O-GlcNAcylation: The Underestimated Emerging Regulators of Skeletal Muscle Physiology
Abstract
1. Introduction
- 1. Phosphorylation
- 2. Acetylation
- 3. Ubiquitylation
- 4. Methylation
2. Dynamic O-GlcNAcylation Cycle and Hexosamine Biosynthesis Pathway
2.1. OGT and OGA Are the Only Antagonistic Enzymes for Precisely Regulating the O-GlcNAcylation Cycle in Space-Time Specificity
2.2. Nutrient Availability Drives Global O-GlcNAcylation through HBP
3. O-GlcNAcylation, Energy Metabolism and Insulin Sensitivity in Skeletal Muscle
3.1. O-GlcNAcylation Is the Key Regulator of Glucose Metabolism in Skeletal Muscle
3.2. O-GlcNAcylation-Mediated Insulin Sensitivity in Skeletal Muscle
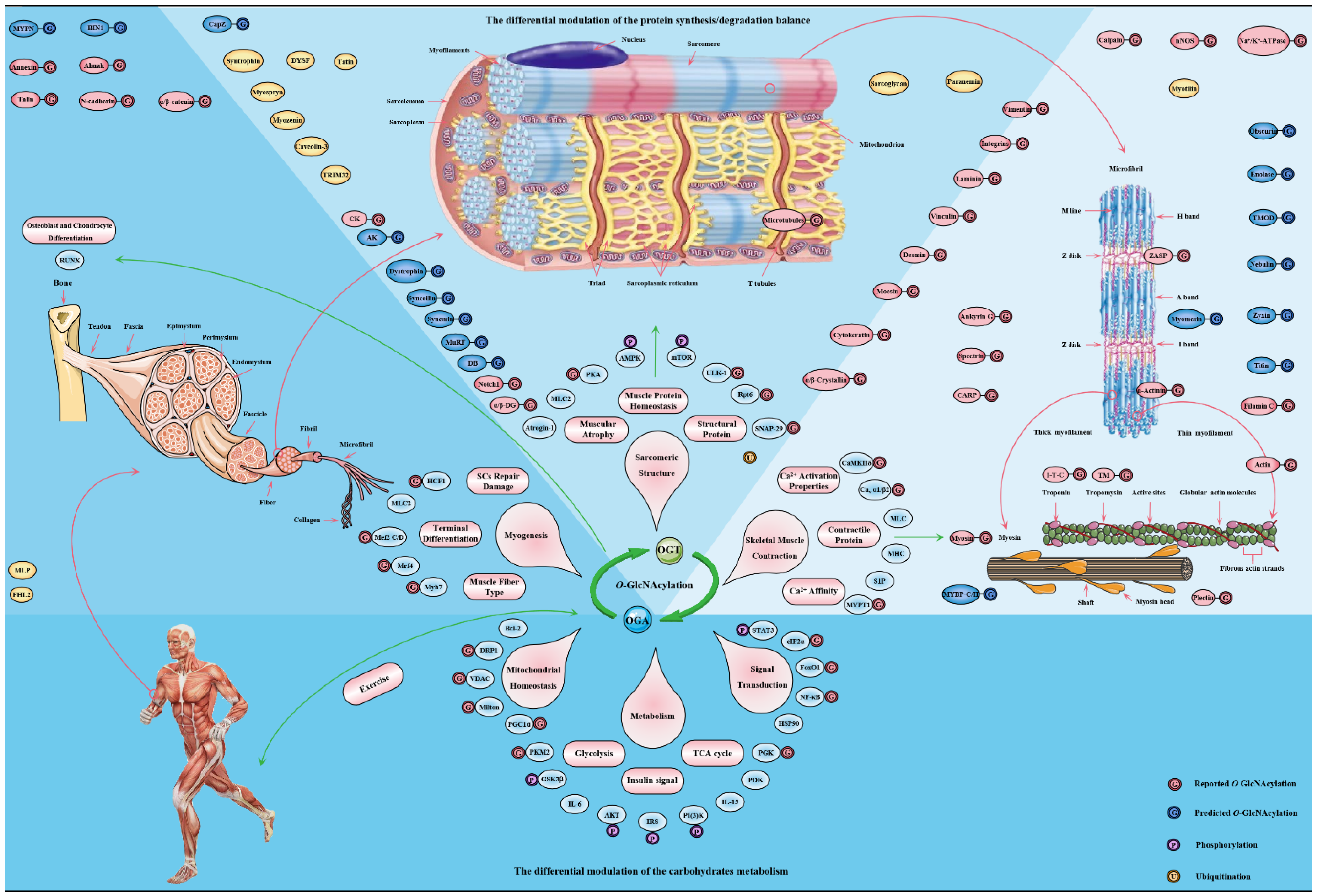
4. O-GlcNAcylation Is an Emerging Mediator of Contractile and Structural Properties in Skeletal Muscle
4.1. O-GlcNAcylation Is an Essential Regulator of Contractile Properties in Skeletal Muscle
4.2. O-GlcNAcylation Is an Emerging Maintainer of the Structural Properties in Skeletal Muscle
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 1,3BPG | 1,3-bisphosphoglycerate |
| 2PG | 2-phosphoglycerate |
| 3PG | 3-phosphoglycerate |
| 5mC | 5-methylcytosine |
| 5S-GlcNAc | 5-thio-N-acetylglucosamine |
| 5S-GlcNHex | 2-deoxy-2-N-hexanamide-5-thio-d-glucopyranoside |
| 6PGD | 6-phosphogluconate dehydrogenase |
| 6-PGL | 6-phosphogluconolactone |
| AGM | phosphoacetylglucosamine mutase |
| AH | aconitate hydratase |
| AK | adenylate kinase |
| AMPK | AMP-activated protein kinase |
| Ara | L-arabinose |
| BAG | benzyl-2-acetamido-2-deoxy-D-galactopyranoside or GalNAc-α-O-benzyl for short |
| BAG3 | BCL2-related athanogene 3 |
| BIN1 | bridging integrator 1 |
| BZX | 4-methoxyphenyl 6-acetyl-2-oxo-2,3-dihydro-1,3-benzoxazole-3-carboxylate; |
| CaMKIIδ | calcium-calmodulin-dependent protein kinase 2 delta |
| CaMKIV | calcium-dependent protein kinase type IV |
| CARP | cardiac ankyrin repeat protein |
| Cav α1/β2 | L-type Ca2+ channel subunit alpha 1 and beta 2 |
| CBP | CREB-binding protein |
| CD I | catalytic domain I |
| CD II | catalytic domain II |
| CK | creatine kinase |
| COLGALT | collagen O-Gal transferase |
| CREB | cyclic AMP-response element-binding protein |
| CS | citrate synthase |
| DB | dystrobrevin |
| Dia | DiActrideoxyhexose |
| DOGT | GlcNAc transferase |
| Dol-P-Man | dolichol phosphate mannose |
| Dol-PP-Oligo | dolichylpyrophosphate Glc3Man9GlcNAc2 |
| DON | 6-Diazo-5-oxo-L-norleucine |
| DYSF | dysferlin |
| E4P | erythrose-4-phosphate |
| EGF | epidermal growth factor-like repeat |
| ENO | beta-enolase |
| ER | endoplasmic reticulum |
| EthNP | ethanolamine phosphate |
| EZH2 | enhancer of zeste homolog 2 |
| FBA | fructose-1,6-bisphosphate aldolase |
| FBP | fructose-1,6-bisphosphate |
| FH | fumarate |
| FHL2 | four and a half LIM domains protein 2 |
| Fru-6-P | fructose-6-phosphate |
| Fuc | L-fucose |
| FucNAc | N-Acetyl-fucosamine |
| G6PD | glucose-6-phosphate dehydrogenase |
| Gal | D-galactose |
| GALE | UDP-galactose-4-epimerase |
| GalNAc | N-acetyl-D-galactosamine |
| GALNT | polypeptide GalNAc transferase |
| GALP | glyceraldehyde-3-phosphate; |
| GAPDH | glyceraldehyde phosphate dehydrogenase |
| GBE | Glycogen-branching enzyme |
| GDP-Fuc | GDP-fucose |
| GDP-Man | GDP-Mannose |
| GFAT | glutamine Fru-6-P aminotransferase |
| GH84 | glycoside hydrolase family 84 |
| Glc | D-glucose; |
| Glc-6-P | glucose-6-phosphate |
| GlcN-6-P | glucosamine-6-phosphate |
| GlcNAc | N-acetylglucosamine |
| GlcNAc | N-acetyl-D-glucosamine; |
| GlcNAc-1-P | N-acetylglucosamine-1-phosphate |
| GlcNAc-6-P | N-acetylglucosamine-6-phosphate |
| GN | glycogenin |
| GNA | glucosamine-6-phosphate N-acetyltransferase 1 |
| GNE | UDP-GlcNAc 2-epimerase/ManNAc kinase |
| GPI | Glc-6-P isomerase |
| GPI | glycosylphosphatidylinositol |
| GS | glycogen synthase |
| GYG | glycogenin |
| HAT | histone acetyltransferase |
| HBP | hexosamine biosynthesis pathway |
| HDAC4 | histone deacetylase 4 |
| HK | hexokinase |
| Hyl | hydroxylysine |
| IDH | isocitrate dehydrogenase |
| Int-D | intervention domain |
| I-T-C | troponin complex (Tn I, Tn T, Tn C) |
| KGD | ketoglutarate dehydrogenase |
| LA | lactate |
| LDH | lactate dehydrogenase |
| Man | Mannose |
| ManNAc | N-acetylmannosamine |
| MDH | malate dehydrogenase |
| Mef2 C/D | myocyte-specific enhancer factor 2 C/D |
| MHC | myosin heavy chain |
| MLC2 | myosin light chain 2 |
| MLP | muscle LIM protein |
| mOGT | mitochondrial OGT |
| MyBP-C | Myosin-binding protein C |
| MYPN | myopalladin |
| MYPT1 | myosin phosphatase target subunit 1 |
| NButGT | 1,2-dideoxy-2’-propyl-alpha-D-glucopyranoso-[2,1-D]-Delta 2’-thiazoline |
| ncOGA | nucleocytoplasmic OGA |
| ncOGT | nucleocytoplasmic OGT |
| OGA | O-GlcNAcase |
| OGT | O-GlcNAc transferase |
| OST | oligosaccharyltransferase |
| PA | pyruvate |
| PDC | pyruvate dehydrogenase complex |
| PEP | phosphoenolpyruvate |
| PFK | phosphofructokinse |
| PGK | phosphoglycerate kinase |
| PGM | phosphoglucomutase |
| PIP3 | phosphatidylinositol (3,4,5)—triphosphate |
| PK | pyruvate kinase |
| PKA | cAMP-dependent protein kinase |
| POFUT | protein O-fucosyltransferase |
| POGLUT | protein O-Glc transferase |
| POMT | protein O-Man transferase |
| PP1 | protein phosphatase 1 |
| PP2A | protein phosphatase 2α |
| PPi | inorganic phosphate |
| PPO | Phosphoinositide-binding domain |
| Pse | pseudaminic |
| PTase | phosphoglycosyltransferase |
| PTM | post-translational modification |
| PUGNAc | O-(2-acetamido-2-deoxy-D-glucopyranosylidene) amino N-phenyl carbamate |
| PY19L | dpy-19 like C-Man transferase |
| R5P | ribose-5-phosphate |
| RER | rough endoplasmic reticulum |
| Rha | L-rhamnose |
| RPE | Ru5P epimerase |
| RPI | Ru5P isomerase |
| Ru5P | ribulose-5-phosphate |
| S1P | sphingosine-1-phosphate |
| S7P | sedoheptulose-7-phosphate |
| SC | satellite cell |
| SDH | succinate dehydrogenase |
| SIN3A | SIN3transcription regulator family member A |
| SIRT1 | sirtuin 1 |
| SL | succinyl-CoA ligase |
| sOGA | short OGA |
| sOGT | short OGT |
| STZ | streptozotocin |
| TALDO | transaldolase |
| TET | ten-eleven translocation protein |
| Thiamet G | O-(2-acetamido-2-deoxy-D-glucopyranoseylidene) |
| TKT | transketolase |
| TM | tropomyosin |
| TMOD | tropomodulin |
| TPR | tetratripeptide repeat domain |
| TRIM32 | tripartite motif protein |
| TT04 | 3-[2-(1-adamantyl)ethyl]-2-(4-chlorophenyl)imino-4-oxo-1,3-thiazinane-6-carboxylic acid |
| UAP | UDP-N-acetylglucosamine pyrophosphorylase |
| UDP-5S-GlcNAc | uridine diphospho-5-thio-N-acetylglucosamine or Ac4-5SGlcNAc |
| UDPG | UDP-Glucose |
| UDP-Gal | UDP-galactose |
| UDP-GalNAc | UDP-N-acetylgalactosamine |
| UDP-GlcNAc | UDP-N-acetylglucosamine |
| UDP-Xyl | UDP-xylose |
| UP | UDP-Glucose pyrophosphosphprylase |
| UTP | uridine triphosphate |
| Xu5P | xylulose-5-phosphate |
| Xyl | D-xylose |
| XYLT | protein O-Xyl transferase |
| ZASP | Z-band alternatively spliced PDZ-motif protein |
| α/β-DG | α/β-dystroglycan |
| α-GlcNAc Thiolsulfonate | 2-acetamido-2-deoxy-1-S-(4-methylbenzenesulfonyl)-1-thio-α-D-glucopyranose |
References
- Conibear, A.C. Deciphering protein post-translational modifications using chemical biology tools. Nat. Rev. Chem. 2020, 4, 674–695. [Google Scholar] [CrossRef]
- Liu, J.; Qian, C.; Cao, X. Post-Translational Modification Control of Innate Immunity. Immunity 2016, 45, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Yang, X.; Qian, A.K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef]
- Chatham, J.C.; Zhang, J.; Wende, A.R. Role of O-Linked N-Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol. Rev. 2021, 101, 427–493. [Google Scholar] [CrossRef]
- Hu, C.W.; Worth, M.; Li, H.; Jiang, J. Chemical and Biochemical Strategies To Explore the Substrate Recognition of O-GlcNAc-Cycling Enzymes. Chembiochem 2019, 20, 312–318. [Google Scholar] [CrossRef]
