Reovirus Activated Cell Death Pathways
Abstract
1. Introduction
2. ReoV Structure, Genome, and Replication
3. ReoV and Innate Immunity
4. ReoV-Induced Non-Necroptotic Cell Death Pathways
4.1. Apoptosis
4.2. Autophagy
4.3. Pyroptosis
5. ReoV and Necroptosis
6. Implications for ReoV Oncolysis
7. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dermody, T.S.; Parker, J.S.; Barbara, S. Orthoreoviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 1304–1346. [Google Scholar]
- Bouziat, R.; Hinterleitner, R.; Brown, J.J.; Stencel-Baerenwald, J.E.; Ikizler, M.; Mayassi, T.; Meisel, M.; Kim, S.M.; Discepolo, V.; Pruijssers, A.J.; et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 2017, 356, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, D.M.; Aravamudhan, P.; Dietrich, M.H.; Stehle, T.; Dermody, T.S. Reovirus Neurotropism and Virulence Are Dictated by Sequences in the Head Domain of the Viral Attachment Protein. J. Virol. 2018, 92, e00974-18. [Google Scholar] [CrossRef] [PubMed]
- Gauvin, L.; Bennett, S.; Liu, H.; Hakimi, M.; Schlossmacher, M.; Majithia, J.; Brown, E.G. Respiratory infection of mice with mammalian reoviruses causes systemic infection with age and strain dependent pneumonia and encephalitis. Virol. J. 2013, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Konopka-Anstadt, J.L.; Mainou, B.A.; Sutherland, D.M.; Sekine, Y.; Strittmatter, S.M.; Dermody, T.S. The Nogo receptor NgR1 mediates infection by mammalian reovirus. Cell Host Microbe 2014, 15, 681–691. [Google Scholar] [CrossRef] [PubMed]
- DeBiasi, R.L.; Robinson, B.A.; Sherry, B.; Bouchard, R.; Brown, R.D.; Rizeq, M.; Long, C.; Tyler, K.L. Caspase inhibition protects against reovirus-induced myocardial injury in vitro and in vivo. J. Virol. 2004, 78, 11040–11050. [Google Scholar] [CrossRef] [PubMed]
- Sherry, B.; Baty, C.J.; Blum, M.A. Reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than generation of infectious virus in cardiac myocytes. J. Virol. 1996, 70, 6709–6715. [Google Scholar] [CrossRef]
- Coffey, M.C.; Strong, J.E.; Forsyth, P.A.; Lee, P.W.K. Reovirus Therapy of Tumors with Activated Ras Pathway. Science 1998, 282, 1332–1334. [Google Scholar] [CrossRef]
- Phillips, M.B.; Stuart, J.D.; Rodriguez Stewart, R.M.; Berry, J.T.; Mainou, B.A.; Boehme, K.W. Current understanding of reovirus oncolysis mechanisms. Oncolytic Virotherapy 2018, 7, 53–63. [Google Scholar] [CrossRef]
- Strong, J.E.; Tang, D.; Lee, P.W. Evidence that the epidermal growth factor receptor on host cells confers reovirus infection efficiency. Virology 1993, 197, 405–411. [Google Scholar] [CrossRef]
- Marelli, G.; Howells, A.; Lemoine, N.R.; Wang, Y. Oncolytic Viral Therapy and the Immune System: A Double-Edged Sword Against Cancer. Front. Immunol. 2018, 9, 866. [Google Scholar] [CrossRef]
- Chaurasiya, S.; Fong, Y.; Warner, S.G. Oncolytic Virotherapy for Cancer: Clinical Experience. Biomedicines 2021, 9, 419. [Google Scholar] [CrossRef]
- Gong, J.; Sachdev, E.; Mita, A.C.; Mita, M.M. Clinical development of reovirus for cancer therapy: An oncolytic virus with immune-mediated antitumor activity. World J. Methodol. 2016, 6, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.K.; Danthi, P. Reovirus activates a caspase-independent cell death pathway. MBio 2013, 4, e00178-00113. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, S.; Rall, G.F. Benefits and Perils of Necroptosis in Influenza Virus Infection. J. Virol. 2020, 94, e01101-01119. [Google Scholar] [CrossRef] [PubMed]
