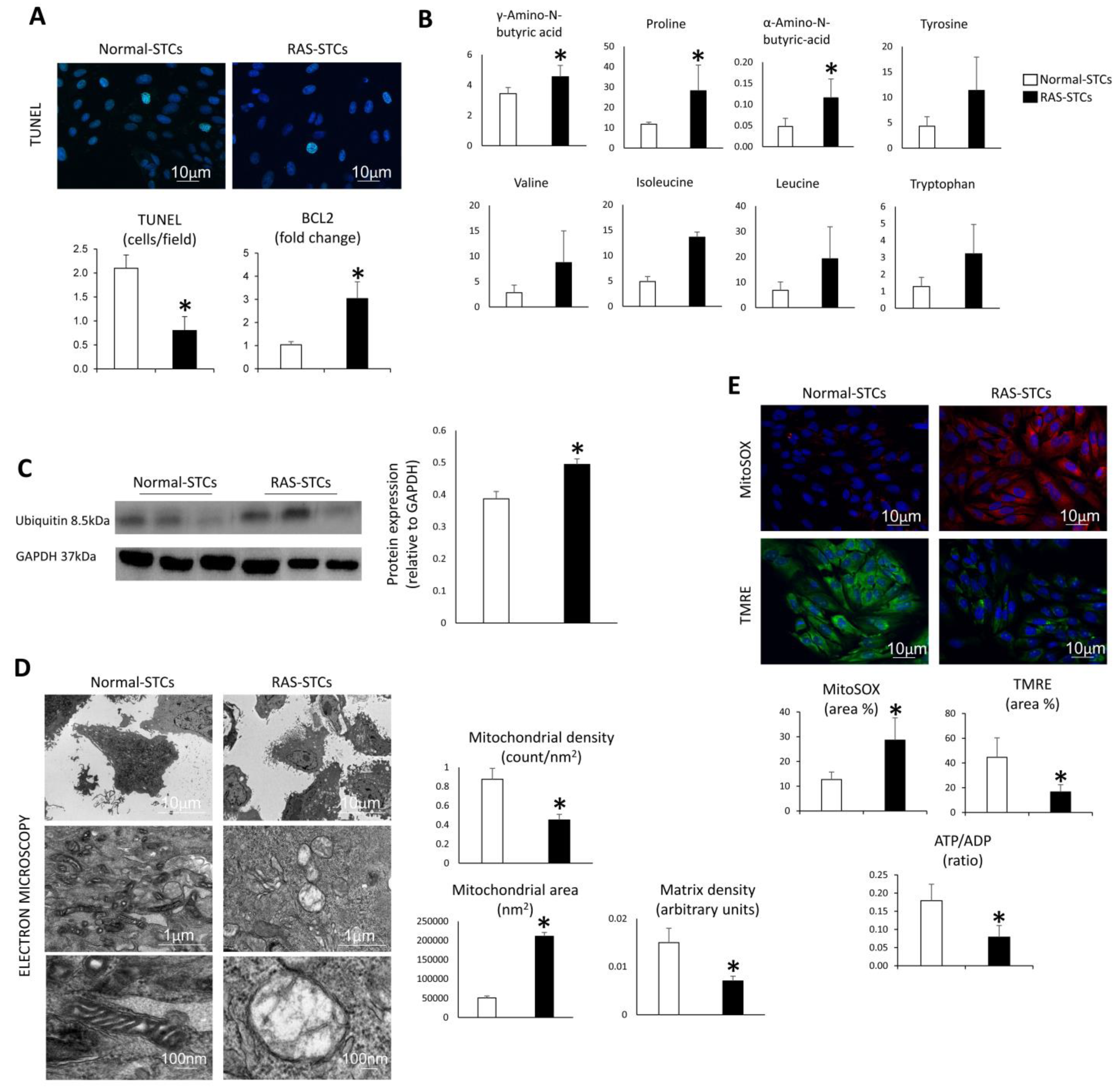
3.4. RAS Modulates Apoptosis, Proteolysis, and Mitochondrial Function in Swine STCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansen, K.J.; Edwards, M.S.; Craven, T.E.; Cherr, G.S.; Jackson, S.A.; Appel, R.G.; Burke, G.L.; Dean, R.H. Prevalence of renovascular disease in the elderly: A population-based study. J. Vasc. Surg. 2002, 36, 443–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzu, T.; Takeji, M.; Yamada, N.; Fujii, T.; Yamauchi, A.; Takishita, S.; Kimura, G. Prevalence and outcome of renal artery stenosis in atherosclerotic patients with renal dysfunction. Hypertens. Res. 2002, 25, 537–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garovic, V.; Textor, S.C. Renovascular Hypertension and Ischemic Nephropathy. Circulation 2005, 112, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Eirin, A.; Lerman, L.O. Darkness at the End of the Tunnel: Poststenotic Kidney Injury. Physiology 2013, 28, 245–253. [Google Scholar] [CrossRef] [Green Version]
- Eirin, A.; Textor, S.C.; Lerman, L.O. Emerging Paradigms in Chronic Kidney Ischemia. Hypertension 2018, 72, 1023–1030. [Google Scholar] [CrossRef]
- Nargesi, A.A.; Zhu, X.Y.; Conley, S.M.; Woollard, J.R.; Saadiq, I.M.; Lerman, L.O.; Eirin, A. Renovascular disease induces mitochondrial damage in swine scattered tubular cells. Am. J. Physiol. Renal Physiol. 2019, 317, F1142–F1153. [Google Scholar] [CrossRef]
- Bonventre, J.V. Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J. Am. Soc. Nephrol. 2003, 14 (Suppl. 1), S55–S61. [Google Scholar] [CrossRef] [Green Version]
- Romagnani, P.; Remuzzi, G. CD133+ renal stem cells always co-express CD24 in adult human kidney tissue. Stem Cell Res. 2014, 12, 828–829. [Google Scholar] [CrossRef] [Green Version]
- Smeets, B.; Boor, P.; Dijkman, H.; Sharma, S.V.; Jirak, P.; Mooren, F.; Berger, K.; Bornemann, J.; Gelman, I.H.; Floege, J. Proximal tubular cells contain a phenotypically distinct, scattered cell population involved in tubular regeneration. J. Pathol. 2013, 229, 645–659. [Google Scholar] [CrossRef]
- Lindgren, D.; Bostrom, A.K.; Nilsson, K.; Hansson, J.; Sjolund, J.; Moller, C.; Jirstrom, K.; Nilsson, E.; Landberg, G.; Axelson, H. Isolation and characterization of progenitor-like cells from human renal proximal tubules. Am. J. Pathol. 2011, 178, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Farahani, R.A.; Zhu, X.Y.; Tang, H.; Jordan, K.L.; Lerman, L.O.; Eirin, A. Renal ischemia alters expression of mitochondria-related genes and impairs mitochondrial structure and function in swine scattered tubular-like cells. Am. J. Physiol. Renal Physiol. 2020, 319, F19–F28. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Kim, S.R.; Jiang, K.; Ferguson, C.M.; Tang, H.; Zhu, X.Y.; Lerman, A.; Eirin, A.; Lerman, L.O. Renovascular Disease Induces Senescence in Renal Scattered Tubular-Like Cells and Impairs Their Reparative Potency. Hypertension 2021, 77, 507–518. [Google Scholar] [CrossRef]