- Sharma, N.S.; Saluja, A.K.; Banerjee, S. “Nutrient-sensing” and self-renewal: O-GlcNAc in a new role. J. Bioenerg. Biomembr. 2018, 50, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shang, J.; Yao, Y.; Yin, X.; Liu, M.; Liu, H.; Zhou, Y. O-GlcNAcylation: A bridge between glucose and cell differentiation. J. Cell Mol. Med. 2016, 20, 769–781. [Google Scholar] [CrossRef]
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43r–56r. [Google Scholar] [CrossRef] [PubMed]
- Itkonen, H.M.; Loda, M.; Mills, I.G. O-GlcNAc Transferase—An Auxiliary Factor or a Full-blown Oncogene? Mol Cancer Res. 2021, 19, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Wulff-Fuentes, E.; Berendt, R.R.; Massman, L.; Danner, L.; Malard, F.; Vora, J.; Kahsay, R.; Olivier-Van Stichelen, S. The human O-GlcNAcome database and meta-analysis. Sci. Data 2021, 8, 25. [Google Scholar] [CrossRef]
- Morino, K.; Maegawa, H. Role of O-linked N-acetylglucosamine in the homeostasis of metabolic organs, and its potential links with diabetes and its complications. J. Diabetes Investig. 2021, 12, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Eustice, M.; Bond, M.R.; Hanover, J.A. O-GlcNAc cycling and the regulation of nucleocytoplasmic dynamics. Biochem. Soc. Trans. 2017, 45, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Sakaidani, Y.; Nomura, T.; Matsuura, A.; Ito, M.; Suzuki, E.; Murakami, K.; Nadano, D.; Matsuda, T.; Furukawa, K.; Okajima, T. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2011, 2, 583. [Google Scholar] [CrossRef]
- Li, Y.; Xie, M.; Men, L.; Du, J. O-GlcNAcylation in immunity and inflammation: An intricate system (Review). Int J. Mol. Med. 2019, 44, 363–374. [Google Scholar] [CrossRef]
- Malard, F.; Wulff-Fuentes, E.; Berendt, R.R.; Didier, G.; Olivier-Van Stichelen, S. Automatization and self-maintenance of the O-GlcNAcome catalog: A smart scientific database. Database 2021, 2021, baab039. [Google Scholar] [CrossRef]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef]
- Yuan, A.; Tang, X.; Li, J. Centrosomes: Til O-GlcNAc Do Us Apart. Front. Endocrinol. 2020, 11, 621888. [Google Scholar] [CrossRef]
- Cui, Z.; Scruggs, S.B.; Gilda, J.E.; Ping, P.; Gomes, A.V. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. J. Mol. Cell. Cardiol. 2014, 71, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Ong, Q.; Han, W.; Yang, X. O-GlcNAc as an Integrator of Signaling Pathways. Front. Endocrinol. 2018, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hart, G.W. Targeting O-GlcNAcylation to develop novel therapeutics. Mol. Aspects Med. 2021, 79, 100885. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yao, R.Z.; Lian, S.; Liu, P.; Hu, Y.J.; Shi, H.Z.; Lv, H.M.; Yang, Y.Y.; Xu, B.; Li, S.Z. O-GlcNAcylation: The “stress and nutrition receptor” in cell stress response. Cell Stress Chaperones 2021, 26, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Chi, J.T.; Boyce, M. Functional crosstalk among oxidative stress and O-GlcNAc signaling pathways. Glycobiology 2018, 28, 556–564. [Google Scholar] [CrossRef]
- Han, C.; Gu, Y.; Shan, H.; Mi, W.; Sun, J.; Shi, M.; Zhang, X.; Lu, X.; Han, F.; Gong, Q.; et al. O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat. Commun. 2017, 8, 1491. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, B.; Hu, Y.; Liu, P.; Lian, S.; Lv, H.; Yang, Y.; Ji, H.; Yang, H.; Liu, J.; et al. O-GlcNAc / Akt pathway regulates glucose metabolism and reduces apoptosis in liver of piglets with acute cold stress. Cryobiology 2021, 100, 125–132. [Google Scholar] [CrossRef]
- Ma, Z.; Vosseller, K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem 2014, 289, 34457–34465. [Google Scholar] [CrossRef]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef]
- Qian, K.; Wang, S.; Fu, M.; Zhou, J.; Singh, J.P.; Li, M.D.; Yang, Y.; Zhang, K.; Wu, J.; Nie, Y.; et al. Transcriptional regulation of O-GlcNAc homeostasis is disrupted in pancreatic cancer. J. Biol. Chem. 2018, 293, 13989–14000. [Google Scholar] [CrossRef]
- Schwein, P.A.; Woo, C.M. The O-GlcNAc Modification on Kinases. ACS Chem. Biol. 2020, 15, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, Q.; Shi, X.; Cheng, Y.; Li, X.; Xu, H.; Duan, X.; Hsieh-Wilson, L.C.; Chu, J.; Pelletier, J.; et al. O-GlcNAcylation of core components of the translation initiation machinery regulates protein synthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 7857–7866. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Liang, Q.; Li, L.; Hu, Z.; Wu, F.; Zhang, P.; Ma, Y.; Zhao, B.; Kovács, A.L.; Zhang, Z.; et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 2014, 16, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, Y.; Pak, J.W.; Kim, H.; Choi, H.; Kim, J.W.; Roth, J.; Cho, J.W. O-GlcNAc modification is essential for the regulation of autophagy in Drosophila melanogaster. Cell. Mol. Life Sci. 2015, 72, 3173–3183. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gong, W.; Wang, H.; Li, T.; Attri, K.S.; Lewis, R.E.; Kalil, A.C.; Bhinderwala, F.; Powers, R.; Yin, G.; et al. O-GlcNAc Transferase Suppresses Inflammation and Necroptosis by Targeting Receptor-Interacting Serine/Threonine-Protein Kinase 3. Immunity 2019, 50, 576–590.e6. [Google Scholar] [CrossRef]
- Chang, Y.H.; Weng, C.L.; Lin, K.I. O-GlcNAcylation and its role in the immune system. J. Biomed. Sci. 2020, 27, 57. [Google Scholar] [CrossRef]
- Machacek, M.; Slawson, C.; Fields, P.E. O-GlcNAc: A novel regulator of immunometabolism. J. Bioenerg. Biomembr. 2018, 50, 223–229. [Google Scholar] [CrossRef]
- Dong, H.; Liu, Z.; Wen, H. Protein O-GlcNAcylation Regulates Innate Immune Cell Function. Front. Immunol. 2022, 13, 805018. [Google Scholar] [CrossRef]
- Baudoin, L.; Issad, T. O-GlcNAcylation and Inflammation: A Vast Territory to Explore. Front. Endocrinol. 2014, 5, 235. [Google Scholar] [CrossRef]
- Leturcq, M.; Lefebvre, T.; Vercoutter-Edouart, A.S. O-GlcNAcylation and chromatin remodeling in mammals: An up-to-date overview. Biochem. Soc. Trans. 2017, 45, 323–338. [Google Scholar] [CrossRef]
- Ruan, H.B.; Singh, J.P.; Li, M.D.; Wu, J.; Yang, X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 2013, 24, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Yi, W. O-GlcNAcylation, a sweet link to the pathology of diseases. J. Zhejiang Univ. Sci. B 2019, 20, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.P.; Zhang, K.; Wu, J.; Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015, 356, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.P.; Peterson, K.R.; Slawson, C. O-GlcNAcylation and O-GlcNAc Cycling Regulate Gene Transcription: Emerging Roles in Cancer. Cancers 2021, 13, 1666. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Duan, X.; Mao, W.; Li, X.; Li, Z.; Li, Q.; Zheng, Z.; Xu, H.; Chen, M.; Wang, P.G.; et al. O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat. Commun. 2015, 6, 8468. [Google Scholar] [CrossRef]
- Vaidyanathan, K.; Niranjan, T.; Selvan, N.; Teo, C.F.; May, M.; Patel, S.; Weatherly, B.; Skinner, C.; Opitz, J.; Carey, J. Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 2017, 292, 8948–8963. [Google Scholar] [CrossRef]
- Akan, I.; Olivier-Van Stichelen, S.; Bond, M.R.; Hanover, J.A. Nutrient-driven O-GlcNAc in proteostasis and neurodegeneration. J. Neurochem. 2018, 144, 7–34. [Google Scholar] [CrossRef]
- Hwang, H.; Rhim, H. Functional significance of O-GlcNAc modification in regulating neuronal properties. Pharmacol. Res. 2018, 129, 295–307. [Google Scholar] [CrossRef]
- Yang, Y.; Fu, M.; Li, M.D.; Zhang, K.; Zhang, B.; Wang, S.; Liu, Y.; Ni, W.; Ong, Q.; Mi, J.; et al. O-GlcNAc transferase inhibits visceral fat lipolysis and promotes diet-induced obesity. Nat. Commun. 2020, 11, 181. [Google Scholar] [CrossRef]