- Shubina, M.; Tummers, B.; Boyd, D.F.; Zhang, T.; Yin, C.; Gautam, A.; Guo, X.-z.J.; Rodriguez, D.A.; Kaiser, W.J.; Vogel, P.; et al. Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J. Exp. Med. 2020, 217, e20191259. [Google Scholar] [CrossRef]
- Pasparakis, M.; Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar] [CrossRef]
- Snyder, A.G.; Hubbard, N.W.; Messmer, M.N.; Kofman, S.B.; Hagan, C.E.; Orozco, S.L.; Chiang, K.; Daniels, B.P.; Baker, D.; Oberst, A. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci. Immunol. 2019, 4, eaaw2004. [Google Scholar] [CrossRef]
- Danthi, P.; Holm, G.H.; Stehle, T.; Dermody, T.S. Reovirus receptors, cell entry, and proapoptotic signaling. Adv. Exp. Med. Biol. 2013, 790, 42–71. [Google Scholar] [CrossRef]
- Duncan, R.; Horne, D.; Cashdollar, L.W.; Joklik, W.K.; Lee, P.W. Identification of conserved domains in the cell attachment proteins of the three serotypes of reovirus. Virology 1990, 174, 399–409. [Google Scholar] [CrossRef]
- Nibert, M.L.; Dermody, T.S.; Fields, B.N. Structure of the reovirus cell-attachment protein: A model for the domain organization of sigma 1. J. Virol. 1990, 64, 2976–2989. [Google Scholar] [CrossRef]
- Attoui, H.; Biagini, P.; Stirling, J.; Mertens, P.P.; Cantaloube, J.F.; Meyer, A.; de Micco, P.; de Lamballerie, X. Sequence characterization of Ndelle virus genome segments 1, 5, 7, 8, and 10: Evidence for reassignment to the genus Orthoreovirus, family Reoviridae. Biochem. Biophys. Res. Commun. 2001, 287, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, K.M.; Nibert, M.L.; Harrison, S.C. Structure of the reovirus core at 3.6?Å resolution. Nature 2000, 404, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Broering, T.J.; Kim, J.; Higgins, D.E.; Nibert, M.L. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 2002, 76, 4483–4496. [Google Scholar] [CrossRef] [PubMed]
- Tenorio, R.; Fernandez de Castro, I.; Knowlton, J.J.; Zamora, P.F.; Lee, C.H.; Mainou, B.A.; Dermody, T.S.; Risco, C. Reovirus sigmaNS and muNS Proteins Remodel the Endoplasmic Reticulum to Build Replication Neo-Organelles. mBio 2018, 9, e01253-18. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.M.; Goral, M.I.; Hazelton, P.R.; Baer, G.S.; Rodgers, S.E.; Brown, E.G.; Coombs, K.M.; Dermody, T.S. Reovirus sigmaNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 2001, 75, 1459–1475. [Google Scholar] [CrossRef]
- Starnes, M.C.; Joklik, W.K. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology 1993, 193, 356–366. [Google Scholar] [CrossRef]
- Chappell, J.D.; Duong, J.L.; Wright, B.W.; Dermody, T.S. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 2000, 74, 8472–8479. [Google Scholar] [CrossRef]
- Roth, A.N.; Aravamudhan, P.; Fernández de Castro, I.; Tenorio, R.; Risco, C.; Dermody, T.S. Ins and Outs of Reovirus: Vesicular Trafficking in Viral Entry and Egress. Trends Microbiol. 2021, 29, 363–375. [Google Scholar] [CrossRef]
- Sutherland, D.M.; Aravamudhan, P.; Dermody, T.S. An Orchestra of Reovirus Receptors: Still Searching for the Conductor. Adv. Virus Res. 2018, 100, 223–246. [Google Scholar] [CrossRef]
