Spatially and Temporally Distributed Complexity—A Refreshed Framework for the Study of GRN Evolution
Abstract
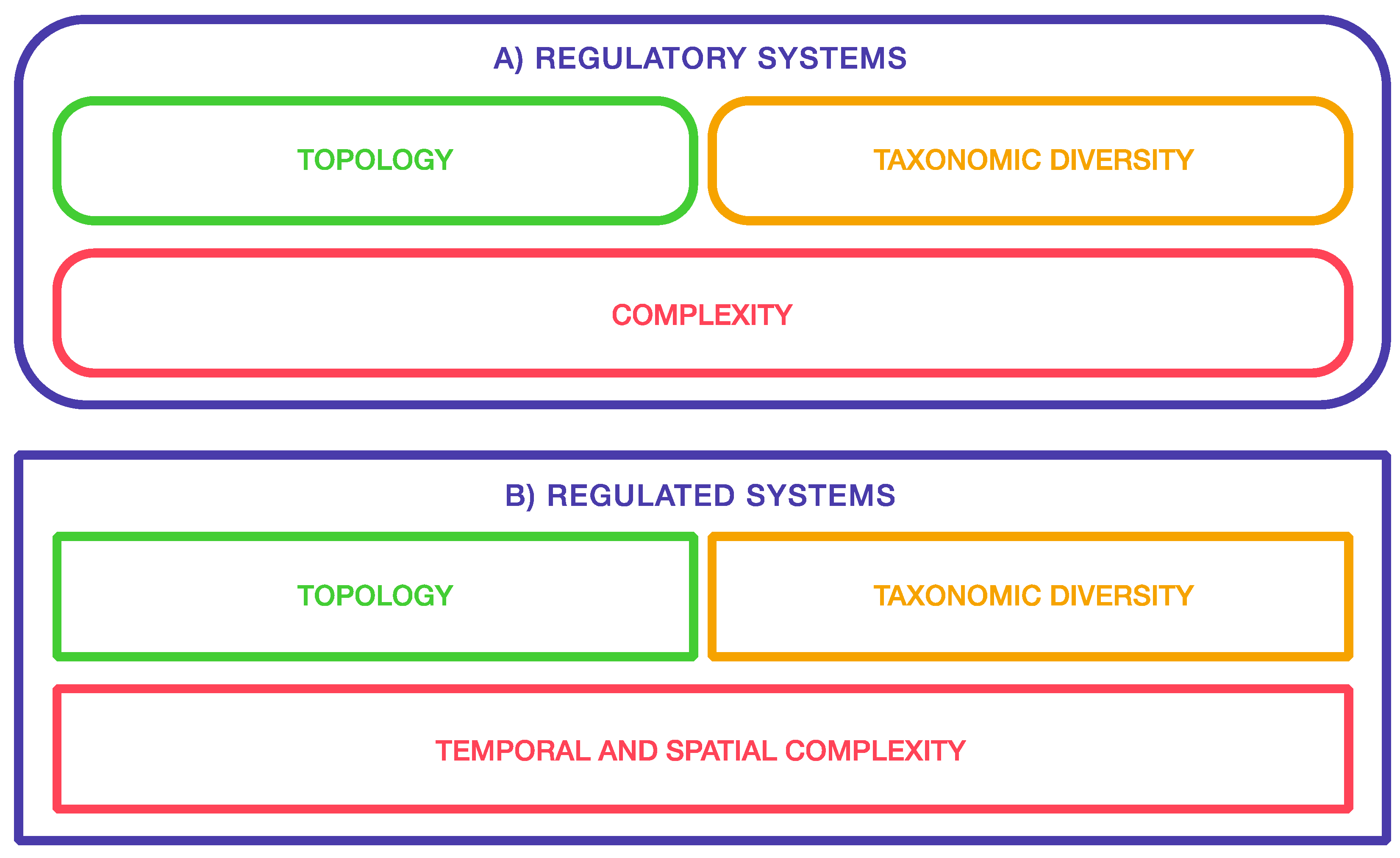
:1. Introduction
- Developmental genes?
- Developmental patterns vs. processes.
- Adaptational and teleological views.
- The development of unicells and its regulation.
- Structural and functional hierarchies.
- Spatial and temporal dimensions of development.