- Chatham, J.C.; Young, M.E.; Zhang, J. Reprint of: Role of O-linked N-acetylglucosamine (O-GlcNAc) modification of proteins in diabetic cardiovascular complications. Curr. Opin. Pharmacol. 2020, 54, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Issad, T.; Masson, E.; Pagesy, P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 2010, 36 6 Pt 1, 423–435. [Google Scholar] [CrossRef]
- Banerjee, P.S.; Lagerlöf, O.; Hart, G.W. Roles of O-GlcNAc in chronic diseases of aging. Mol. Aspects Med. 2016, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.B.; Hart, G.W. New insights: A role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Gurel, Z.; Sheibani, N. O-Linked β-N-acetylglucosamine (O-GlcNAc) modification: A new pathway to decode pathogenesis of diabetic retinopathy. Clin. Sci. 2018, 132, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Stevens, L.; Bastide, B.; Hedou, J.; Cieniewski-Bernard, C.; Montel, V.; Cochon, L.; Dupont, E.; Mounier, Y. Potential regulation of human muscle plasticity by MLC2 post-translational modifications during bed rest and countermeasures. Arch. Biochem. Biophys. 2013, 540, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Nagy, T.; Kátai, E.; Fisi, V.; Takács, T.T.; Stréda, A.; Wittmann, I.; Miseta, A. Protein O-GlcNAc Modification Increases in White Blood Cells After a Single Bout of Physical Exercise. Front. Immunol. 2018, 9, 970. [Google Scholar] [CrossRef]
- Peternelj, T.T.; Marsh, S.A.; Strobel, N.A.; Matsumoto, A.; Briskey, D.; Dalbo, V.J.; Tucker, P.S.; Coombes, J.S. Glutathione depletion and acute exercise increase O-GlcNAc protein modification in rat skeletal muscle. Mol. Cell. Biochem. 2015, 400, 265–275. [Google Scholar] [CrossRef]
- Murata, K.; Morino, K.; Ida, S.; Ohashi, N.; Lemecha, M.; Park, S.Y.; Ishikado, A.; Kume, S.; Choi, C.S.; Sekine, O.; et al. Lack of O-GlcNAcylation enhances exercise-dependent glucose utilization potentially through AMP-activated protein kinase activation in skeletal muscle. Biochem. Biophys. Res. Commun. 2018, 495, 2098–2104. [Google Scholar] [CrossRef]
- Lambert, M.; Bastide, B.; Cieniewski-Bernard, C. Involvement of O-GlcNAcylation in the Skeletal Muscle Physiology and Physiopathology: Focus on Muscle Metabolism. Front. Endocrinol. 2018, 9, 578. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Z.; Wang, X.; Yang, L.; Han, S.; Cao, K.; Xu, J.; Zhao, L.; Zhang, Y.; Liu, J. O-GlcNAcase deficiency suppresses skeletal myogenesis and insulin sensitivity in mice through the modulation of mitochondrial homeostasis. Diabetologia 2016, 59, 1287–1296. [Google Scholar] [CrossRef]
- Pedowitz, N.J.; Batt, A.R.; Darabedian, N.; Pratt, M.R. MYPT1 O-GlcNAc modification regulates sphingosine-1-phosphate mediated contraction. Nat. Chem. Biol. 2021, 17, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Munk, A.; Nielsen, T.S.; Daughtry, M.R.; Larsson, L.; Li, S.; Høyer, K.F.; Geisler, H.W.; Sulek, K.; Kjøbsted, R.; et al. Skeletal muscle O-GlcNAc transferase is important for muscle energy homeostasis and whole-body insulin sensitivity. Mol. Metab. 2018, 11, 160–177. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, D.M.; Al-Sajee, D.; Hawke, T.J. Diabetic myopathy: Impact of diabetes mellitus on skeletal muscle progenitor cells. Front. Physiol. 2013, 4, 379. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Nakano, S.; Nishii, M.; Kaneko, S.; Kusaka, H. Localization of O-GlcNAc-modified proteins in neuromuscular diseases. Med. Mol. Morphol. 2012, 45, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Cieniewski-Bernard, C.; Lambert, M.; Dupont, E.; Montel, V.; Stevens, L.; Bastide, B. O-GlcNAcylation, contractile protein modifications and calcium affinity in skeletal muscle. Front. Physiol. 2014, 5, 421. [Google Scholar] [CrossRef]
- Akimoto, Y.; Yan, K.; Miura, Y.; Tsumoto, H.; Toda, T.; Fukutomi, T.; Sugahara, D.; Kudo, A.; Arai, T.; Chiba, Y.; et al. O-GlcNAcylation and phosphorylation of β-actin Ser(199) in diabetic nephropathy. Am. J. Physiol. Renal. Physiol. 2019, 317, F1359–F1374. [Google Scholar] [CrossRef]
- Lambert, M.; Claeyssen, C.; Bastide, B.; Cieniewski-Bernard, C. O-GlcNAcylation as a regulator of the functional and structural properties of the sarcomere in skeletal muscle: An update review. Acta Physiol. 2020, 228, e13301. [Google Scholar] [CrossRef]
- Ng, Y.H.; Okolo, C.A.; Erickson, J.R.; Baldi, J.C.; Jones, P.P. Protein O-GlcNAcylation in the heart. Acta Physiol. 2021, 233, e13696. [Google Scholar] [CrossRef]
- Ngoh, G.A.; Facundo, H.T.; Zafir, A.; Jones, S.P. O-GlcNAc signaling in the cardiovascular system. Circ. Res. 2010, 107, 171–185. [Google Scholar] [CrossRef]
- Buse, M.G. Hexosamines, insulin resistance, and the complications of diabetes: Current status. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E1–E8. [Google Scholar] [CrossRef]
- Bolanle, I.O.; Riches-Suman, K.; Loubani, M.; Williamson, R.; Palmer, T.M. Revascularisation of type 2 diabetics with coronary artery disease: Insights and therapeutic targeting of O-GlcNAcylation. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Joiner, C.M.; Li, H.; Jiang, J.; Walker, S. Structural characterization of the O-GlcNAc cycling enzymes: Insights into substrate recognition and catalytic mechanisms. Curr. Opin. Struct. Biol. 2019, 56, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Meoz, R.F.; Jiang, J.; Lazarus, M.B.; Orman, M.; Janetzko, J.; Fan, C.; Duveau, D.Y.; Tan, Z.W.; Thomas, C.J.; Walker, S. A small molecule that inhibits OGT activity in cells. ACS Chem. Biol. 2015, 10, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Alteen, M.G.; Tan, H.Y.; Vocadlo, D.J. Monitoring and modulating O-GlcNAcylation: Assays and inhibitors of O-GlcNAc processing enzymes. Curr. Opin. Struct. Biol. 2021, 68, 157–165. [Google Scholar] [CrossRef]
- Mariappa, D.; Zheng, X.; Schimpl, M.; Raimi, O.; Ferenbach, A.T.; Müller, H.A.; van Aalten, D.M. Dual functionality of O-GlcNAc transferase is required for Drosophila development. Open Biol. 2015, 5, 150234. [Google Scholar] [CrossRef]
- Trapannone, R.; Rafie, K.; van Aalten, D.M. O-GlcNAc transferase inhibitors: Current tools and future challenges. Biochem. Soc. Trans. 2016, 44, 88–93. [Google Scholar] [CrossRef]
- Fehl, C.; Hanover, J.A. Tools, tactics and objectives to interrogate cellular roles of O-GlcNAc in disease. Nat. Chem. Biol. 2022, 18, 8–17. [Google Scholar] [CrossRef]
- Kim, E.J.; Abramowitz, L.K.; Bond, M.R.; Love, D.C.; Kang, D.W.; Leucke, H.F.; Kang, D.W.; Ahn, J.S.; Hanover, J.A. Versatile O-GlcNAc transferase assay for high-throughput identification of enzyme variants, substrates, and inhibitors. Bioconjug. Chem. 2014, 25, 1025–1030. [Google Scholar] [CrossRef]
- Shafi, R.; Iyer, S.P.; Ellies, L.G.; O’Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739. [Google Scholar] [CrossRef]
- Olivier-Van Stichelen, S.; Hanover, J.A. X-inactivation normalizes O-GlcNAc transferase levels and generates an O-GlcNAc-depleted Barr body. Front Genet. 2014, 5, 256. [Google Scholar] [CrossRef][Green Version]
- Nolte, D.; Müller, U. Human O-GlcNAc transferase (OGT): Genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm. Genome 2002, 13, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, M.B.; Nam, Y.; Jiang, J.; Sliz, P.; Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469, 564–567. [Google Scholar] [CrossRef]
- Iyer, S.P.; Hart, G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 2003, 278, 24608–24616. [Google Scholar] [CrossRef]
- Ramirez, D.H.; Yang, B.; D’Souza, A.K.; Shen, D.; Woo, C.M. Truncation of the TPR domain of OGT alters substrate and glycosite selection. Anal. Bioanal. Chem. 2021, 413, 7385–7399. [Google Scholar] [CrossRef] [PubMed]
- Levine, Z.G.; Walker, S. The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells? Annu. Rev. Biochem. 2016, 85, 631–657. [Google Scholar] [CrossRef] [PubMed]
- Joiner, C.M.; Levine, Z.G.; Aonbangkhen, C.; Woo, C.M.; Walker, S. Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. J. Am. Chem. Soc. 2019, 141, 12974–12978. [Google Scholar] [CrossRef] [PubMed]