- Koehler, M.; Aravamudhan, P.; Guzman-Cardozo, C.; Dumitru, A.C.; Yang, J.; Gargiulo, S.; Soumillion, P.; Dermody, T.S.; Alsteens, D. Glycan-mediated enhancement of reovirus receptor binding. Nat. Commun. 2019, 10, 4460. [Google Scholar] [CrossRef]
- Dietrich, M.H.; Ogden, K.M.; Katen, S.P.; Reiss, K.; Sutherland, D.M.; Carnahan, R.H.; Goff, M.; Cooper, T.; Dermody, T.S.; Stehle, T. Structural Insights into Reovirus sigma1 Interactions with Two Neutralizing Antibodies. J. Virol. 2017, 91, e01621-16. [Google Scholar] [CrossRef] [PubMed]
- Barton, E.S.; Connolly, J.L.; Forrest, J.C.; Chappell, J.D.; Dermody, T.S. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 2001, 276, 2200–2211. [Google Scholar] [CrossRef] [PubMed]
- Maginnis, M.S.; Forrest, J.C.; Kopecky-Bromberg, S.A.; Dickeson, S.K.; Santoro, S.A.; Zutter, M.M.; Nemerow, G.R.; Bergelson, J.M.; Dermody, T.S. Beta1 integrin mediates internalization of mammalian reovirus. J. Virol. 2006, 80, 2760–2770. [Google Scholar] [CrossRef] [PubMed]
- Mainou, B.A.; Dermody, T.S. Transport to late endosomes is required for efficient reovirus infection. J. Virol. 2012, 86, 8346–8358. [Google Scholar] [CrossRef]
- Aravamudhan, P.; Raghunathan, K.; Konopka-Anstadt, J.; Pathak, A.; Sutherland, D.M.; Carter, B.D.; Dermody, T.S. Reovirus uses macropinocytosis-mediated entry and fast axonal transport to infect neurons. PLoS Pathog. 2020, 16, e1008380. [Google Scholar] [CrossRef]
- Maginnis, M.S.; Mainou, B.A.; Derdowski, A.; Johnson, E.M.; Zent, R.; Dermody, T.S. NPXY motifs in the beta1 integrin cytoplasmic tail are required for functional reovirus entry. J. Virol. 2008, 82, 3181–3191. [Google Scholar] [CrossRef]
- Schulz, W.L.; Haj, A.K.; Schiff, L.A. Reovirus uses multiple endocytic pathways for cell entry. J. Virol. 2012, 86, 12665–12675. [Google Scholar] [CrossRef]
- Liemann, S.; Chandran, K.; Baker, T.S.; Nibert, M.L.; Harrison, S.C. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell 2002, 108, 283–295. [Google Scholar] [CrossRef]
- Mainou, B.A.; Dermody, T.S. Src kinase mediates productive endocytic sorting of reovirus during cell entry. J. Virol. 2011, 85, 3203–3213. [Google Scholar] [CrossRef]
- Bujnicki, J.M.; Rychlewski, L. Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda 2 protein. Genome Biol. 2001, 2, 1–6. [Google Scholar] [CrossRef]
- Seliger, L.S.; Zheng, K.; Shatkin, A.J. Complete nucleotide sequence of reovirus L2 gene and deduced amino acid sequence of viral mRNA guanylyltransferase. J. Biol. Chem. 1987, 262, 16289–16293. [Google Scholar] [CrossRef]
- Murray, K.E.; Nibert, M.L. Guanidine hydrochloride inhibits mammalian orthoreovirus growth by reversibly blocking the synthesis of double-stranded RNA. J. Virol. 2007, 81, 4572–4584. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.M.; Mainou, B.A.; Kim, K.S.; Dermody, T.S. Directional release of reovirus from the apical surface of polarized endothelial cells. mBio 2013, 4, e00049-00013. [Google Scholar] [CrossRef] [PubMed]
- Excoffon, K.J.; Guglielmi, K.M.; Wetzel, J.D.; Gansemer, N.D.; Campbell, J.A.; Dermody, T.S.; Zabner, J. Reovirus preferentially infects the basolateral surface and is released from the apical surface of polarized human respiratory epithelial cells. J. Infect. Dis. 2008, 197, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Lemay, G. Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions. Viruses 2018, 10, 671. [Google Scholar] [CrossRef]