2. Developmental Genes?
3. Developmental Patterns vs. Processes
3.1. Adultocentrism
Development progresses from phase to phase, and this fundamental phenomenon reflects the underlying sequential hierarchy of the GRN control system. In the earliest embryonic phases, the function of the developmental GRN is establishment of specific regulatory states in the spatial domains of the developing organism. In this way, the design of the future body plan is mapped out in regional regulatory landscapes, which differentially endow the potentialities of the future parts. Lower down in the hierarchy, GRN apparatus continues regional regulatory specification on finer scales. Ultimately, precisely confined regulatory states determine how the differentiation and morphogenetic gene batteries at the terminal periphery of the GRN will be deployed.[24] (p. 970)
3.2. GRN in Blastogenesis
4. Adaptational and Teleological Views
5. The Development of Unicells and Its Regulation
6. Structural and Functional Hierarchies
6.1. The Traditional View
These processes of differentiation consist in the more or less similar morphological elements (cells) which represent the organism, acquiring, in larger or smaller groups, distinct characters: in their being differentiated, and forming the rudiments (first stages) of organs, by taking a definite order and arrangement. These organs then are made up of cells, which form their tissues. We thus arrive at the essence of the architecture of organisms; we have tissues, which make up organs, and are themselves composed of form-elements—the cells.[57] (p. 20)
Development of animal body plans proceeds by the progressive installation of transcriptional regulatory states, transiently positioned in embryonic space. The underlying mechanism is the localized expression of genes encoding sequence-specific transcription factors at specific times and places.[59] (p. 4835)
Developmental programs comprise the sets of stepwise changes in cells, tissues, and organs that ultimately produce phenotypes. The developmental program of a given phenotype is generally controlled by one or more GRNs.[7] (p. 2)
The GRNs controlling embryonic development of the body plan are intrinsically hierarchical, essentially because of the number of successive spatial regulatory states that must be installed in the course of pattern formation, cell-type specification, and differentiation. If the regulatory state defines a progenitor field for a given organ, then all the subsequent stages in the development of that organ must take place within that domain.[24] (p. 973)
The morphogenetic tree is a diagrammatic construct that represents ontogeny as a tree, with causal connections between hierarchical levels represented as the branches. … This model is extremely simple and understandable, and it allows predictions to be made. But heuristic elegance comes at a high cost. To achieve simplicity, the nature of developmental processes is ignored, and some very static (and demonstrably incorrect) assumptions about development have to be built in. The most serious explicit assumption is that genes acting early in development have larger effects on adult phenotype than those acting later. Such a view … is a gross oversimplification, and it leads to a number of misleading predictions about development and how development must evolve.[61] (p. 518)
6.2. I “Know” What Needs to Be Done, So There Has to Be a Mechanism to Do It
6.3. From Single-Cell Sequencing to Modeling the Topology of Developmental Systems
7. Spatial and Temporal Dimensions
7.1. Time and Space, Intertwined
A distinction between temporal and spatial aspects of the molecular control of development is often artificial. This is true, in particular, in the context of the collinearity between the spatial organization of the Hox genes along the chromosome, the temporal sequence of their activation and the spatial order of the regions along the animal’s main body axis, in which each of these genes is expressed. Therefore, it is wise to identify a search for correspondence between spatial (morphological) and temporal (developmental) units and patterns as a primary target of developmental (and evo-devo) biology.
7.2. Trade-Off between Spatial and Temporal Differentiation
8. Concluding Remarks
8.1. GRNs and Phenotypes
8.2. No Universal Regulatory Metaphor