- Gloster, T.M.; Vocadlo, D.J. Mechanism, Structure, and Inhibition of O-GlcNAc Processing Enzymes. Curr. Signal Transduct. Ther. 2010, 5, 74–91. [Google Scholar] [CrossRef]
- Clarke, A.J.; Hurtado-Guerrero, R.; Pathak, S.; Schüttelkopf, A.W.; Borodkin, V.; Shepherd, S.M.; Ibrahim, A.F.; van Aalten, D.M. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008, 27, 2780–2788. [Google Scholar] [CrossRef]
- Rafie, K.; Raimi, O.; Ferenbach, A.T.; Borodkin, V.S.; Kapuria, V.; van Aalten, D.M.F. Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Pathak, S.; Alonso, J.; Schimpl, M.; Rafie, K.; Blair, D.E.; Borodkin, V.S.; Albarbarawi, O.; van Aalten, D.M.F. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 2015, 22, 744–750. [Google Scholar] [CrossRef]
- Shi, J.; Ruijtenbeek, R.; Pieters, R.J. Demystifying O-GlcNAcylation: Hints from peptide substrates. Glycobiology 2018, 28, 814–824. [Google Scholar] [CrossRef] [PubMed]
- She, N.; Zhao, Y.; Hao, J.; Xie, S.; Wang, C. Uridine diphosphate release mechanism in O-N-acetylglucosamine (O-GlcNAc) transferase catalysis. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, B.D.; Roos, M.D.; Hanover, J.A. Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase. J. Biol. Chem. 2005, 280, 35537–35544. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Moen, A.; Anonsen, J.H.; Bindesbøll, C.; Sæther, T.; Carlson, C.R.; Grønning-Wang, L.M. O-GlcNAc site-mapping of liver X receptor-α and O-GlcNAc transferase. Biochem. Biophys. Res. Commun. 2018, 499, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar] [CrossRef]
- Moore, M.; Avula, N.; Jo, S.; Beetch, M.; Alejandro, E.U. Disruption of O-Linked N-Acetylglucosamine Signaling in Placenta Induces Insulin Sensitivity in Female Offspring. Int. J. Mol. Sci. 2021, 22, 6918. [Google Scholar] [CrossRef]
- Gorelik, A.; van Aalten, D.M.F. Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chem. Biol. 2020, 1, 98–109. [Google Scholar] [CrossRef]
- Hurtado-Guerrero, R.; Dorfmueller, H.C.; van Aalten, D.M. Molecular mechanisms of O-GlcNAcylation. Curr. Opin. Struct. Biol. 2008, 18, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Hardivillé, S.; Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014, 20, 208–213. [Google Scholar] [CrossRef]
- Kaasik, K.; Kivimäe, S.; Allen, J.J.; Chalkley, R.J.; Huang, Y.; Baer, K.; Kissel, H.; Burlingame, A.L.; Shokat, K.M.; Ptáček, L.J.; et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013, 17, 291–302. [Google Scholar] [CrossRef]
- Bullen, J.W.; Balsbaugh, J.L.; Chanda, D.; Shabanowitz, J.; Hunt, D.F.; Neumann, D.; Hart, G.W. Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK). J. Biol. Chem. 2014, 289, 10592–10606. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, L.; Ma, C.; Shi, Y.; Liu, C.; Xiao, Z.; Zhang, Y.; Tian, F.; Gao, Y.; Zhang, J.; et al. O-GlcNAcylation of Thr(12)/Ser(56) in short-form O-GlcNAc transferase (sOGT) regulates its substrate selectivity. J. Biol. Chem. 2019, 294, 16620–16633. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Nai, S.; Geng, Q.; Liao, J.; Xu, X.; Li, J. Checkpoint kinase 1-induced phosphorylation of O-linked β-N-acetylglucosamine transferase regulates the intermediate filament network during cytokinesis. J. Biol. Chem. 2017, 292, 19548–19555. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.B.; Ma, Y.; Torres, S.; Zhang, B.; Feriod, C.; Heck, R.M.; Qian, K.; Fu, M.; Li, X.; Nathanson, M.H.; et al. Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 2017, 31, 1655–1665. [Google Scholar] [CrossRef]
- Seo, H.G.; Kim, H.B.; Kang, M.J.; Ryum, J.H.; Yi, E.C.; Cho, J.W. Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci. Rep. 2016, 6, 34614. [Google Scholar] [CrossRef]
- Alonso, J.; Schimpl, M.; van Aalten, D.M. O-GlcNAcase: Promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J. Biol. Chem. 2014, 289, 34433–34439. [Google Scholar] [CrossRef]
- Kim, E.J. Chemical arsenal for the study of O-GlcNAc. Molecules 2011, 16, 1987–2022. [Google Scholar] [CrossRef]
- Elbatrawy, A.A.; Kim, E.J.; Nam, G. Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase. Nat. Commun. 2017, 8, 666. [Google Scholar]
- Elbatrawy, A.A.; Kim, E.J.; Nam, G. O-GlcNAcase: Emerging Mechanism, Substrate Recognition and Small-Molecule Inhibitors. ChemMedChem 2020, 15, 1244–1257. [Google Scholar] [CrossRef]
- Elsen, N.L.; Patel, S.B.; Ford, R.E.; Hall, D.L.; Hess, F.; Kandula, H.; Kornienko, M.; Reid, J.; Selnick, H.; Shipman, J.M.; et al. Insights into activity and inhibition from the crystal structure of human O-GlcNAcase. Nat. Chem. Biol. 2017, 13, 613–615. [Google Scholar] [CrossRef]
- Macauley, M.S.; Vocadlo, D.J. Increasing O-GlcNAc levels: An overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 2010, 1800, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Kohler, J.J. Carb cutting works better with a partner. Nat. Struct. Mol. Biol. 2017, 24, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.V.; Schüttelkopf, A.W.; Dorfmueller, H.C.; Ferenbach, A.T.; Navratilova, I.; van Aalten, D.M. Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol. 2013, 3, 130021. [Google Scholar] [CrossRef]
- He, Y.; Roth, C.; Turkenburg, J.P.; Davies, G.J. Three-dimensional structure of a Streptomyces sviceus GNAT acetyltransferase with similarity to the C-terminal domain of the human GH84 O-GlcNAcase. Acta Crystallogr. D Biol. Crystallogr. 2014, 70 Pt 1, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.C.; Fadda, E.; Ito, K.; Woods, R.J. Defining the structural origin of the substrate sequence independence of O-GlcNAcase using a combination of molecular docking and dynamics simulation. Glycobiology 2014, 24, 85–96. [Google Scholar] [CrossRef]
- de Alencar, N.A.; Sousa, P.R.; Silva, J.R.; Lameira, J.; Alves, C.N.; Martí, S.; Moliner, V. Computational analysis of human OGA structure in complex with PUGNAc and NAG-thiazoline derivatives. J. Chem. Inf. Model 2012, 52, 2775–2783. [Google Scholar] [CrossRef]
- Roth, C.; Chan, S.; Offen, W.A.; Hemsworth, G.R.; Willems, L.I.; King, D.T.; Varghese, V.; Britton, R.; Vocadlo, D.J.; Davies, G.J. Structural and functional insight into human O-GlcNAcase. Nat. Chem. Biol. 2017, 13, 610–612. [Google Scholar] [CrossRef]
- Schimpl, M.; Schüttelkopf, A.W.; Borodkin, V.S.; van Aalten, D.M. Human OGA binds substrates in a conserved peptide recognition groove. Biochem. J. 2010, 432, 1–7. [Google Scholar] [CrossRef]
- Kim, E.J. In Vitro Biochemical Assays for O-GlcNAc-Processing Enzymes. Chembiochem 2017, 18, 1462–1472. [Google Scholar] [CrossRef]
- Wu, D.; Cai, Y.; Jin, J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell 2017, 8, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Balsollier, C.; Pieters, R.J.; Anderluh, M. Overview of the Assays to Probe O-Linked β-N-Acetylglucosamine Transferase Binding and Activity. Molecules 2021, 26, 1037. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Flores-Opazo, M.; McGee, S.L.; Hargreaves, M. Exercise and GLUT4. Exerc. Sport Sci. Rev. 2020, 48, 110–118. [Google Scholar] [CrossRef]
- Richter, E.A.; Hargreaves, M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol. Rev. 2013, 93, 993–1017. [Google Scholar] [CrossRef]
- Hanover, J.A.; Krause, M.W.; Love, D.C. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 2010, 1800, 80–95. [Google Scholar] [CrossRef]
- Adeva-Andany, M.M.; Pérez-Felpete, N.; Fernández-Fernández, C.; Donapetry-García, C.; Pazos-García, C. Liver glucose metabolism in humans. Biosci Rep. 2016, 36, e00416. [Google Scholar] [CrossRef]
- Filhoulaud, G.; Guillemain, G.; Scharfmann, R. The hexosamine biosynthesis pathway is essential for pancreatic beta cell development. J. Biol. Chem. 2009, 284, 24583–24594. [Google Scholar] [CrossRef]
- Love, D.C.; Hanover, J.A. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci. STKE 2005, 2005, re13. [Google Scholar] [CrossRef]