- Tenorio, R.; Fernandez de Castro, I.; Knowlton, J.J.; Zamora, P.F.; Sutherland, D.M.; Risco, C.; Dermody, T.S. Function, Architecture, and Biogenesis of Reovirus Replication Neoorganelles. Viruses 2019, 11, 288. [Google Scholar] [CrossRef]
- Roebke, K.E.; Danthi, P. Cell Entry-Independent Role for the Reovirus mu1 Protein in Regulating Necroptosis and the Accumulation of Viral Gene Products. J. Virol. 2019, 93, e00199-19. [Google Scholar] [CrossRef]
- Knowlton, J.J.; Fernandez de Castro, I.; Ashbrook, A.W.; Gestaut, D.R.; Zamora, P.F.; Bauer, J.A.; Forrest, J.C.; Frydman, J.; Risco, C.; Dermody, T.S. The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat. Microbiol. 2018, 3, 481–493. [Google Scholar] [CrossRef]
- Fernández de Castro, I.; Tenorio, R.; Ortega-González, P.; Knowlton, J.J.; Zamora, P.F.; Lee, C.H.; Fernández, J.J.; Dermody, T.S.; Risco, C. A modified lysosomal organelle mediates nonlytic egress of reovirus. J. Cell Biol. 2020, 219, e201910131. [Google Scholar] [CrossRef]
- Koehler, M.; Petitjean, S.J.L.; Yang, J.; Aravamudhan, P.; Somoulay, X.; Lo Giudice, C.; Poncin, M.A.; Dumitru, A.C.; Dermody, T.S.; Alsteens, D. Reovirus directly engages integrin to recruit clathrin for entry into host cells. Nat. Commun. 2021, 12, 2149. [Google Scholar] [CrossRef]
- Gummersheimer, S.L.; Danthi, P. Reovirus Core Proteins λ1 and σ2 Promote Stability of Disassembly Intermediates and Influence Early Replication Events. J. Virol. 2020, 94, 83. [Google Scholar] [CrossRef]
- Brentano, L.; Noah, D.L.; Brown, E.G.; Sherry, B. The reovirus protein mu2, encoded by the M1 gene, is an RNA-binding protein. J. Virol. 1998, 72, 8354–8357. [Google Scholar] [CrossRef] [PubMed]
- Zurney, J.; Kobayashi, T.; Holm, G.H.; Dermody, T.S.; Sherry, B. Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J. Virol. 2009, 83, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Lanoie, D.; Boudreault, S.; Bisaillon, M.; Lemay, G. How Many Mammalian Reovirus Proteins are involved in the Control of the Interferon Response? Pathogens 2019, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chandran, K.; Nibert, M.L.; Harrison, S.C. Reovirus mu1 structural rearrangements that mediate membrane penetration. J. Virol. 2006, 80, 12367–12376. [Google Scholar] [CrossRef]
- Coffey, C.M.; Sheh, A.; Kim, I.S.; Chandran, K.; Nibert, M.L.; Parker, J.S. Reovirus outer capsid protein micro1 induces apoptosis and associates with lipid droplets, endoplasmic reticulum, and mitochondria. J. Virol. 2006, 80, 8422–8438. [Google Scholar] [CrossRef]
- Lee, P.W.; Hayes, E.C.; Joklik, W.K. Protein sigma 1 is the reovirus cell attachment protein. Virology 1981, 108, 156–163. [Google Scholar] [CrossRef]
- Barton, E.S.; Forrest, J.C.; Connolly, J.L.; Chappell, J.D.; Liu, Y.; Schnell, F.J.; Nusrat, A.; Parkos, C.A.; Dermody, T.S. Junction adhesion molecule is a receptor for reovirus. Cell 2001, 104, 441–451. [Google Scholar] [CrossRef]
- Poggioli, G.J.; Keefer, C.; Connolly, J.L.; Dermody, T.S.; Tyler, K.L. Reovirus-induced G(2)/M cell cycle arrest requires sigma1s and occurs in the absence of apoptosis. J. Virol. 2000, 74, 9562–9570. [Google Scholar] [CrossRef]
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; García-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef]