8.3. Spatially and Temporally Distributed Complexity
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McQueen, E.; Rebeiz, M. On the specificity of gene regulatory networks: How does network co-option affect subsequent evolution? Curr. Top. Dev. Biol. 2020, 139, 375–405. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S. Bacterial regulation: Global regulatory networks. Annu. Rev. Genet. 1984, 18, 415–441. [Google Scholar] [CrossRef] [PubMed]
- Rebeiz, M.; Patel, N.H.; Hinman, V.F. Unraveling the tangled skein: The evolution of transcriptional regulatory networks in development. Annu. Rev. Genom. Hum. Genet. 2015, 16, 103–131. [Google Scholar] [CrossRef] [PubMed]
- Britten, R.J.; Davidson, E.H. Gene regulation for higher cells: A theory. Science 1969, 165, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Desplan, C. Gene regulatory networks during the development of the Drosophila visual system. Curr. Top. Dev. Biol. 2020, 139, 89–125. [Google Scholar] [CrossRef]
- Davidson, E.H. The Regulatory Genome; Academic Press: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Feigin, C.; Li, S.; Moreno, J.; Mallarino, R. The GRN concept as a guide for evolutionary developmental biology. J. Exp. Zool. B Mol. Dev. Evol. 2022. [Google Scholar] [CrossRef]
- Keller, E.F. The Century of the Gene; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hall, B.K. The gene is not dead, merely orphaned and seeking a home. Evol. Dev. 2001, 3, 225–228. [Google Scholar] [CrossRef]
- Nijhout, H.F. Metaphors and the role of genes in development. BioEssays 1990, 12, 441–446. [Google Scholar] [CrossRef]
- Oyama, S. The Ontogeny of Information, 2nd ed.; Duke University Press: Durham, NC, USA, 2000. [Google Scholar] [CrossRef]
- Pigliucci, M.; Murren, C.J.; Schlichting, C.D. Phenotypic plasticity and evolution by genetic assimilation. J. Exp. Biol. 2006, 209, 2362–2367. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, S.F.; Epel, D. Ecological Developmental Biology: Integrating Epigenetics, Medicine, and Evolution; Sinauer: Sunderland, UK, 2008. [Google Scholar]
- Fusco, G.; Minelli, A. Phenotypic plasticity in development and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 547–556. [Google Scholar] [CrossRef] [Green Version]
- Lidgard, S.; Carter, M.C.; Dick, M.H.; Gordon, D.P.; Ostrovsky, A.N. Division of labor and recurrent evolution of polymorphisms in a group of colonial animals. Evol. Ecol. 2012, 26, 233–257. [Google Scholar] [CrossRef]
- Anderson, K.E.; Linksvayer, T.A.; Smith, C.R. The causes and consequences of genetic caste determination in ants (Hymenoptera: Formicidae). Myrmecol. News 2008, 11, 119–132. [Google Scholar]
- Nijhout, H.F. Hormonal control in larval development and evolution. In The Origin and Evolution of Larval Forms; Hall, B., Wake, M., Eds.; Academic Press: New York, NY, USA, 1999; pp. 217–254. [Google Scholar] [CrossRef]
- Minelli, A.; Fusco, G. Developmental plasticity and the evolution of animal complex life cycles. Phil. Trans. R. Soc. B 2010, 365, 631–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, P.B. Seasonal polyphenism in butterflies: A hormonally controlled phenomenon of pattern formation. Zool. Jahrbücher Abt. Allg. Zool. Physiol. Tiere 1992, 96, 227–240. [Google Scholar]
- Schack, C.R.; Gordon, D.P.; Ryan, K.G. Modularity is the mother of invention: A review of polymorphism in bryozoans. Biol. Rev. 2019, 94, 773–809. [Google Scholar] [CrossRef]
- Hiebert, L.S.; Simpson, C.; Tiozzo, S. Coloniality, clonality, and modularity in animals: The elephant in the room. J. Exp. Zool. B Mol. Dev. Evol. 2021, 336, 198–211. [Google Scholar] [CrossRef]
- Nijhout, F.H.; Davidowitz, G. The developmental-physiological basis of phenotypic plasticity. In Phenotypic Plasticity of Insects: Mechanism and Consequences; Whitman, D.W., Ananthakrishnan, T.N., Eds.; Science Publishers: Enfield, NH, USA, 2009; pp. 589–608. [Google Scholar] [CrossRef]
- Minelli, A. The Development of Animal Form; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar] [CrossRef]
- Peter, I.S.; Davidson, E.H. Evolution of gene regulatory networks controlling body plan development. Cell 2011, 144, 970–985. [Google Scholar] [CrossRef] [Green Version]
- Bernard, C. Leçons sur les Phénomènes de la vie Communs aux Animaux et aux Végétaux; Baillière: Paris, France, 1878; Volume 1. [Google Scholar] [CrossRef] [Green Version]