- Zibrova, D.; Vandermoere, F.; Göransson, O.; Peggie, M.; Mariño, K.V.; Knierim, A.; Spengler, K.; Weigert, C.; Viollet, B.; Morrice, N.A.; et al. GFAT1 phosphorylation by AMPK promotes VEGF-induced angiogenesis. Biochem. J. 2017, 474, 983–1001. [Google Scholar] [CrossRef]
- Ruegenberg, S.; Horn, M.; Pichlo, C.; Allmeroth, K.; Baumann, U.; Denzel, M.S. Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis. Nat. Commun. 2020, 11, 687. [Google Scholar] [CrossRef] [PubMed]
- Leto, D.; Saltiel, A.R. Regulation of glucose transport by insulin: Traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 2012, 13, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, S.; Oshiro, N.; Miyamoto, T.; Yoshino, K.; Okamoto, S.; Ono, T.; Kikkawa, U.; Yonezawa, K. AMP-activated protein kinase phosphorylates glutamine: Fructose-6-phosphate amidotransferase 1 at Ser243 to modulate its enzymatic activity. Genes Cells 2009, 14, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Ruegenberg, S.; Mayr, F.; Atanassov, I.; Baumann, U.; Denzel, M.S. Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1. Nat. Commun. 2021, 12, 2176. [Google Scholar] [CrossRef] [PubMed]
- Al-Mukh, H.; Baudoin, L.; Bouaboud, A.; Sanchez-Salgado, J.L.; Maraqa, N.; Khair, M.; Pagesy, P.; Bismuth, G.; Niedergang, F.; Issad, T. Lipopolysaccharide Induces GFAT2 Expression to Promote O-Linked β-N-Acetylglucosaminylation and Attenuate Inflammation in Macrophages. J. Immunol. 2020, 205, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.; Denzel, S.I.; Srinivasan, B.; Allmeroth, K.; Schiffer, I.; Karthikaisamy, V.; Miethe, S.; Breuer, P.; Antebi, A.; Denzel, M.S. Hexosamine Pathway Activation Improves Protein Homeostasis through the Integrated Stress Response. iScience 2020, 23, 100887. [Google Scholar] [CrossRef]
- Denzel, M.S.; Antebi, A. Hexosamine pathway and (ER) protein quality control. Curr. Opin. Cell Biol. 2015, 33, 14–18. [Google Scholar] [CrossRef]
- Zachara, N.E.; Hart, G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 2004, 1673, 13–28. [Google Scholar] [CrossRef]
- Biwi, J.; Biot, C.; Guerardel, Y.; Vercoutter-Edouart, A.S.; Lefebvre, T. The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations. Molecules 2018, 23, 2858. [Google Scholar] [CrossRef]
- Lam, C.; Low, J.Y.; Tran, P.T.; Wang, H. The hexosamine biosynthetic pathway and cancer: Current knowledge and future therapeutic strategies. Cancer Lett. 2021, 503, 11–18. [Google Scholar] [CrossRef]
- Olivier-Van Stichelen, S.; Hanover, J.A. You are what you eat: O-linked N-acetylglucosamine in disease, development and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Coussement, P.; Bauwens, D.; Peters, G.; Maertens, J.; De Mey, M. Mapping and refactoring pathway control through metabolic and protein engineering: The hexosamine biosynthesis pathway. Biotechnol. Adv. 2020, 40, 107512. [Google Scholar] [CrossRef] [PubMed]
- Vincenz, L.; Hartl, F.U. Sugarcoating ER Stress. Cell 2014, 156, 1125–1127. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Bello, D.; Fernández-Tejada, A. Advances in chemical probing of protein O-GlcNAc glycosylation: Structural role and molecular mechanisms. Chem. Soc. Rev. 2021, 50, 10451–10485. [Google Scholar] [CrossRef]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019, 17, 52. [Google Scholar] [CrossRef]
- Chatham, J.C.; Nöt, L.G.; Fülöp, N.; Marchase, R.B. Hexosamine biosynthesis and protein O-glycosylation: The first line of defense against stress, ischemia, and trauma. Shock 2008, 29, 431–440. [Google Scholar] [CrossRef]
- Vasconcelos-Dos-Santos, A.; Oliveira, I.A.; Lucena, M.C.; Mantuano, N.R.; Whelan, S.A.; Dias, W.B.; Todeschini, A.R. Biosynthetic Machinery Involved in Aberrant Glycosylation: Promising Targets for Developing of Drugs Against Cancer. Front. Oncol. 2015, 5, 138. [Google Scholar] [CrossRef]
- Denzel, M.S.; Storm, N.J.; Gutschmidt, A.; Baddi, R.; Hinze, Y.; Jarosch, E.; Sommer, T.; Hoppe, T.; Antebi, A. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell 2014, 156, 1167–1178. [Google Scholar] [CrossRef]
- Groves, J.A.; Lee, A.; Yildirir, G.; Zachara, N.E. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones 2013, 18, 535–558. [Google Scholar] [CrossRef]
- Bolanle, I.O.; Palmer, T.M. Targeting Protein O-GlcNAcylation, a Link between Type 2 Diabetes Mellitus and Inflammatory Disease. Cells 2022, 11, 705. [Google Scholar] [CrossRef]
- Ma, Z.; Vosseller, K. O-GlcNAc in cancer biology. Amino Acids 2013, 45, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Bacigalupa, Z.A.; Bhadiadra, C.H.; Reginato, M.J. O-GlcNAcylation: Key regulator of glycolytic pathways. J. Bioenerg. Biomembr. 2018, 50, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Ju, H.; Fan, J.; Shi, X.; Cheng, Y.; Cang, X.; Zheng, Z.; Duan, X.; Yi, W. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun. 2020, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.P.; Qian, K.; Lee, J.S.; Zhou, J.; Han, X.; Zhang, B.; Ong, Q.; Ni, W.; Jiang, M.; Ruan, H.B.; et al. O-GlcNAcase targets pyruvate kinase M2 to regulate tumor growth. Oncogene 2020, 39, 560–573. [Google Scholar] [CrossRef]
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A., 3rd; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980. [Google Scholar] [CrossRef]
- Maury, J.J.; Ng, D.; Bi, X.; Bardor, M.; Choo, A.B. Multiple reaction monitoring mass spectrometry for the discovery and quantification of O-GlcNAc-modified proteins. Anal. Chem. 2014, 86, 395–402. [Google Scholar] [CrossRef]
- Ou, W.; Liang, Y.; Qin, Y.; Wu, W.; Xie, M.; Zhang, Y.; Zhang, Y.; Ji, L.; Yu, H.; Li, T. Hypoxic acclimation improves cardiac redox homeostasis and protects heart against ischemia-reperfusion injury through upregulation of O-GlcNAcylation. Redox Biol. 2021, 43, 101994. [Google Scholar] [CrossRef]
- Baldini, S.F.; Lefebvre, T. O-GlcNAcylation and the Metabolic Shift in High-Proliferating Cells: All the Evidence Suggests that Sugars Dictate the Flux of Lipid Biogenesis in Tumor Processes. Front. Oncol. 2016, 6, 6. [Google Scholar] [CrossRef]
- Benhamed, F.; Filhoulaud, G.; Caron, S.; Lefebvre, P.; Staels, B.; Postic, C. O-GlcNAcylation Links ChREBP and FXR to Glucose-Sensing. Front. Endocrinol. 2014, 5, 230. [Google Scholar] [CrossRef]
- Lim, K.; Yoon, B.H.; Ha, C.H. O-Linked N-acetylglucosaminylation of Sp1 interferes with Sp1 activation of glycolytic genes. Biochem. Biophys. Res. Commun. 2015, 468, 349–353. [Google Scholar] [CrossRef]
- Kuo, M.; Zilberfarb, V.; Gangneux, N.; Christeff, N.; Issad, T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008, 582, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Gabius, H.J.; Aureli, M.; Römer, W.; Sonnino, S.; Eskelinen, E.L. Glycans in autophagy, endocytosis and lysosomal functions. Glycoconj J. 2021, 38, 625–647. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Cao, J.; Huang, J.; Yao, J.; Yan, G.; Xu, H.; Yang, P. Discovery and confirmation of O-GlcNAcylated proteins in rat liver mitochondria by combination of mass spectrometry and immunological methods. PLoS ONE 2013, 8, e76399. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.S. Insulin-stimulated glucose uptake in healthy and insulin-resistant skeletal muscle. Horm. Mol. Biol. Clin. Investig. 2016, 26, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, S.B.; Richter, E.A.; Wojtaszewski, J.F. Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. J. Physiol. 2006, 574, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Ojuka, E.O.; Goyaram, V.; Smith, J.A. The role of CaMKII in regulating GLUT4 expression in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E322–E331. [Google Scholar] [CrossRef]
- Merz, K.E.; Thurmond, D.C. Role of Skeletal Muscle in Insulin Resistance and Glucose Uptake. Compr. Physiol. 2020, 10, 785–809. [Google Scholar]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef]