- Thirukkumaran, C.; Shi, Z.-Q.; Thirukkumaran, P.; Luider, J.; Kopciuk, K.; Spurrell, J.; Elzinga, K.; Morris, D. PUMA and NF-kB Are Cell Signaling Predictors of Reovirus Oncolysis of Breast Cancer. PLoS ONE 2017, 12, e0168233. [Google Scholar] [CrossRef] [PubMed]
- Maitra, R.; Augustine, T.; Dayan, Y.; Chandy, C.; Coffey, M.; Goel, S. Toll like receptor 3 as an immunotherapeutic target for KRAS mutated colorectal cancer. Oncotarget 2017, 8, 35138–35153. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, K.H.; Richardson-Burns, S.; Alexopoulou, L.; Tyler, K.L.; Flavell, R.A.; Oldstone, M.B. Does Toll-like receptor 3 play a biological role in virus infections? Virology 2004, 322, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Antiviral Actions of Interferons. J. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Goubau, D.; Schlee, M.; Deddouche, S.; Pruijssers, A.J.; Zillinger, T.; Goldeck, M.; Schuberth, C.; Van der Veen, A.G.; Fujimura, T.; Rehwinkel, J.; et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 2014, 514, 372–375. [Google Scholar] [CrossRef]
- Abad, A.T.; Danthi, P. Recognition of Reovirus RNAs by the Innate Immune System. Viruses 2020, 12, 667. [Google Scholar] [CrossRef]
- Holm, G.H.; Zurney, J.; Tumilasci, V.; Leveille, S.; Danthi, P.; Hiscott, J.; Sherry, B.; Dermody, T.S. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 2007, 282, 21953–21961. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Berger, A.K.; Hiller, B.E.; Thete, D.; Snyder, A.J.; Perez, E., Jr.; Upton, J.W.; Danthi, P. Viral RNA at Two Stages of Reovirus Infection Is Required for the Induction of Necroptosis. J. Virol. 2017, 91, e02404-16. [Google Scholar] [CrossRef]
- Johansson, C.; Wetzel, J.D.; He, J.; Mikacenic, C.; Dermody, T.S.; Kelsall, B.L. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J. Exp. Med. 2007, 204, 1349–1358. [Google Scholar] [CrossRef]
- Zhang, Z.; Kim, T.; Bao, M.; Facchinetti, V.; Jung, S.Y.; Ghaffari, A.A.; Qin, J.; Cheng, G.; Liu, Y.J. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 2011, 34, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Mitoma, H.; Hanabuchi, S.; Kim, T.; Bao, M.; Zhang, Z.; Sugimoto, N.; Liu, Y.-J. The DHX33 RNA Helicase Senses Cytosolic RNA and Activates the NLRP3 Inflammasome. Immunity 2013, 39, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Yamazaki, T.; Koshiba, T.; Yanagi, Y. Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc. Natl. Acad. Sci. USA 2013, 110, 17963–17968. [Google Scholar] [CrossRef] [PubMed]
- Danthi, P.; Pruijssers, A.J.; Berger, A.K.; Holm, G.H.; Zinkel, S.S.; Dermody, T.S. Bid Regulates the Pathogenesis of Neurotropic Reovirus. PLoS Pathog. 2010, 6, e1000980. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, A.; Minze, L.J.; Li, X.C.; Zhang, Z. TRIM29 Negatively Regulates the Type I IFN Production in Response to RNA Virus. J. Immunol. 2018, 201, 183–192. [Google Scholar] [CrossRef]
- Xing, J.; Weng, L.; Yuan, B.; Wang, Z.; Jia, L.; Jin, R.; Lu, H.; Li, X.C.; Liu, Y.J.; Zhang, Z. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat. Immunol. 2016, 17, 1373–1380. [Google Scholar] [CrossRef]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell. Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef]