- Sultan, S.E.; Moczek, A.P.; Walsh, D. Bridging the explanatory gaps: What can we learn from a biological agency perspective? BioEssays 2022, 44, 2100185. [Google Scholar] [CrossRef]
- Gerhart, J.C.; Kirschner, M.W. The theory of facilitated variation. Proc. Natl. Acad. Sci. USA 2007, 104, 8582–8589. [Google Scholar] [CrossRef] [Green Version]
- Kirschner, M.W.; Gerhart, J.C. Facilitated variation. In Evolution: The Extended Synthesis; Pigliucci, M., Mueller, G.B., Eds.; MIT Press: Cambridge, MA, USA, 2010; pp. 253–280. [Google Scholar] [CrossRef]
- Minelli, A. Development, an open-ended segment of life. Biol. Theory 2011, 6, 4–15. [Google Scholar] [CrossRef]
- Minelli, A. Understanding Development; Cambridge University Press: Cambridge, UK, 2021. [Google Scholar] [CrossRef]
- Alié, A.; Hiebert, L.S.; Scelzo, M.; Tiozzo, S. The eventful history of nonembryonic development in tunicates. J. Exp. Zool. B Mol. Dev. Evol. 2021, 336, 250–266. [Google Scholar] [CrossRef] [PubMed]
- Tiozzo, S.; Christiaen, L.; Deyts, C.; Manni, L.; Joly, J.; Burighel, P. Embryonic vs. blastogenetic development in the compound ascidian Botryllus schlosseri: Insights from Pitx expression patterns. Dev. Dyn. 2005, 232, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, A.; Setar, L.; Tiozzo, S.; De Tomaso, A.W. Wnt affects symmetry and morphogenesis during post-embryonic development in colonial chordates. EvoDevo 2015, 6, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, L.; Cabrera, F.; Lotito, S.; Tiozzo, S. Redeployment of germ layers related TFs shows regionalized expression during two non-embryonic developments. Dev. Biol. 2016, 416, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Prünster, M.M.; Ricci, L.; Brown, F.; Tiozzo, S. Modular co-option of cardiopharyngeal genes during non-embryonic myogenesis. EvoDevo 2019, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Kowarsky, M.; Anselmi, C.; Hotta, K.; Burighel, P.; Zaniolo, G.; Caicci, F.; Rosental, B.; Neff, N.F.; Ishizuka, K.J.; Palmeri, K.J.; et al. Sexual and asexual development: Two distinct programs producing the same tunicate. Cell Rep. 2021, 34, 108681. [Google Scholar] [CrossRef]
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Peterson, K.J.; Cameron, R.A.; Davidson, E.H. Set-aside cells in maximal indirect development: Evolutionary and developmental significance. BioEssays 1997, 19, 623–631. [Google Scholar] [CrossRef]
- Strathmann, R.R. Functional design in the evolution of embryos and larvae. Semin. Cell Dev. Biol. 2000, 11, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Bird, R.J. Chaos and Life; Columbia University Press: New York, NY, USA, 2003. [Google Scholar]
- Bonner, J.T. First Signals: The Evolution of Multicellular Development; Princeton University Press: Princeton, NJ, USA, 2001. [Google Scholar] [CrossRef]
- Mani, S.; Tlusty, T. A topological look into the evolution of developmental programs. Biophys. J. 2021, 120, 4193–4201. [Google Scholar] [CrossRef]
- Matthews, K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005, 118, 283–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabani, S.; Fenn, K.; Ross, A.; Ivens, A.; Smith, T.K.; Ghazal, P.; Matthews, K. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genom. 2009, 10, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haile, S.; Papadopoulou, B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007, 10, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkaufer, G.M.; Haque, R.; Hackney, J.A.; Eichinger, D.J.; Singh, U. Identification of developmentally regulated genes in Entamoeba histolytica: Insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. 2007, 9, 1426–1444. [Google Scholar] [CrossRef]
- Bozdech, Z.; Llinas, M.; Pulliam, B.L.; Wong, E.D.; Zhu, J.; DeRisi, J.L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003, 1, E5. [Google Scholar] [CrossRef] [Green Version]
- Arbeitman, M.N.; Furlong, E.E.; Imam, F.; Johnson, E.; Null, B.H.; Baker, B.S.; Krasnow, M.A.; Scott, M.P.; Davis, R.W.; White, K.P. Gene expression during the life cycle of Drosophila melanogaster. Science 2002, 297, 2270–2275. [Google Scholar] [CrossRef]