- Hortemo, K.H.; Lunde, P.K.; Anonsen, J.H.; Kvaløy, H.; Munkvik, M.; Rehn, T.A.; Sjaastad, I.; Lunde, I.G.; Aronsen, J.M.; Sejersted, O.M. Exercise training increases protein O-GlcNAcylation in rat skeletal muscle. Physiol. Rep. 2016, 4, e12896. [Google Scholar] [CrossRef]
- Teo, C.F.; Wollaston-Hayden, E.E.; Wells, L. Hexosamine flux, the O-GlcNAc modification, and the development of insulin resistance in adipocytes. Mol. Cell. Endocrinol. 2010, 318, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.M.; Ribeiro, S.B.; Gaya, A.R.; Appell, H.J.; Duarte, J.A. Skeletal muscle pathways of contraction-enhanced glucose uptake. Int. J. Sports Med. 2008, 29, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Jaldin-Fincati, J.R.; Pavarotti, M.; Frendo-Cumbo, S.; Bilan, P.J.; Klip, A. Update on GLUT4 Vesicle Traffic: A Cornerstone of Insulin Action. Trends. Endocrinol. Metab. 2017, 28, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Sun, W.; Luo, G.; Wu, L.; Xu, G.; Hou, D.; Hou, Y.; Guo, X.; Mu, X.; Qin, L.; et al. Panax notoginseng saponins alleviate skeletal muscle insulin resistance by regulating the IRS1-PI3K-AKT signaling pathway and GLUT4 expression. FEBS Open Bio. 2019, 9, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Buse, M.G.; Robinson, K.A.; Marshall, B.A.; Hresko, R.C.; Mueckler, M.M. Enhanced O-GlcNAc protein modification is associated with insulin resistance in GLUT1-overexpressing muscles. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E241–E250. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, C.M.; Lynch, T.P.; Sodi, V.L.; Falcone, J.N.; Schwab, L.P.; Peacock, D.L.; Vocadlo, D.J.; Seagroves, T.N.; Reginato, M.J. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 2014, 54, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Baldini, S.F.; Steenackers, A.; Olivier-Van Stichelen, S.; Mir, A.M.; Mortuaire, M.; Lefebvre, T.; Guinez, C. Glucokinase expression is regulated by glucose through O-GlcNAc glycosylation. Biochem. Biophys. Res. Commun. 2016, 478, 942–948. [Google Scholar] [CrossRef]
- Dong, X.; Li, Y.; Chang, P.; Tang, H.; Hess, K.R.; Abbruzzese, J.L.; Li, D. Glucose metabolism gene variants modulate the risk of pancreatic cancer. Cancer Prev. Res. 2011, 4, 758–766. [Google Scholar] [CrossRef]
- Qian, X.; Li, X.; Shi, Z.; Xia, Y.; Cai, Q.; Xu, D.; Tan, L.; Du, L.; Zheng, Y.; Zhao, D.; et al. PTEN Suppresses Glycolysis by Dephosphorylating and Inhibiting Autophosphorylated PGK1. Mol. Cell 2019, 76, 516–527.e7. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, T.; Li, Y.; Shang, M.; Zhang, Y.; Jin, Y.; Yu, Q.; Guo, F.; Wang, T. O-GlcNAcylation of PFKFB3 is required for tumor cell proliferation under hypoxia. Oncogenesis 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, H.; Yang, J.J.; Liu, X.; Liu, Z.R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 2012, 45, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Chaiyawat, P.; Chokchaichamnankit, D.; Lirdprapamongkol, K.; Srisomsap, C.; Svasti, J.; Champattanachai, V. Alteration of O-GlcNAcylation affects serine phosphorylation and regulates gene expression and activity of pyruvate kinase M2 in colorectal cancer cells. Oncol. Rep. 2015, 34, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Jóźwiak, P.; Forma, E.; Bryś, M.; Krześlak, A. O-GlcNAcylation and Metabolic Reprograming in Cancer. Front. Endocrinol. 2014, 5, 145. [Google Scholar]
- Woolbright, B.L.; Rajendran, G.; Harris, R.A.; Taylor, J.A. 3rd. Metabolic Flexibility in Cancer: Targeting the Pyruvate Dehydrogenase Kinase:Pyruvate Dehydrogenase Axis. Mol. Cancer Ther. 2019, 18, 1673–1681. [Google Scholar] [CrossRef]
- Ande, S.R.; Padilla-Meier, G.P.; Mishra, S. Mutually exclusive acetylation and ubiquitylation among enzymes involved in glucose metabolism. Adipocyte 2013, 2, 256–261. [Google Scholar] [CrossRef]
- Tan, E.P.; Villar, M.T.; Lezi, E.; Lu, J.; Selfridge, J.E.; Artigues, A.; Swerdlow, R.H.; Slawson, C. Altering O-linked β-N-acetylglucosamine cycling disrupts mitochondrial function. J. Biol. Chem. 2014, 289, 14719–14730. [Google Scholar] [CrossRef]
- Jóźwiak, P.; Ciesielski, P.; Zakrzewski, P.K.; Kozal, K.; Oracz, J.; Budryn, G.; Żyżelewicz, D.; Flament, S.; Vercoutter-Edouart, A.S.; Bray, F.; et al. Mitochondrial O-GlcNAc Transferase Interacts with and Modifies Many Proteins and Its Up-Regulation Affects Mitochondrial Function and Cellular Energy Homeostasis. Cancers 2021, 13, 2956. [Google Scholar] [CrossRef]
- Ohashi, N.; Morino, K.; Ida, S.; Sekine, O.; Lemecha, M.; Kume, S.; Park, S.Y.; Choi, C.S.; Ugi, S.; Maegawa, H. Pivotal Role of O-GlcNAc Modification in Cold-Induced Thermogenesis by Brown Adipose Tissue Through Mitochondrial Biogenesis. Diabetes 2017, 66, 2351–2362. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Q.; Zhang, N.; Zhang, K.; Dou, T.; Cao, Y.; Liu, Y.; Li, K.; Hao, X.; Xie, X.; et al. Proteomic profiling and genome-wide mapping of O-GlcNAc chromatin-associated proteins reveal an O-GlcNAc-regulated genotoxic stress response. Nat. Commun. 2020, 11, 5898. [Google Scholar] [CrossRef]
- Gizak, A.; Duda, P.; Wisniewski, J.; Rakus, D. Fructose-1,6-bisphosphatase: From a glucose metabolism enzyme to multifaceted regulator of a cell fate. Adv. Biol. Regul. 2019, 72, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Rakus, D.; Mamczur, P.; Gizak, A.; Dus, D.; Dzugaj, A. Colocalization of muscle FBPase and muscle aldolase on both sides of the Z-line. Biochem. Biophys. Res. Commun. 2003, 311, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Gizak, A.; Maciaszczyk, E.; Dzugaj, A.; Eschrich, K.; Rakus, D. Evolutionary conserved N-terminal region of human muscle fructose 1,6-bisphosphatase regulates its activity and the interaction with aldolase. Proteins 2008, 72, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Issad, T. O-GlcNAc glycosylation and regulation of cell signaling. Med. Sci. 2010, 26, 753–759. [Google Scholar]
- Martinez, M.; Renuse, S.; Kreimer, S.; O’Meally, R.; Natov, P.; Madugundu, A.K.; Nirujogi, R.S.; Tahir, R.; Cole, R.; Pandey, A. Quantitative Proteomics Reveals that the OGT Interactome Is Remodeled in Response to Oxidative Stress. Mol. Cell. Proteom. 2021, 20, 100069. [Google Scholar] [CrossRef]
- Munemoto, M.; Mukaisho, K.I.; Miyashita, T.; Oyama, K.; Haba, Y.; Okamoto, K.; Kinoshita, J.; Ninomiya, I.; Fushida, S.; Taniura, N.; et al. Roles of the hexosamine biosynthetic pathway and pentose phosphate pathway in bile acid-induced cancer development. Cancer Sci. 2019, 110, 2408–2420. [Google Scholar] [CrossRef]
- Su, Z.; Gao, A.; Li, X.; Zou, S.; He, C.; Wu, J.; Ding, W.Q.; Zhou, J. DNA Polymerase Iota Promotes Esophageal Squamous Cell Carcinoma Proliferation Through Erk-OGT-Induced G6PD Overactivation. Front. Oncol. 2021, 11, 706337. [Google Scholar] [CrossRef]
- Zumbaugh, M.D.; Yen, C.N.; Bodmer, J.S.; Shi, H.; Gerrard, D.E. Skeletal Muscle O-GlcNAc Transferase Action on Global Metabolism Is Partially Mediated Through Interleukin-15. Front. Physiol. 2021, 12, 682052. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, Y.; Yang, Y.; Lv, H.; Lian, S.; Xu, B.; Li, S. OGT upregulates myogenic IL-6 by mediating O-GlcNAcylation of p65 in mouse skeletal muscle under cold exposure. J. Cell. Physiol. 2022, 237, 1341–1352. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Bastide, B.; Lefebvre, T.; Lemoine, J.; Mounier, Y.; Michalski, J.C. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol. Cell. Proteom. 2004, 3, 577–585. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, J.; Tian, L.; Liu, Q.; Fu, Y.; Garvey, W.T. TRIB3 mediates glucose-induced insulin resistance via a mechanism that requires the hexosamine biosynthetic pathway. Diabetes 2013, 62, 4192–4200. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Ghani, M.A.; DeFronzo, R.A. Pathogenesis of insulin resistance in skeletal muscle. J. Biomed. Biotechnol. 2010, 2010, 476279. [Google Scholar] [CrossRef] [PubMed]