- Danthi, P. Viruses and the Diversity of Cell Death. Annu. Rev. Virol. 2016, 3, 533–553. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Chawla-Sarkar, M.; Lindner, D.J.; Liu, Y.F.; Williams, B.R.; Sen, G.C.; Silverman, R.H.; Borden, E.C. Apoptosis and interferons: Role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 2003, 8, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.; Tyler, K.L. Apoptosis in animal models of virus-induced disease. Nat. Rev. Microbiol. 2009, 7, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Rajan, J.V.; Rodriguez, D.; Miao, E.A.; Aderem, A. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J. Virol. 2011, 85, 4167–4172. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Jiang, K.; Zhang, X.; Zhang, M.; Zhou, Z.; Hu, M.; Yang, R.; Sun, C.; Wu, Y. Avian reovirus triggers autophagy in primary chicken fibroblast cells and Vero cells to promote virus production. Arch. Virol. 2012, 157, 661–668. [Google Scholar] [CrossRef]
- Kemp, V.; Dautzenberg, I.J.C.; Limpens, R.W.; van den Wollenberg, D.J.M.; Hoeben, R.C. Oncolytic Reovirus Infection Is Facilitated by the Autophagic Machinery. Viruses 2017, 9, 266. [Google Scholar] [CrossRef]
- Knowlton, J.J.; Dermody, T.S.; Holm, G.H. Apoptosis Induced by Mammalian Reovirus Is Beta Interferon (IFN) Independent and Enhanced by IFN Regulatory Factor 3- and NF-κB-Dependent Expression of Noxa. J. Virol. 2012, 86, 1650–1660. [Google Scholar] [CrossRef]
- Danthi, P.; Kobayashi, T.; Holm, G.H.; Hansberger, M.W.; Abel, T.W.; Dermody, T.S. Reovirus Apoptosis and Virulence Are Regulated by Host Cell Membrane Penetration Efficiency. J. Virol. 2008, 82, 161. [Google Scholar] [CrossRef]
- Wisniewski, M.L.; Werner, B.G.; Hom, L.G.; Anguish, L.J.; Coffey, C.M.; Parker, J.S.L. Reovirus Infection or Ectopic Expression of Outer Capsid Protein μ1 Induces Apoptosis Independently of the Cellular Proapoptotic Proteins Bax and Bak. J. Virol. 2011, 85, 296–304. [Google Scholar] [CrossRef]
- Brown, J.J.; Short, S.P.; Stencel-Baerenwald, J.; Urbanek, K.; Pruijssers, A.J.; McAllister, N.; Ikizler, M.; Taylor, G.; Aravamudhan, P.; Khomandiak, S.; et al. Reovirus-Induced Apoptosis in the Intestine Limits Establishment of Enteric Infection. J. Virol. 2018, 92, e02062-17. [Google Scholar] [CrossRef]
- Danthi, P.; Guglielmi, K.M.; Kirchner, E.; Mainou, B.; Stehle, T.; Dermody, T.S. From touchdown to transcription: The reovirus cell entry pathway. Curr. Top. Microbiol. Immunol. 2010, 343, 91–119. [Google Scholar] [CrossRef]
- Duan, S.; Cheng, J.; Li, C.; Yu, L.; Zhang, X.; Jiang, K.; Wang, Y.; Xu, J.; Wu, Y. Autophagy inhibitors reduce avian-reovirus-mediated apoptosis in cultured cells and in chicken embryos. Arch. Virol. 2015, 160, 1679–1685. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Tian, S.; Pan, Y.; Li, W.; Wang, Q.; Tang, Y.; Yu, T.; Wu, X.; Shi, Y.; Ma, P.; et al. Pyroptosis: A new frontier in cancer. Biomed. Pharm. 2020, 121, 109595. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94. [Google Scholar] [CrossRef]
- Yu, P.; Zhang, X.; Liu, N.; Tang, L.; Peng, C.; Chen, X. Pyroptosis: Mechanisms and diseases. Signal. Transduct. Target. Ther. 2021, 6, 128. [Google Scholar] [CrossRef]
- Beckham, J.D.; Tuttle, K.D.; Tyler, K.L. Caspase-3 activation is required for reovirus-induced encephalitis in vivo. J. Neurovirol. 2010, 16, 306–317. [Google Scholar] [CrossRef][Green Version]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Thorburn, A. Autophagy and disease. J. Biol. Chem. 2018, 293, 5425–5430. [Google Scholar] [CrossRef]