- Hertel, L.A.; Bayne, C.J.; Loker, E.S. The symbiont Capsaspora owczarzaki, nov. gen. nov. sp., isolated from three strains of the pulmonate snail Biomphalaria glabrata is related to members of the Mesomycetozoea. Int. J. Parasitol. 2002, 32, 1183–1191. [Google Scholar] [CrossRef]
- Owczarzak, A.; Stibbs, H.H.; Bayne, C.J. The destruction of Schistosoma mansoni mother sporocysts in vitro by amoebae isolated from Biomphalaria glabrata: An ultrastructural study. J. Invertebr. Pathol. 1980, 35, 26–33. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Irimia, M.; del Campo, J.; Parra-Acero, H.; Russ, C.; Nusbaum, C.; Blencowe, B.J.; Ruiz-Trillo, I. Regulated aggregative multicellularity in a close unicellular relative of Metazoa. eLife 2013, 2, e01287. [Google Scholar] [CrossRef] [Green Version]
- Sebé-Pedrós, A.; Peña, M.I.; Capella-Gutiérrez, S.; Antó, M.; Gabaldón, T.; Ruiz-Trillo, I.; Sabidó, E. High-throughput proteomics reveals the unicellular roots of animal phosphosignaling and cell differentiation. Dev. Cell 2016, 39, 186–197. [Google Scholar] [CrossRef] [Green Version]
- Dayel, M.J.; Alegado, R.A.; Fairclough, S.R.; Nichols, S.A.; McDonald, K.; King, N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 2011, 357, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairclough, S.R.; Chen, Z.; Kramer, E.; Zeng, Q.; Young, S.; Robertson, H.M.; Begovic, E.; Richter, D.J.; Russ, C.; Westbrook, M.J.; et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013, 14, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumann, B.; Burkhardt, P. Spatial cell disparity in the colonial choanoflagellate Salpingoeca rosetta. Front. Cell Dev. Biol. 2019, 7, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, E.H. Evolutionary bioscience as regulatory systems biology. Dev. Biol. 2011, 357, 35–40. [Google Scholar] [CrossRef] [Green Version]
- Gegenbaur, C. Elements of Comparative Anatomy; Macmillan: London, UK, 1878. [Google Scholar]
- Erwin, D.H.; Davidson, E.H. The evolution of hierarchical gene regulatory networks. Nat. Rev. Genet. 2009, 10, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Levine, M.; Davidson, E.H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 2005, 102, 4936–4942. [Google Scholar] [CrossRef] [Green Version]
- Arthur, W. A Theory of the Evolution of Development; Wiley: New York, NY, USA, 1988. [Google Scholar]
- Raff, R.A. The resurrection of recapitulation. Nature 1989, 337, 518. [Google Scholar] [CrossRef]
- Alberch, P. From genes to phenotype: Dynamical systems and evolvability. Genetica 1991, 84, 5–11. [Google Scholar] [CrossRef]
- DiFrisco, J.; Love, A.C.; Wagner, G.P. Character identity mechanisms: A conceptual model for comparative-mechanistic biology. Biol. Philos. 2020, 35, 44. [Google Scholar] [CrossRef]
- Deutsch, J. Hox and wings. BioEssays 2005, 27, 673–675. [Google Scholar] [CrossRef]
- Wagner, G.P. The developmental genetics of homology. Nat. Rev. Genet. 2007, 8, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Minelli, A. Molecules, developmental modules and phenotypes: A combinatorial approach to homology. Mol. Phylogenet. Evol. 1998, 9, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Zhang, M.Q.; Qian, H. Emergent Levy behavior in single-cell stochastic gene expression. Phys. Rev. E 2017, 96, 040402. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Wang, S.; Li, T.; Nie, Q. Dissecting transition cells from single-cell transcriptome data through multiscale stochastic dynamics. Nat. Commun. 2021, 12, 5609. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lin, F.; Zhong, C.-H.; Wang, S.; Xue, X.; Shao, Y. Single-cell sequencing to unveil the mystery of embryonic development. Adv. Biol. 2022, 6, 2101151. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Han, J.-D.J. Regulatory network characterization in development: Challenges and opportunities. F1000Research 2018, 7, 1477. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Loeffler-Wirth, H.; Binder, H. Developmental scRNAseq trajectories in gene- and cell-state space—The flatworm example. Genes 2020, 11, 1214. [Google Scholar] [CrossRef]
- Babtie, A.C.; Kirk, P.; Stumpf, M.P.H. Topological sensitivity analysis for systems biology. Proc. Natl. Acad. Sci. USA 2014, 111, 18507–18512. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Albergante, L.; Hsu, J.Y.; Lareau, C.A.; Bosco, G.L.; Guan, J.; Zhou, S.; Gorban, A.N.; Bauer, D.E.; Aryee, M.J.; et al. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat. Commun. 2019, 10, 1903. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Ernberg, I.; Kauffman, S. Cancer attractors: A systems view of tumors from a gene network dynamics and developmental perspective. Sem. Cell Dev. Biol. 2009, 20, 869–876. [Google Scholar] [CrossRef] [Green Version]