- Sermikli, B.P.; Aydogdu, G.; Yilmaz, E. Role of the O-GlcNAc modification on insulin resistance and endoplasmic reticulum stress in 3T3-L1 cells. Mol. Biol. Rep. 2020, 47, 5927–5942. [Google Scholar] [CrossRef] [PubMed]
- Argilés, J.M.; Campos, N.; Lopez-Pedrosa, J.M.; Rueda, R.; Rodriguez-Mañas, L. Skeletal Muscle Regulates Metabolism via Interorgan Crosstalk: Roles in Health and Disease. J. Am. Med Dir. Assoc. 2016, 17, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.L.; Gu, J.H.; Liu, F.; Iqbal, K.; Gong, C.X. Neuronal O-GlcNAc transferase regulates appetite, body weight, and peripheral insulin resistance. Neurobiol. Aging 2018, 70, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.A.; Emerald, B.S. The Role of Insulin Resistance and Protein O-GlcNAcylation in Neurodegeneration. Front. Neurosci. 2019, 13, 473. [Google Scholar] [CrossRef] [PubMed]
- Kebede, M.; Ferdaoussi, M.; Mancini, A.; Alquier, T.; Kulkarni, R.N.; Walker, M.D.; Poitout, V. Glucose activates free fatty acid receptor 1 gene transcription via phosphatidylinositol-3-kinase-dependent O-GlcNAcylation of pancreas-duodenum homeobox-1. Proc. Natl. Acad. Sci. USA 2012, 109, 2376–2381. [Google Scholar] [CrossRef]
- Kaleem, A.; Javed, S.; Rehman, N.; Abdullah, R.; Iqtedar, M.; Aftab, M.N.; Hoessli, D.C.; Haq, I.U. Phosphorylated and O-GlcNAc Modified IRS-1 (Ser1101) and -2 (Ser1149) Contribute to Human Diabetes Type II. Protein Pept. Lett. 2021, 28, 333–339. [Google Scholar] [CrossRef]
- Park, S.Y.; Ryu, J.; Lee, W. O-GlcNAc modification on IRS-1 and Akt2 by PUGNAc inhibits their phosphorylation and induces insulin resistance in rat primary adipocytes. Exp. Mol. Med. 2005, 37, 220–229. [Google Scholar] [CrossRef]
- Wang, S.; Huang, X.; Sun, D.; Xin, X.; Pan, Q.; Peng, S.; Liang, Z.; Luo, C.; Yang, Y.; Jiang, H.; et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates Akt signaling. PLoS ONE 2012, 7, e37427. [Google Scholar] [CrossRef]
- Copeland, R.J.; Bullen, J.W.; Hart, G.W. Cross-talk between GlcNAcylation and phosphorylation: Roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E17–E28. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Correa, G.A.; Jin, W.; Wang, Z.; Zhong, X.; Gao, W.D.; Dias, W.B.; Vecoli, C.; Hart, G.W.; Murphy, A.M. O-linked GlcNAc modification of cardiac myofilament proteins: A novel regulator of myocardial contractile function. Circ. Res. 2008, 103, 1354–1358. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Correa, G.A.; Ma, J.; Slawson, C.; Zeidan, Q.; Lugo-Fagundo, N.S.; Xu, M.; Shen, X.; Gao, W.D.; Caceres, V.; Chakir, K.; et al. Removal of Abnormal Myofilament O-GlcNAcylation Restores Ca2+ Sensitivity in Diabetic Cardiac Muscle. Diabetes 2015, 64, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- Hedou, J.; Cieniewski-Bernard, C.; Leroy, Y.; Michalski, J.C.; Mounier, Y.; Bastide, B. O-linked N-acetylglucosaminylation is involved in the Ca2+ activation properties of rat skeletal muscle. J. Biol. Chem. 2007, 282, 10360–10369. [Google Scholar] [CrossRef]
- Salhi, H.E.; Hassel, N.C.; Siddiqui, J.K.; Brundage, E.A.; Ziolo, M.T.; Janssen, P.M.; Davis, J.P.; Biesiadecki, B.J. Myofilament Calcium Sensitivity: Mechanistic Insight into TnI Ser-23/24 and Ser-150 Phosphorylation Integration. Front. Physiol. 2016, 7, 567. [Google Scholar] [CrossRef]
- Metzger, J.M.; Westfall, M.V. Covalent and noncovalent modification of thin filament action: The essential role of troponin in cardiac muscle regulation. Circ. Res. 2004, 94, 146–158. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Montel, V.; Berthoin, S.; Bastide, B. Increasing O-GlcNAcylation level on organ culture of soleus modulates the calcium activation parameters of muscle fibers. PLoS ONE 2012, 7, e48218. [Google Scholar] [CrossRef]
- Bayliss, C.R.; Jacques, A.M.; Leung, M.C.; Ward, D.G.; Redwood, C.S.; Gallon, C.E.; Copeland, O.; McKenna, W.J.; Dos Remedios, C.; Marston, S.B.; et al. Myofibrillar Ca2+ sensitivity is uncoupled from troponin I phosphorylation in hypertrophic obstructive cardiomyopathy due to abnormal troponin T. Cardiovasc. Res. 2013, 97, 500–508. [Google Scholar] [CrossRef]
- Manstein, D.J.; Meiring, J.C.M.; Hardeman, E.C.; Gunning, P.W. Actin-tropomyosin distribution in non-muscle cells. J. Muscle Res. Cell Motil. 2020, 41, 11–22. [Google Scholar] [CrossRef]
- Hédou, J.; Bastide, B.; Page, A.; Michalski, J.C.; Morelle, W. Mapping of O-linked beta-N-acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics 2009, 9, 2139–2148. [Google Scholar] [CrossRef]
- Basu, H.; Pekkurnaz, G.; Falk, J.; Wei, W.; Chin, M.; Steen, J.; Schwarz, T.L. FHL2 anchors mitochondria to actin and adapts mitochondrial dynamics to glucose supply. J. Cell Biol. 2021, 220, e201912077. [Google Scholar] [CrossRef] [PubMed]
- Butkinaree, C.; Park, K.; Hart, G.W. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 2010, 1800, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Deruy, E.; Belliard, A.; Mulder, P.; Bouvet, M.; Smet-Nocca, C.; Janel, S.; Lafont, F.; Beseme, O.; Amouyel, P.; Richard, V.; et al. Interplay between troponin T phosphorylation and O-N-acetylglucosaminylation in ischaemic heart failure. Cardiovasc. Res. 2015, 107, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- Zhi, G.; Ryder, J.W.; Huang, J.; Ding, P.; Chen, Y.; Zhao, Y.; Kamm, K.E.; Stull, J.T. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc. Natl. Acad. Sci. USA 2005, 102, 17519–17524. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Dupont, E.; Richard, E.; Bastide, B. Phospho-GlcNAc modulation of slow MLC2 during soleus atrophy through a multienzymatic and sarcomeric complex. Pflugers Arch. 2014, 466, 2139–2151. [Google Scholar] [CrossRef]
- Ryder, J.W.; Lau, K.S.; Kamm, K.E.; Stull, J.T. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J. Biol. Chem. 2007, 282, 20447–20454. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Y.; Wang, T.; Hawke, D.H.; Zheng, Y.; Li, X.; Zhou, Q.; Majumder, S.; Bi, E.; Liu, D.X.; et al. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat. Commun. 2014, 5, 5566. [Google Scholar] [CrossRef]
- Hartshorne, D.J.; Ito, M.; Erdödi, F. Role of protein phosphatase type 1 in contractile functions: Myosin phosphatase. J. Biol. Chem. 2004, 279, 37211–37214. [Google Scholar] [CrossRef]
- Kiss, A.; Erdődi, F.; Lontay, B. Myosin phosphatase: Unexpected functions of a long-known enzyme. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 2–15. [Google Scholar] [CrossRef]
- Stull, J.T.; Kamm, K.E.; Vandenboom, R. Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch. Biochem. Biophys. 2011, 510, 120–128. [Google Scholar] [CrossRef]
- Agarwal, P.; Zaidel-Bar, R. Principles of Actomyosin Regulation In Vivo. Trends Cell Biol. 2019, 29, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Lin, J.; Jiang, J.; He, M.; Sun, D.; Zhao, Z.; Shen, Y.; Xue, A. Phosphorylated Myosin Light Chain 2 (p-MLC2) as a Molecular Marker of Antemortem Coronary Artery Spasm. Med Sci. Monit. 2016, 22, 3316–3327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kanaya, N.; Gable, B.; Murray, P.A.; Damron, D.S. Propofol increases phosphorylation of troponin I and myosin light chain 2 via protein kinase C activation in cardiomyocytes. Anesthesiology 2003, 98, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Hortemo, K.H.; Aronsen, J.M.; Lunde, I.G.; Sjaastad, I.; Lunde, P.K.; Sejersted, O.M. Exhausting treadmill running causes dephosphorylation of sMLC2 and reduced level of myofilament MLCK2 in slow twitch rat soleus muscle. Physiol. Rep. 2015, 3, e12285. [Google Scholar] [CrossRef]
- Cai, L.X.; Tanada, Y.; Bello, G.D.; Fleming, J.C.; Alkassis, F.F.; Ladd, T.; Golde, T.; Koh, J.; Chen, S.; Kasahara, H. Cardiac MLC2 kinase is localized to the Z-disc and interacts with α-actinin2. Sci. Rep. 2019, 9, 12580. [Google Scholar] [CrossRef]
- Wang, L.; Geist, J.; Grogan, A.; Hu, L.R.; Kontrogianni-Konstantopoulos, A. Thick Filament Protein Network, Functions, and Disease Association. Compr. Physiol. 2018, 8, 631–709. [Google Scholar]