- Tang, J.; Zhuang, S. Epigenetics in acute kidney injury. Curr. Opin. Nephrol. Hypertens. 2015, 24, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Bomsztyk, K.; Denisenko, O. Epigenetic Alterations in Acute Kidney Injury. Semin. Nephrol. 2013, 33, 327–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifian, R.; Okamura, D.M.; Denisenko, O.; Zager, R.A.; Johnson, A.; Gharib, S.A.; Bomsztyk, K. Distinct patterns of transcriptional and epigenetic alterations characterize acute and chronic kidney injury. Sci. Rep. 2018, 8, 17870. [Google Scholar] [CrossRef]
- Breiling, A.; Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 2015, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [Green Version]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef]
- Shi, D.Q.; Ali, I.; Tang, J.; Yang, W.C. New Insights into 5hmC DNA Modification: Generation, Distribution and Function. Front Genet. 2017, 8, 100. [Google Scholar] [CrossRef]
- Lerman, L.O.; Schwartz, R.S.; Grande, J.P.; Sheedy, P.F.; Rommero, J.C. Noninvasive evaluation of a novel swine model of renal artery stenosis. J. Am. Soc. Nephrol. 1999, 10, 1455–1465. [Google Scholar] [CrossRef]
- Chade, A.R.; Zhu, X.; Lavi, R.; Krier, J.D.; Pislaru, S.; Simari, R.D.; Napoli, C.; Lerman, A.; Lerman, L.O. Endothelial progenitor cells restore renal function in chronic experimental renovascular disease. Circulation 2009, 119, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Chade, A.R.; Rodriguez-Porcel, M.; Grande, J.P.; Krier, J.D.; Lerman, A.; Romero, J.C.; Napoli, C.; Lerman, L.O. Distinct renal injury in early atherosclerosis and renovascular disease. Circulation 2002, 106, 1165–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krier, J.D.; Ritman, E.L.; Bajzer, Z.; Romero, J.C.; Lerman, A.; Lerman, L.O. Noninvasive measurement of concurrent single-kidney perfusion, glomerular filtration, and tubular function. Am. J. Physiol Renal Physiol. 2001, 281, F630-8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eirin, A.; Ebrahimi, B.; Zhang, X.; Zhu, X.Y.; Tang, H.; Crane, J.A.; Lerman, A.; Textor, S.C.; Lerman, L.O. Changes in glomerular filtration rate after renal revascularization correlate with microvascular hemodynamics and inflammation in Swine renal artery stenosis. Circ. Cardiovasc Interv. 2012, 5, 720–728. [Google Scholar] [CrossRef] [Green Version]