- Marr, C.; Zhou, J.X.; Huang, S. Single-cell gene expression profiling and cell state dynamics: Collecting data, correlating data points and connecting the dots. Curr. Opin. Biotechnol. 2016, 39, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saelens, W.; Cannoodt, R.; Todorov, H.; Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019, 37, 547. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. Canalization of development and the inheritance of acquired characters. Nature 1942, 150, 563–565. [Google Scholar] [CrossRef]
- Guillemin, A.; Stumpf, M.P. Noise and the molecular processes underlying cell fate decision-making. Phys. Biol. 2020, 17, 065013. [Google Scholar] [CrossRef] [PubMed]
- Moris, N.; Pina, C.; Martinez-Arias, A. Transition states and cell fate decisions in epigenetic landscapes. Nat. Rev. Genet. 2016, 17, 693–703. [Google Scholar] [CrossRef]
- Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 2015, 25, 1491–1498. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, H.; Kiryu, H. SCOUP: A probabilistic model based on the Ornstein-Uhlenbeck process to analyze single-cell expression data during differentiation. BMC Bioinf. 2016, 17, 232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Nie, Q.; Zhou, T. Revealing dynamic mechanisms of cell fate decisions from single-cell transcriptomic data. Front. Genet. 2019, 10, 1280. [Google Scholar] [CrossRef] [Green Version]
- Peng, G.; Cui, G.; Ke, J.; Jing, N. Using single-cell and spatial transcriptomes to understand stem cell lineage specification during early embryo development. Ann. Rev. Genom. Hum. Genet. 2020, 21, 163–181. [Google Scholar] [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Tritschler, S.; Büttner, M.; Fischer, D.S.; Lange, M.; Bergen, V.; Lickert, H.; Theis, F.J. Concepts and limitations for learning developmental trajectories from single cell genomics. Development 2019, 146, 170506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, S.; Tlusty, T. A comprehensive survey of developmental programs reveals a dearth of tree-like lineage graphs and ubiquitous regeneration. BMC Biol. 2021, 19, 111. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.E.; Weinreb, C.; Collins, Z.M.; Briggs, J.A.; Megason, S.G.; Klein, A.M. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 2018, 360, 981–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebert, S.; Farrell, J.A.; Juliano, C.E. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 2019, 365, eaav9314. [Google Scholar] [CrossRef]
- Minelli, A. A principle of developmental inertia. In Epigenetics: Linking Genotype and Phenotype in Development and Evolution; Hallgrímsson, B., Hall, B.K., Eds.; University of California Press: Berkeley, CA, USA, 2011; pp. 116–133. [Google Scholar]
- Bely, A.E. Evolutionary loss of animal regeneration: Pattern and process. Integr. Comp. Biol. 2010, 50, 515–527. [Google Scholar] [CrossRef] [Green Version]
- Duboule, D. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 1994, 1994 (Suppl. S135), 135–142. [Google Scholar] [CrossRef]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef] [Green Version]
- Gaunt, S.J. The significance of Hox gene collinearity. Int. J. Dev. Biol. 2015, 59, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Barmina, O.; Gonzalo, M.; McIntyre, L.; Kopp, A. Sex- and segment-specific modulation of gene expression profiles in Drosophila. Dev. Biol. 2005, 288, 528–544. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.M.; Baker, B.S. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell 2002, 109, 651–661. [Google Scholar] [CrossRef] [Green Version]