- Wende, A.R. Unsticking the Broken Diabetic Heart: O-GlcNAcylation and Calcium Sensitivity. Diabetes 2015, 64, 3339–3341. [Google Scholar] [CrossRef]
- Rassier, D.E. Sarcomere mechanics in striated muscles: From molecules to sarcomeres to cells. Am. J. Physiol. Cell Physiol. 2017, 313, C134–C145. [Google Scholar] [CrossRef]
- Galińska-Rakoczy, A.; Engel, P.; Xu, C.; Jung, H.; Craig, R.; Tobacman, L.S.; Lehman, W. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J. Mol. Biol. 2008, 379, 929–935. [Google Scholar] [CrossRef]
- Khaitlina, S.Y. Tropomyosin as a Regulator of Actin Dynamics. Int. Rev. Cell Mol. Biol. 2015, 318, 255–291. [Google Scholar] [PubMed]
- Leung, M.C.; Hitchen, P.G.; Ward, D.G.; Messer, A.E.; Marston, S.B. Z-band alternatively spliced PDZ motif protein (ZASP) is the major O-linked β-N-acetylglucosamine-substituted protein in human heart myofibrils. J. Biol. Chem. 2013, 288, 4891–4898. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the Muscle Cytoskeleton. Compr. Physiol. 2017, 7, 891–944. [Google Scholar]
- Wang, Z.; Grange, M.; Wagner, T.; Kho, A.L.; Gautel, M.; Raunser, S. The molecular basis for sarcomere organization in vertebrate skeletal muscle. Cell 2021, 184, 2135–2150.e13. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Hoessli, D.C.; Walker-Nasir, E.; Choudhary, M.I.; Rafik, S.M.; Shakoori, A.R. Phosphorylation and glycosylation interplay: Protein modifications at hydroxy amino acids and prediction of signaling functions of the human beta3 integrin family. J. Cell Biochem. 2006, 99, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Laczy, B.; Marsh, S.A.; Brocks, C.A.; Wittmann, I.; Chatham, J.C. Inhibition of O-GlcNAcase in perfused rat hearts by NAG-thiazolines at the time of reperfusion is cardioprotective in an O-GlcNAc-dependent manner. Am. J. Physiol. Heart Circ. Physiol. 2010, 299, H1715–H1727. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Richard, E.; Duban-Deweer, S.; Krzewinski, F.; Deracinois, B.; Dupont, E.; Bastide, B.; Cieniewski-Bernard, C. O-GlcNAcylation is a key modulator of skeletal muscle sarcomeric morphometry associated to modulation of protein-protein interactions. Biochim. Biophys. Acta 2016, 1860, 2017–2030. [Google Scholar] [CrossRef]
- von Nandelstadh, P.; Ismail, M.; Gardin, C.; Suila, H.; Zara, I.; Belgrano, A.; Valle, G.; Carpen, O.; Faulkner, G. A A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: A common link for Z-disc myopathies. Mol. Cell. Biol. 2009, 29, 822–834. [Google Scholar] [CrossRef]
- Deracinois, B.; Camoin, L.; Lambert, M.; Boyer, J.B.; Dupont, E.; Bastide, B.; Cieniewski-Bernard, C. O-GlcNAcylation site mapping by (azide-alkyne) click chemistry and mass spectrometry following intensive fractionation of skeletal muscle cells proteins. J. Proteom. 2018, 186, 83–97. [Google Scholar] [CrossRef]
- Hnia, K.; Ramspacher, C.; Vermot, J.; Laporte, J. Desmin in muscle and associated diseases: Beyond the structural function. Cell Tissue Res. 2015, 360, 591–608. [Google Scholar] [CrossRef]
- Srikanth, B.; Vaidya, M.M.; Kalraiya, R.D. O-GlcNAcylation determines the solubility, filament organization, and stability of keratins 8 and 18. J. Biol. Chem. 2010, 285, 34062–34071. [Google Scholar] [CrossRef] [PubMed]
- Ise, H.; Kobayashi, S.; Goto, M.; Sato, T.; Kawakubo, M.; Takahashi, M.; Ikeda, U.; Akaike, T. Vimentin and desmin possess GlcNAc-binding lectin-like properties on cell surfaces. Glycobiology 2010, 20, 843–864. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Kang, J.G.; Park, S.Y.; Lee, J.; Oh, Y.J.; Cho, J.W. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids 2011, 40, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.L.; Song, T.; Sadayappan, S. Myofilaments: Movers and Rulers of the Sarcomere. Compr. Physiol. 2017, 7, 675–692. [Google Scholar] [PubMed]
- Ma, J.; Wang, W.H.; Li, Z.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. O-GlcNAc Site Mapping by Using a Combination of Chemoenzymatic Labeling, Copper-Free Click Chemistry, Reductive Cleavage, and Electron-Transfer Dissociation Mass Spectrometry. Anal. Chem. 2019, 91, 2620–2625. [Google Scholar] [CrossRef]
- Krishnamoorthy, V.; Donofrio, A.J.; Martin, J.L. O-GlcNAcylation of αB-crystallin regulates its stress-induced translocation and cytoprotection. Mol. Cell. Biochem. 2013, 379, 59–68. [Google Scholar] [CrossRef]
- Sziklai, D.; Sallai, J.; Papp, Z.; Kellermayer, D.; Mártonfalvi, Z.; Pires, R.H.; Kellermayer, M.S.Z. Nanosurgical Manipulation of Titin and Its M-Complex. Nanomaterials 2022, 12, 178. [Google Scholar] [CrossRef]
- Kontrogianni-Konstantopoulos, A.; Ackermann, M.A.; Bowman, A.L.; Yap, S.V.; Bloch, R.J. Muscle giants: Molecular scaffolds in sarcomerogenesis. Physiol. Rev. 2009, 89, 1217–1267. [Google Scholar] [CrossRef]
- Li, Y.; Lang, P.; Linke, W.A. Titin stiffness modifies the force-generating region of muscle sarcomeres. Sci. Rep. 2016, 6, 24492. [Google Scholar] [CrossRef]
- Canault, M.; Tellier, E.; Bonardo, B.; Mas, E.; Aumailley, M.; Juhan-Vague, I.; Nalbone, G.; Peiretti, F. FHL2 interacts with both ADAM-17 and the cytoskeleton and regulates ADAM-17 localization and activity. J. Cell. Physiol. 2006, 208, 363–372. [Google Scholar] [CrossRef]
- Massaccesi, L.; Goi, G.; Tringali, C.; Barassi, A.; Venerando, B.; Papini, N. Dexamethasone-Induced Skeletal Muscle Atrophy Increases O-GlcNAcylation in C2C12 Cells. J. Cell. Biochem. 2016, 117, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Ho, S.R.; Wang, K.; Roessler, B.C.; Zhang, F.; Hu, Y.; Bowe, D.B.; Kudlow, J.E.; Paterson, A.J. Muscle-specific overexpression of NCOATGK, splice variant of O-GlcNAcase, induces skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2011, 300, C456–C465. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Mizofuchi, H.; Kobayashi, Y.; Tsuzuki, G.; Yamamoto, M.; Wada, S.; Kamemura, K. erminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta 2012, 1820, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Sakakibara, Y.; Kamemura, K. Requirement of decreased O-GlcNAc glycosylation of Mef2D for its recruitment to the myogenin promoter. Biochem. Biophys. Res. Commun. 2013, 433, 558–562. [Google Scholar] [CrossRef]
- Joumaa, V.; Bertrand, F.; Liu, S.; Poscente, S.; Herzog, W. Does partial titin degradation affect sarcomere length nonuniformities and force in active and passive myofibrils? Am. J. Physiol. Cell Physiol. 2018, 315, C310–C318. [Google Scholar] [CrossRef]
- Neel, B.A.; Lin, Y.; Pessin, J.E. Skeletal muscle autophagy: A new metabolic regulator. Trends Endocrinol. Metab. 2013, 24, 635–643. [Google Scholar] [CrossRef]
- Byon, C.H.; Kim, S.W. Regulatory Effects of O-GlcNAcylation in Vascular Smooth Muscle Cells on Diabetic Vasculopathy. J. Lipid. Atheroscler. 2020, 9, 243–254. [Google Scholar] [CrossRef]
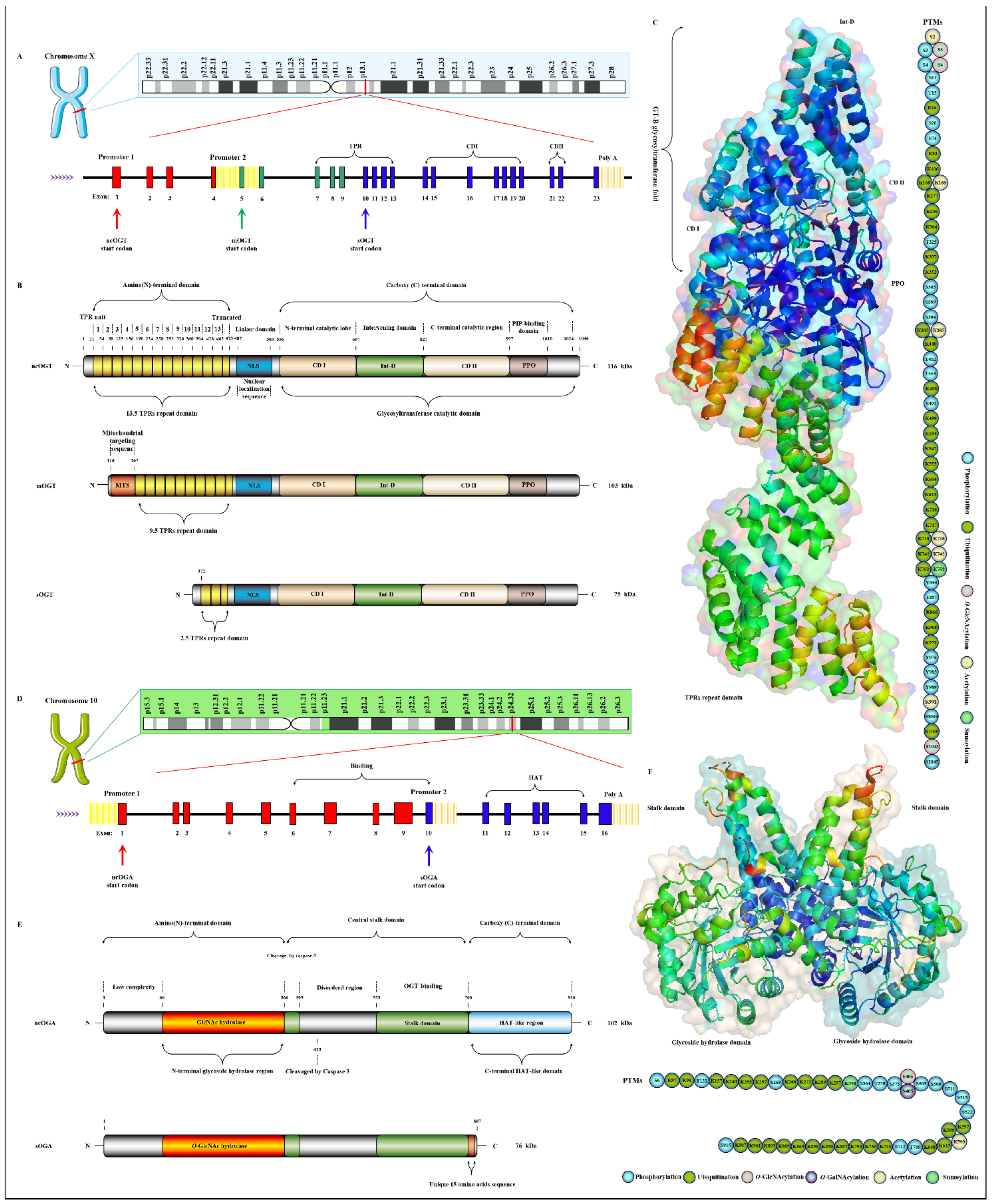
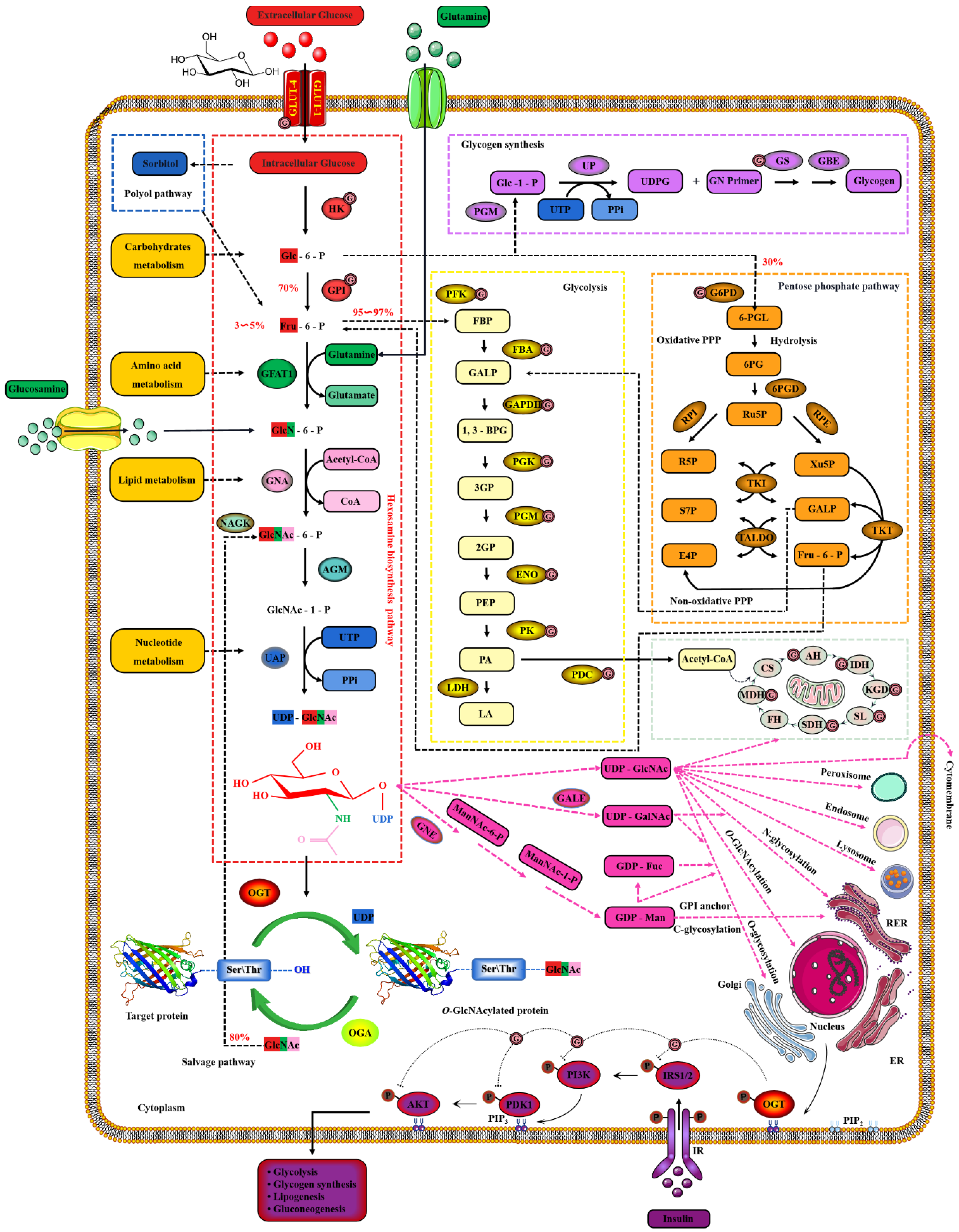
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Hu, Y.-J.; Fan, W.-X.; Quan, X.; Xu, B.; Li, S.-Z. O-GlcNAcylation: The Underestimated Emerging Regulators of Skeletal Muscle Physiology. Cells 2022, 11, 1789. https://doi.org/10.3390/cells11111789
Liu Y, Hu Y-J, Fan W-X, Quan X, Xu B, Li S-Z. O-GlcNAcylation: The Underestimated Emerging Regulators of Skeletal Muscle Physiology. Cells. 2022; 11(11):1789. https://doi.org/10.3390/cells11111789
Chicago/Turabian StyleLiu, Yang, Ya-Jie Hu, Wen-Xuan Fan, Xin Quan, Bin Xu, and Shi-Ze Li. 2022. "O-GlcNAcylation: The Underestimated Emerging Regulators of Skeletal Muscle Physiology" Cells 11, no. 11: 1789. https://doi.org/10.3390/cells11111789
APA StyleLiu, Y., Hu, Y.-J., Fan, W.-X., Quan, X., Xu, B., & Li, S.-Z. (2022). O-GlcNAcylation: The Underestimated Emerging Regulators of Skeletal Muscle Physiology. Cells, 11(11), 1789. https://doi.org/10.3390/cells11111789