- Aghajani Nargesi, A.; Zhu, X.Y.; Liu, Y.; Tang, H.; Jordan, K.L.; Lerman, L.O.; Eirin, A. Renal Artery Stenosis Alters Gene Expression in Swine Scattered Tubular-Like Cells. Int. J. Mol. Sci. 2019, 20, 5069. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Jia, Y.; Li, C.; Cheng, Q.; Yue, W.; Pei, X. Hyperglycemic Stress Impairs the Stemness Capacity of Kidney Stem Cells in Rats. PLoS ONE 2015, 10, 0139607. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Urbieta-Caceres, V.; Krier, J.D.; Textor, S.C.; Lerman, A.; Lerman, L.O. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem Cells. 2013, 31, 117–125. [Google Scholar] [CrossRef] [Green Version]
- Isik, B.; Thaler, R.; Goksu, B.B.; Conley, S.M.; Al-Khafaji, H.; Mohan, A.; Afarideh, M.; Abumoawad, A.M.; Zhu, X.Y.; Krier, J.D.; et al. preconditioning induces epigenetic changes and modifies swine mesenchymal stem cell angiogenesis and senescence in experimental atherosclerotic renal artery stenosis. Stem Cell Res. Ther. 2021, 12, 240. [Google Scholar] [CrossRef]
- Pollock, K.; Samsonraj, R.M.; Dudakovic, A.; Thaler, R.; Stumbras, A.; McKenna, D.H.; Dosa, P.I.; van Wijnen, A.J.; Hubel, A. Improved Post-Thaw Function and Epigenetic Changes in Mesenchymal Stromal Cells Cryopreserved Using Multicomponent Osmolyte Solutions. Stem Cells Dev. 2017, 26, 828–842. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat.Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Negre, N.; Li, Q.; Mieczkowska, J.O.; Slattery, M.; Liu, T.; Zhang, Y.; Kim, T.K.; He, H.H.; Zieba, J.; et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat. Methods. 2012, 9, 609–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Eirin, A.; Riester, S.M.; Zhu, X.Y.; Tang, H.; Evans, J.M.; O’Brien, D.; van Wijnen, A.J.; Lerman, L.O. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene 2014, 551, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Eirin, A.; Zhu, X.Y.; Tang, H.; Chanana, P.; Lerman, A.; Van Wijnen, A.J.; Lerman, L.O. The metabolic syndrome alters the miRNA signature of porcine adipose tissue-derived mesenchymal stem cells. Cytometry A. 2018, 93, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, D1251–D1257. [Google Scholar] [CrossRef] [Green Version]
- Eirin, A.; Li, Z.; Zhang, X.; Krier, J.D.; Woollard, J.R.; Zhu, X.Y.; Tang, H.; Herrmann, S.M.; Lerman, A.; Textor, S.C.; et al. A mitochondrial permeability transition pore inhibitor improves renal outcomes after revascularization in experimental atherosclerotic renal artery stenosis. Hypertension 2012, 60, 1242–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eirin, A.; Ebrahimi, B.; Zhang, X.; Zhu, X.Y.; Woollard, J.R.; He, Q.; Textor, S.C.; Lerman, A.; Lerman, L.O. Mitochondrial protection restores renal function in swine atherosclerotic renovascular disease. Cardiovasc Res. 2014, 103, 461–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaspar, H.; Dettmer, K.; Chan, Q.; Daniels, S.; Nimkar, S.; Daviglus, M.L.; Stamler, J.; Elliott, P.; Oefner, P.J. Urinary amino acid analysis: A comparison of iTRAQ-LC-MS/MS, GC-MS, and amino acid analyzer. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2009, 877, 1838–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nargesi, A.A.; Zhang, L.; Tang, H.; Jordan, K.L.; Saadiq, I.M.; Textor, S.C.; Lerman, L.O.; Eirin, A. Coexisting renal artery stenosis and metabolic syndrome magnifies mitochondrial damage, aggravating poststenotic kidney injury in pigs. J. Hypertens. 2019, 37, 2061–2073. [Google Scholar] [CrossRef]
- Eirin, A.; Woollard, J.R.; Ferguson, C.M.; Jordan, K.L.; Tang, H.; Textor, S.C.; Lerman, A.; Lerman, L.O. The metabolic syndrome induces early changes in the swine renal medullary mitochondria. Transl. Res. 2017, 184, 45–56.e9. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Rajesh, M.; Yoshihiro, K.; Hasko, G.; Pacher, P. Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 2007, 358, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Farrelly, E.; Amaral, M.C.; Marshall, L.; Huang, S.G. A high-throughput assay for mitochondrial membrane potential in permeabilized yeast cells. Anal. Biochem. 2001, 293, 269–276. [Google Scholar] [CrossRef]
- Rinkevich, Y.; Montoro, D.T.; Contreras-Trujillo, H.; Harari-Steinberg, O.; Newman, A.M.; Tsai, J.M.; Lim, X.; Van-Amerongen, R.; Bowman, A.; Januszyk, M.; et al. In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell Rep. 2014, 7, 1270–1283. [Google Scholar] [CrossRef] [Green Version]
- Hansson, J.; Hultenby, K.; Cramnert, C.; Ponten, F.; Jansson, H.; Lindgren, D.; Axelson, H.; Johansson, M.E. Evidence for a morphologically distinct and functionally robust cell type in the proximal tubules of human kidney. Hum. Pathol. 2014, 45, 382–393. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Kwon, S.H.; Jiang, K.; Ferguson, C.M.; Puranik, A.S.; Zhu, X.; Lerman, L.O. Renal scattered tubular-like cells confer protective effects in the stenotic murine kidney mediated by release of extracellular vesicles. Sci. Rep. 2018, 8, 1263. [Google Scholar] [CrossRef] [Green Version]
- Hata, H.; Tatemichi, M.; Nakadate, T. Involvement of annexin A8 in the properties of pancreatic cancer. Mol. Carcinog. 2014, 53, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Persaud, S.D.; Nhieu, J.; Wei, L.N. Cellular Retinoic Acid-Binding Protein 1 Modulates Stem Cell Proliferation to Affect Learning and Memory in Male Mice. Endocrinology 2017, 158, 3004–3014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doycheva, D.; Xu, N.; Kaur, H.; Malaguit, J.; McBride, D.W.; Tang, J.; Zhang, J.H. Adenoviral TMBIM6 vector attenuates ER-stress-induced apoptosis in a neonatal hypoxic-ischemic rat model. Dis Model. Mech. 2019, 12, dmm040352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haase, M.; Fitze, G. HSP90AB1: Helping the good and the bad. Gene 2016, 575, 171–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Long, M.J.C.; Wang, Y.; Zhang, S.; Aye, Y. Ube2V2 Is a Rosetta Stone Bridging Redox and Ubiquitin Codes, Coordinating DNA Damage Responses. ACS Cent. Sci. 2018, 4, 246–259. [Google Scholar] [CrossRef]