- Belote, J.M.; Baker, B.S. Sex determination in Drosophila melanogaster: Analysis of transformer-2, a sex-transforming locus. Proc. Natl. Acad. Sci. USA 1982, 79, 1568–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopp, A.; Duncan, I.; Carroll, S.B. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 2000, 408, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Godt, D.; Couderc, J.L.; Cramton, S.E.; Laski, F.A. Pattern formation in the limbs of Drosophila: Bric à brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development 1993, 119, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Holguera, I.; Desplan, C. Neuronal specification in space and time. Science 2018, 362, 176–180. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.; Riley-Berger, R.; Harshman, L.; Kopp, A.; Vacha, S.; Nuzhdin, S.; Wayne, M. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 2004, 167, 1791–1799. [Google Scholar] [CrossRef] [Green Version]
- Meiklejohn, C.D.; Parsch, J.; Ranz, J.M.; Hartl, D.L. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 2003, 100, 9894–9899. [Google Scholar] [CrossRef] [Green Version]
- Timberlake, W.E. Temporal and spatial controls of Aspergillus development. Curr. Opin. Genet. Dev. 1991, 1, 351–357. [Google Scholar] [CrossRef]
- Minelli, A. Larva nuova, adulto antico. Ist. Lomb. Accad. Sci. Lett. Rend. Cl. Sci. Mat. Nat. 2008, 139, 223–229. [Google Scholar]
- Zverkov, O.A.; Mikhailov, K.V.; Isaev, S.V.; Rusin, L.Y.; Popova, O.V.; Logacheva, M.D.; Penin, A.A.; Moroz, L.L.; Panchin, Y.V.; Lyubetsky, V.A.; et al. Dicyemida and Orthonectida: Two stories of body plan simplification. Front. Genet. 2019, 10, 443. [Google Scholar] [CrossRef]
- Neves, R.C.; Sørensen, K.J.K.; Kristensen, R.M.; Wanninger, A. Cycliophoran dwarf males break the rule: High complexity with low cell numbers. Biol. Bull. 2009, 217, 2–5. [Google Scholar] [CrossRef] [Green Version]
- Egger, B.; Lapraz, F.; Tomiczek, B.; Müller, S.; Dessimoz, C.; Girstmair, J.; Škunca, N.; Rawlinson, K.A.; Cameron, C.B.; Beli, E.; et al. A transcriptomic-phylogenomic analysis of the evolutionary relationships of flatworms. Curr. Biol. 2015, 25, 1347–1353. [Google Scholar] [CrossRef] [Green Version]
- Vorel, J.; Cwiklinski, K.; Roudnický, P.; Ilgová, J.; Jedličková, L.; Dalton, J.P.; Mikeš, L.; Gelnar, M.; Kašný, M. Eudiplozoon nipponicum (Monogenea, Diplozoidae) and its adaptation to haematophagy as revealed by transcriptome and secretome profiling. BMC Genom. 2021, 22, 274. [Google Scholar] [CrossRef] [PubMed]
- Pomaznoy, M.Y.; Logacheva, M.D.; Young, N.D.; Penin, A.A.; Ershov, N.I.; Katokhin, A.V.; Mordvinov, V.A. Whole transcriptome profiling of adult and infective stages of the trematode Opisthorchis felineus. Parasitol. Int. 2016, 65, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Olson, P.D.; Zarowiecki, M.; James, K.; Baillie, A.; Bartl, G.; Burchell, P.; Chellappoo, A.; Jarero, F.; Tan, L.Y.; Holroyd, N.; et al. Genome-wide transcriptome profiling and spatial expression analyses identify signals and switches of development in tapeworms. EvoDevo 2018, 9, R991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.-H.; Yang, Y.; Zhang, N.-Z.; Wang, J.-K.; Liu, Y.-J.; Li, L.; Yan, H.-B.; Jia, W.-Z.; Fu, B. Comparative transcriptome analyses of the developmental stages of Taenia multiceps. Front. Vet. Sci. 2021, 8, 677045. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, Z.; Jin, L.; Zhu, Y.; Zhao, L.; Shi, B.; Li, J.; Guo, G.; Guo, B.; McManus, D.P.; et al. Dynamic changes in the global transcriptome and microRNAome reveal complex miRNA-mRNA regulation in early stages of the bi-directional development of Echinococcus granulosus protoscoleces. Front. Microbiol. 2020, 11, 654. [Google Scholar] [CrossRef]
- Alberti, C.; Manzenreither, R.A.; Sowemimo, I.; Burkard, T.R.; Wang, J.; Mahofsky, K.; Ameres, S.L.; Cochella, L. Cell-type specific sequencing of microRNAs from complex animal tissues. Nat. Methods 2018, 15, 283–289. [Google Scholar] [CrossRef]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Galagali, H.; Kim, J.K. The multifaceted roles of microRNAs in differentiation. Curr. Opin. Cell Biol. 2020, 67, 118–140. [Google Scholar] [CrossRef]
- Liu, Z.C.; Ambros, V. Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes Dev. 1989, 3, 2039–2049. [Google Scholar] [CrossRef]
- Hammell, C.M.; Lubin, I.; Boag, P.R.; Blackwell, T.K.; Ambros, V. Nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 2009, 136, 926–938. [Google Scholar] [CrossRef] [Green Version]