- Chiu, H.C.; Richart, S.; Lin, F.Y.; Hsu, W.L.; Liu, H.J. The interplay of reovirus with autophagy. Biomed. Res. Int. 2014, 2014, 483657. [Google Scholar] [CrossRef]
- Niu, X.; Zhang, C.; Wang, Y.; Guo, M.; Ruan, B.; Wang, X.; Wu, T.; Zhang, X.; Wu, Y. Autophagy induced by avian reovirus enhances viral replication in chickens at the early stage of infection. BMC Vet. Res. 2019, 15, 173. [Google Scholar] [CrossRef]
- Thirukkumaran, C.M.; Shi, Z.Q.; Luider, J.; Kopciuk, K.; Gao, H.; Bahlis, N.; Neri, P.; Pho, M.; Stewart, D.; Mansoor, A.; et al. Reovirus modulates autophagy during oncolysis of multiple myeloma. Autophagy 2013, 9, 413–414. [Google Scholar] [CrossRef] [PubMed]
- Jiffry, J.; Thavornwatanayong, T.; Rao, D.; Fogel, E.J.; Saytoo, D.; Nahata, R.; Guzik, H.; Chaudhary, I.; Augustine, T.; Goel, S.; et al. Oncolytic Reovirus (pelareorep) Induces Autophagy in KRAS-mutated Colorectal Cancer. Clin. Cancer Res. 2021, 27, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host cell death and inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Burdette, B.E.; Esparza, A.N.; Zhu, H.; Wang, S. Gasdermin D in pyroptosis. Acta Pharm. Sin. B 2021, 11, 2768–2782. [Google Scholar] [CrossRef]
- Zhu, S.; Ding, S.; Wang, P.; Wei, Z.; Pan, W.; Palm, N.W.; Yang, Y.; Yu, H.; Li, H.-B.; Wang, G.; et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 2017, 546, 667–670. [Google Scholar] [CrossRef]
- Degterev, A.; Hitomi, J.; Germscheid, M.; Ch’en, I.L.; Korkina, O.; Teng, X.; Abbott, D.; Cuny, G.D.; Yuan, C.; Wagner, G.; et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008, 4, 313–321. [Google Scholar] [CrossRef]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed Lineage Kinase Domain-like Protein Mediates Necrosis Signaling Downstream of RIP3 Kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef]
- Weinlich, R.; Oberst, A.; Beere, H.M.; Green, D.R. Necroptosis in development, inflammation and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 127–136. [Google Scholar] [CrossRef]
- Upton, J.W.; Shubina, M.; Balachandran, S. RIPK3-driven cell death during virus infections. Immunol. Rev. 2017, 277, 90–101. [Google Scholar] [CrossRef]
- Roebke, K.E.; Guo, Y.; Parker, J.S.L.; Danthi, P. Reovirus σ3 protein limits interferon expression and cell death induction. J. Virol. 2020, 94, 83. [Google Scholar] [CrossRef]
- Kominsky, D.J.; Bickel, R.J.; Tyler, K.L. Reovirus-induced apoptosis requires mitochondrial release of Smac/DIABLO and involves reduction of cellular inhibitor of apoptosis protein levels. J. Virol. 2002, 76, 11414–11424. [Google Scholar] [CrossRef] [PubMed]
- Annibaldi, A.; Wicky John, S.; Vanden Berghe, T.; Swatek, K.N.; Ruan, J.; Liccardi, G.; Bianchi, K.; Elliott, P.R.; Choi, S.M.; Van Coillie, S.; et al. Ubiquitin-Mediated Regulation of RIPK1 Kinase Activity Independent of IKK and MK2. Mol. Cell 2018, 69, 566–580.e565. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.J.; Ingram, J.P.; Ragan, K.B.; Nogusa, S.; Boyd, D.F.; Benitez, A.A.; Sridharan, H.; Kosoff, R.; Shubina, M.; Landsteiner, V.J.; et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe 2016, 20, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yin, C.; Boyd, D.F.; Quarato, G.; Ingram, J.P.; Shubina, M.; Ragan, K.B.; Ishizuka, T.; Crawford, J.C.; Tummers, B.; et al. Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020, 180, 1115–1129.e1113. [Google Scholar] [CrossRef] [PubMed]