- Norton, M.; Ng, A.C.; Baird, S.; Dumoulin, A.; Shutt, T.; Mah, N.; Andrade-Navarro, M.A.; McBride, H.M.; Screaton, R.A. ROMO1 is an essential redox-dependent regulator of mitochondrial dynamics. Sci. Signal. 2014, 7, ra10. [Google Scholar] [CrossRef]
- Kaguni, L.S. DNA polymerase gamma, the mitochondrial replicase. Annu Rev. Biochem. 2004, 73, 293–320. [Google Scholar] [CrossRef]
- Kuwata, H.; Hara, S. Role of acyl-CoA synthetase ACSL4 in arachidonic acid metabolism. Prostaglandins Other Lipid Mediat. 2019, 144, 106363. [Google Scholar] [CrossRef]
- Dupuy, F.; Tabaries, S.; Andrzejewski, S.; Dong, Z.; Blagih, J.; Annis, M.G.; Omeroglu, A.; Gao, D.; Leung, S.; Amir, E.; et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015, 22, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Meng, Y.; Liu, Y.; van Wijnen, A.J.; Eirin, A.; Lerman, L.O. Differentially Expressed Functional LncRNAs in Human Subjects With Metabolic Syndrome Reflect a Competing Endogenous RNA Network in Circulating Extracellular Vesicles. Front. Mol. Biosci. 2021, 8, 667056. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Orian, A.; Schwartz, A.L. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays 2000, 22, 442–451. [Google Scholar] [CrossRef]
- Wojcik, C. Regulation of apoptosis by the ubiquitin and proteasome pathway. J. Cell Mol. Med. 2002, 6, 25–48. [Google Scholar] [CrossRef] [PubMed]
- Colombini, M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol. Cell Biochem. 2004, 256–257, 107–115. [Google Scholar] [CrossRef]
- Kaasik, A.; Safiulina, D.; Zharkovsky, A.; Veksler, V. Regulation of mitochondrial matrix volume. Am. J. Physiol Cell Physiol. 2007, 292, C157–C163. [Google Scholar] [CrossRef]
- Zick, M.; Rabl, R.; Reichert, A.S. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009, 1793, 5–19. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; Avshalumov, M.V.; Patel, J.C.; Lee, C.R.; Miller, E.W.; Chang, C.J.; Rice, M.E. Mitochondria are the source of hydrogen peroxide for dynamic brain-cell signaling. J. Neurosci. 2009, 29, 9002–9010. [Google Scholar] [CrossRef]
- Eirin, A.; Zhu, X.Y.; Urbieta-Caceres, V.H.; Grande, J.P.; Lerman, A.; Textor, S.C.; Lerman, L.O. Persistent kidney dysfunction in swine renal artery stenosis correlates with outer cortical microvascular remodeling. Am. J. Physiol Renal Physiol. 2011, 300, F1394–F1401. [Google Scholar] [CrossRef] [Green Version]