- Bethke, A.; Fielenbach, N.; Wang, Z.; Mangelsdorf, D.J.; Antebi, A. Nuclear hormone receptor regulation of microRNAs control development progression. Science 2009, 324, 95–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karp, X.; Ambros, V. The developmental timing regulator HBL-1 modulates the dauer formation decision in Caenorhabditis elegans. Genetics 2011, 187, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halfon, M.S. Perspectives on gene regulatory network evolution. Trends Genet. 2017, 33, 436–447. [Google Scholar] [CrossRef] [PubMed]
- DiFrisco, J.; Jaeger, J. Beyond networks: Mechanism and process in evo-devo. Biol. Philos. 2019, 34, 54. [Google Scholar] [CrossRef]
- Von Dassow, G.; Munro, E. Modularity in animal development and evolution: Elements of a conceptual framework for EvoDevo. J. Exp. Zool. B Mol. Dev. Evol. 1999, 285, 307–325. [Google Scholar] [CrossRef]
- True, J.R.; Haag, E.S. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 2001, 3, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Haag, E.S.; True, J.R. Developmental system drift. In Evolutionary Developmental Biology; Nuño de la Rosa, L., Müller, G.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 99–110. [Google Scholar] [CrossRef]
- Minelli, A. On the nature of organs and organ systems—A chapter in the history and philosophy of biology. Front. Evol. Dev. Biol. 2021, 9, 745564. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Benitez, M.; Davila, E.B.; Chaos, A.; Espinosa-Soto, C.; Padilla-Longoria, P. Gene regulatory network models for plant development. Curr. Opin. Plant Biol. 2007, 10, 83–91. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Shibata, M.; Rymen, B.; Iwase, A.; Bågman, A.M.; Watt, L.; Coleman, D.; Favero, D.S.; Takahashi, T.; Ahnert, S.E.; et al. A gene regulatory network for cellular reprogramming in plant regeneration. Plant Cell Physiol. 2018, 59, 765–777. [Google Scholar] [CrossRef]
- Pires, N.D.; Yi, K.; Breuninger, H.; Catarino, B.; Menand, B.; Dolan, L. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 9571–9576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das Gupta, M.; Tsiantis, M. Gene networks and the evolution of plant morphology. Curr. Opin. Plant Biol. 2018, 45, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Castelli-Gair Hombría, J.; Bovolenta, P. (Eds.) Organogenetic Gene Networks. Genetic Control of Organ Formation; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Zakhvatkin, A.A. The Comparative Embryology of the Low Invertebrates. Sources and Method of the Origin of Metazoan Development; Soviet Science University Press: Moscow, Russia, 1949. [Google Scholar]
- Mikhailov, K.V.; Konstantinova, A.V.; Nikitin, M.A.; Troshin, P.V.; Rusin, L.Y.; Lyubetsky, V.A.; Panchin, Y.V.; Mylnikov, A.P.; Moroz, L.L.; Kumar, S.; et al. The origin of Metazoa: A transition from temporal to spatial cell differentiation. BioEssays 2009, 31, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Valentine, J.W. On the Origin of Phyla; University of Chicago Press: Chicago, IL, USA, 2004. [Google Scholar]
- Brunet, T.; King, N. The origin of animal multicellularity and cell differentiation. Dev. Cell 2017, 43, 124–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebé-Pedrós, A.; Degnan, B.M.; Ruiz-Trillo, I. The origin of Metazoa: A unicellular perspective. Nat. Rev. Genet. 2017, 18, 498–512. [Google Scholar] [CrossRef]
- Leadbeater, B.S.C. The Choanoflagellates: Evolution, Biology and Ecology; Cambridge University Press: Cambridge, UK, 2015. [Google Scholar]
- Nijhout, H.F. Development and evolution of adaptive polyphenism. Evol. Dev. 2003, 5, 9–18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minelli, A.; Valero-Gracia, A. Spatially and Temporally Distributed Complexity—A Refreshed Framework for the Study of GRN Evolution. Cells 2022, 11, 1790. https://doi.org/10.3390/cells11111790
Minelli A, Valero-Gracia A. Spatially and Temporally Distributed Complexity—A Refreshed Framework for the Study of GRN Evolution. Cells. 2022; 11(11):1790. https://doi.org/10.3390/cells11111790
Chicago/Turabian StyleMinelli, Alessandro, and Alberto Valero-Gracia. 2022. "Spatially and Temporally Distributed Complexity—A Refreshed Framework for the Study of GRN Evolution" Cells 11, no. 11: 1790. https://doi.org/10.3390/cells11111790
APA StyleMinelli, A., & Valero-Gracia, A. (2022). Spatially and Temporally Distributed Complexity—A Refreshed Framework for the Study of GRN Evolution. Cells, 11(11), 1790. https://doi.org/10.3390/cells11111790