- Arce, C.; Lucena, C.; Moreno, A.; Garrido, J.J. Proteomic analysis of intestinal mucosa responses to Salmonella enterica serovar typhimurium in naturally infected pig. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 59–67. [Google Scholar] [CrossRef]
- Shen, S.Q.; Yan, X.W.; Li, P.T.; Ji, X.H. Analysis of differential gene expression by RNA-seq data in ABCG1 knockout mice. Gene 2019, 689, 24–33. [Google Scholar] [CrossRef]
- Shu-Shan, Z.; Jian-Jun, D.; Cai-Feng, W.; Ting-Yu, Z.; De-Fu, Z. Comparative proteomic analysis of hearts of adult SCNT Bama miniature pigs (Sus scrofa). Theriogenology 2014, 81, 901–905. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Normal | RAS |
|---|---|---|
| Body Weight (Kg) | 51.5 ± 0.5 | 54.7 ± 5.5 |
| Degree of stenosis (%) | 0 | 86.7 ± 11.6 * |
| Systolic blood pressure (mmHg) | 96.7 ± 1.5 | 158.3 ± 21.2 * |
| Diastolic blood pressure (mmHg) | 71.7 ± 3.8 | 116 ± 18.5 * |
| Mean arterial pressure (mmHg) | 80.5 ± 2.8 | 131.5 ± 19.2 * |
| Serum creatinine (mg/dL) | 1.1 ± 0.0 | 1.9 ± 0.1 * |
| Cortical volume (mL) | 104.3 ± 4.6 | 59.2 ± 25.8 * |
| Medullary volume (mL) | 20.1 ± 1.0 | 20.6 ± 1.6 |
| Cortical perfusion (mL/min/mL tissue) | 5.6 ± 0.2 | 2.4 ± 0.5 * |
| Medullary perfusion (mL/min/mL tissue) | 2.7 ± 0.3 | 2.5 ± 0.3 |
| RBF (mL/min) | 585.2 ± 65.8 | 343.9 ± 69.1 * |
| GFR (mL/min) | 93.6 ± 6.4 | 56.1 ± 9.1 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajagopalan, K.S.; Glasstetter, L.M.; Zhu, X.-Y.; Thaler, R.; Tang, H.; Jordan, K.L.; Saadiq, I.M.; Herrmann, S.M.; Chade, A.R.; Irazabal, M.V.; et al. Renal Ischemia Induces Epigenetic Changes in Apoptotic, Proteolytic, and Mitochondrial Genes in Swine Scattered Tubular-like Cells. Cells 2022, 11, 1803. https://doi.org/10.3390/cells11111803
Rajagopalan KS, Glasstetter LM, Zhu X-Y, Thaler R, Tang H, Jordan KL, Saadiq IM, Herrmann SM, Chade AR, Irazabal MV, et al. Renal Ischemia Induces Epigenetic Changes in Apoptotic, Proteolytic, and Mitochondrial Genes in Swine Scattered Tubular-like Cells. Cells. 2022; 11(11):1803. https://doi.org/10.3390/cells11111803
Chicago/Turabian StyleRajagopalan, Kamalnath S., Logan M. Glasstetter, Xiang-Yang Zhu, Roman Thaler, Hui Tang, Kyra L. Jordan, Ishran M. Saadiq, Sandra M. Herrmann, Alejandro R. Chade, Maria V. Irazabal, and et al. 2022. "Renal Ischemia Induces Epigenetic Changes in Apoptotic, Proteolytic, and Mitochondrial Genes in Swine Scattered Tubular-like Cells" Cells 11, no. 11: 1803. https://doi.org/10.3390/cells11111803
APA StyleRajagopalan, K. S., Glasstetter, L. M., Zhu, X.-Y., Thaler, R., Tang, H., Jordan, K. L., Saadiq, I. M., Herrmann, S. M., Chade, A. R., Irazabal, M. V., Lerman, L. O., & Eirin, A. (2022). Renal Ischemia Induces Epigenetic Changes in Apoptotic, Proteolytic, and Mitochondrial Genes in Swine Scattered Tubular-like Cells. Cells, 11(11), 1803. https://doi.org/10.3390/cells11111803