- Van Hoecke, L.; Van Lint, S.; Roose, K.; Van Parys, A.; Vandenabeele, P.; Grooten, J.; Tavernier, J.; De Koker, S.; Saelens, X. Treatment with mRNA coding for the necroptosis mediator MLKL induces antitumor immunity directed against neo-epitopes. Nat. Commun. 2018, 9, 3417. [Google Scholar] [CrossRef]
- Huang, Z.; Wu, S.-Q.; Liang, Y.; Zhou, X.; Chen, W.; Li, L.; Wu, J.; Zhuang, Q.; Chen, C.a.; Li, J.; et al. RIP1/RIP3 Binding to HSV-1 ICP6 Initiates Necroptosis to Restrict Virus Propagation in Mice. Cell Host Microbe 2015, 17, 229–242. [Google Scholar] [CrossRef]
- Yue, Z.; Shatkin, A.J. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 1997, 234, 364–371. [Google Scholar] [CrossRef]
- Duncan, M.R.; Stanish, S.M.; Cox, D.C. Differential sensitivity of normal and transformed human cells to reovirus infection. J. Virol. 1978, 28, 444–449. [Google Scholar] [CrossRef]
- Shmulevitz, M.; Pan, L.Z.; Garant, K.; Pan, D.; Lee, P.W. Oncogenic Ras promotes reovirus spread by suppressing IFN-beta production through negative regulation of RIG-I signaling. Cancer Res. 2010, 70, 4912–4921. [Google Scholar] [CrossRef]
- Cristi, F.; Gutiérrez, T.; Hitt, M.M.; Shmulevitz, M. Genetic Modifications That Expand Oncolytic Virus Potency. Front. Mol. Biosci. 2022, 9, 831091. [Google Scholar] [CrossRef]
- Shmulevitz, M.; Gujar, S.A.; Ahn, D.-G.; Mohamed, A.; Lee, P.W.K. Reovirus variants with mutations in genome segments S1 and L2 exhibit enhanced virion infectivity and superior oncolysis. J. Virol. 2012, 86, 7403–7413. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Maitra, R.; Goel, S. Multimodal immune activation abilities and characteristics of reovirus. Am. J. Transl. Res. 2021, 13, 14176–14185. [Google Scholar] [PubMed]
- Goel, S.; Ocean, A.J.; Parakrama, R.Y.; Ghalib, M.H.; Chaudhary, I.; Shah, U.; Viswanathan, S.; Kharkwal, H.; Coffey, M.; Maitra, R. Elucidation of Pelareorep Pharmacodynamics in A Phase I Trial in Patients with KRAS-Mutated Colorectal Cancer. Mol. Cancer Ther. 2020, 19, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Li, L.; Jiang, M.; Qi, L.; Wu, Y.; Song, D.; Gan, J.; Li, Y.; Bai, Y. Pyroptosis, a new bridge to tumor immunity. Cancer Sci. 2021, 112, 3979–3994. [Google Scholar] [CrossRef]
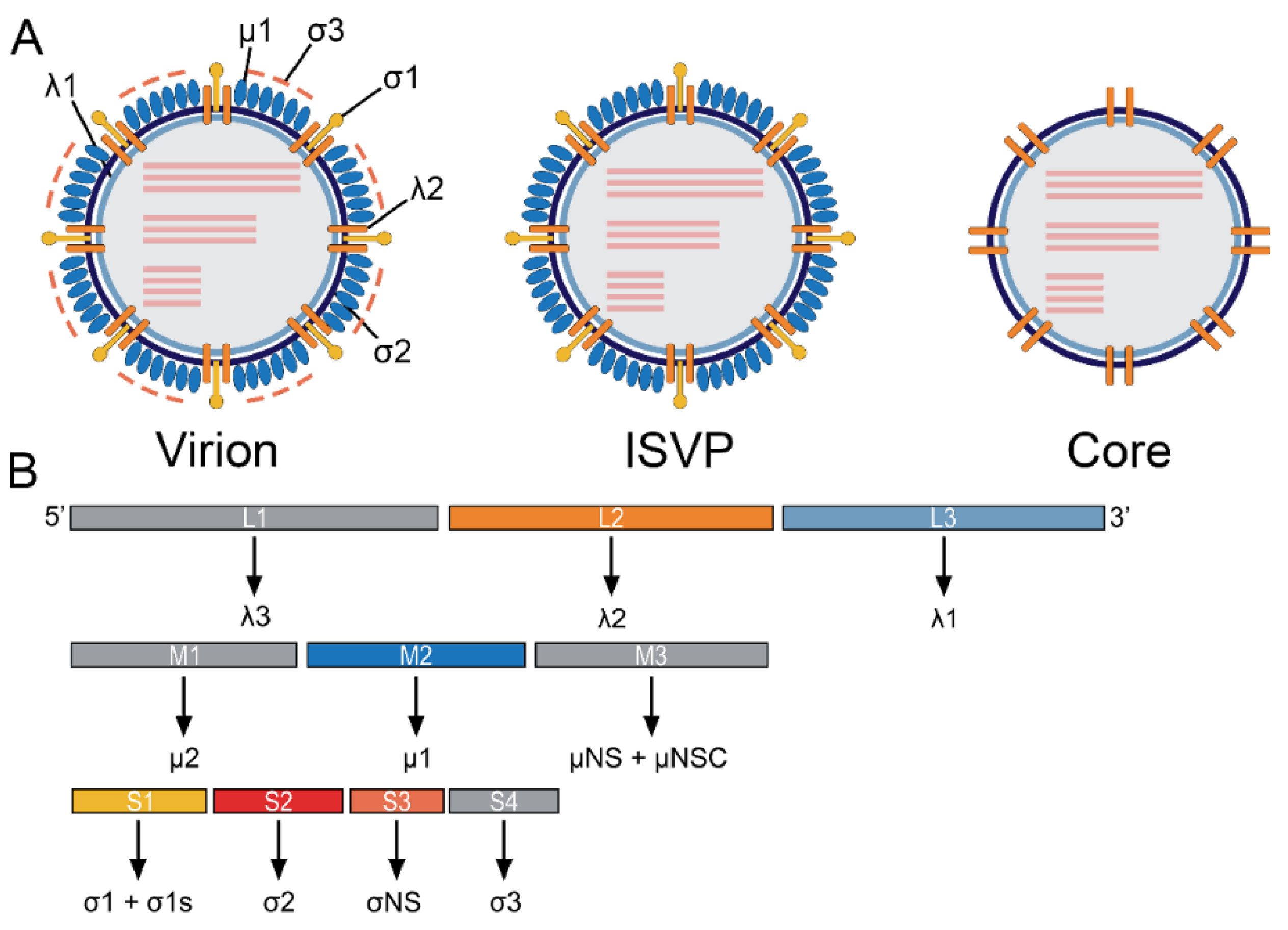
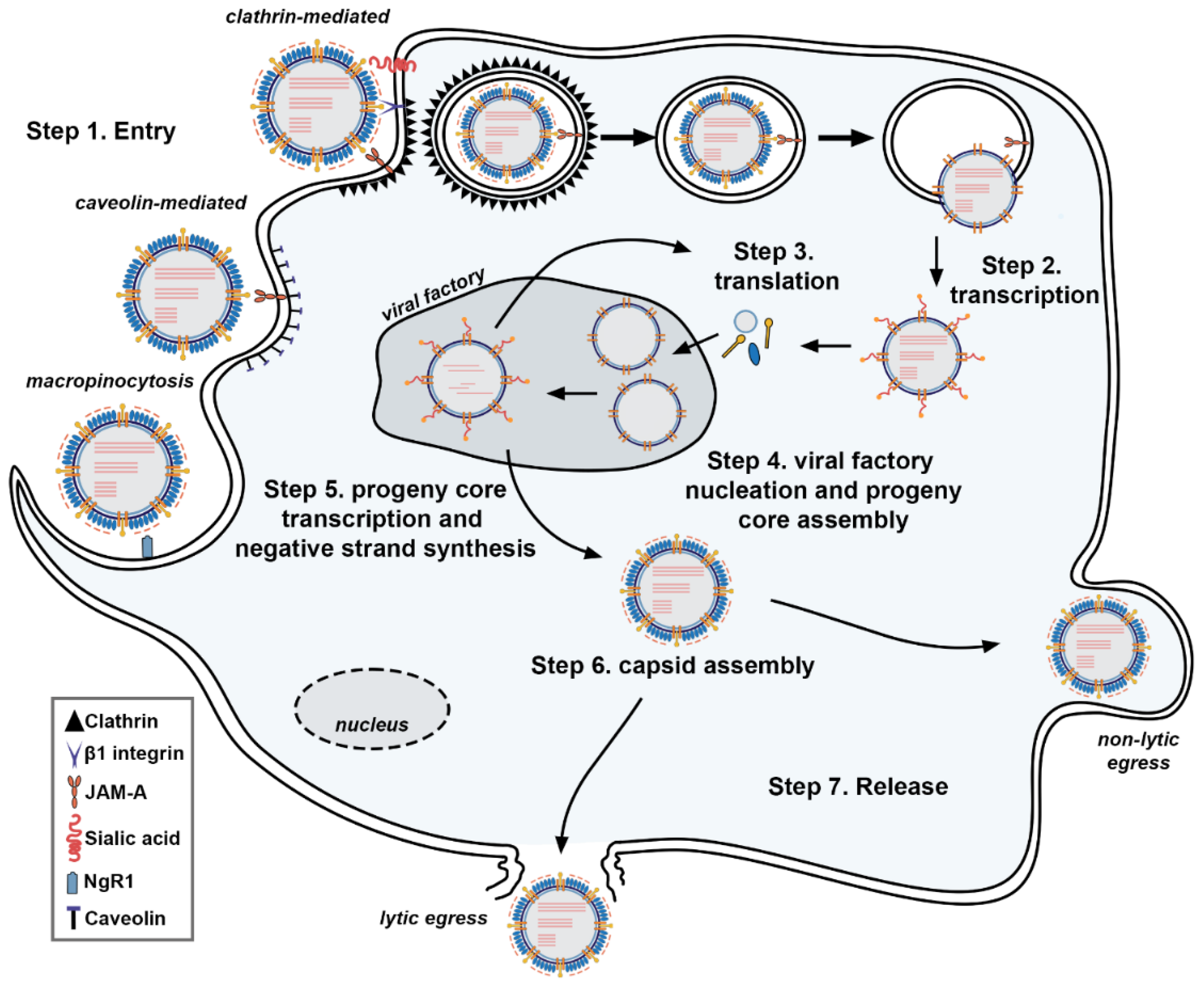
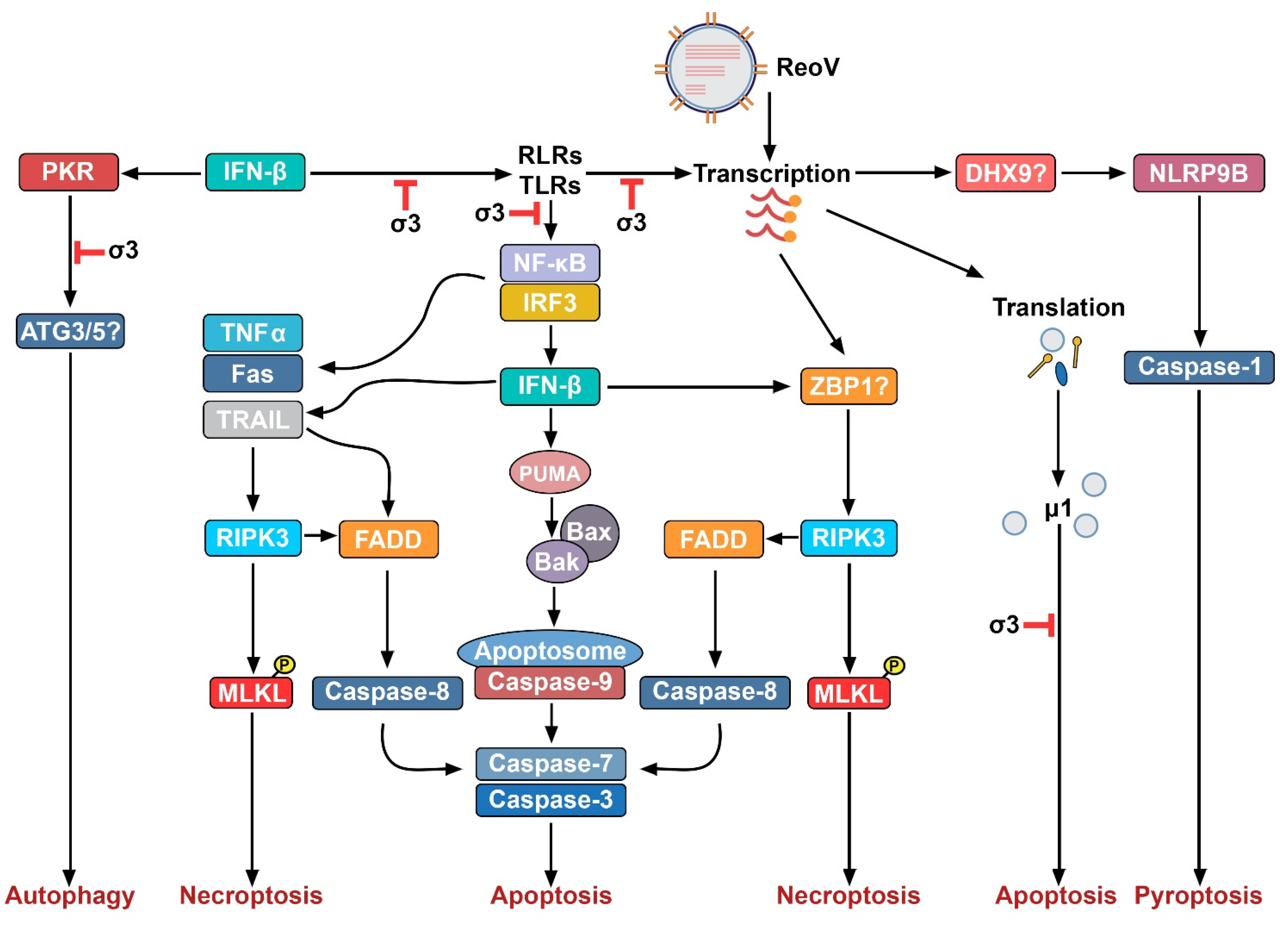
| Encoded Protein | Genome Segment | Role in Viral Life Cycle | Role in the Immune Response | Location | Reference |
|---|---|---|---|---|---|
| λ3 | L1 | RNA-dependent RNA polymerase | Unknown | inner capsid | [27] |
| λ2 | L2 | Capping (methyltransferase and guanylyltransferase activity) Forms interactions with β1 integrin on host cell Forms a channel for the export of viral RNA | Unknown | inner capsid | [41,51] |
| λ1 | L3 | Possible helicase/NTPase Forms the viral core | Unknown | inner capsid | [52] |
| μ2 | M1 | RNA binding NTPase RNA triphosphatase Associates with host microtubules to aid in viral factory formation | Inhibits interferon signaling | inner capsid | [24,53,54,55] |
| μ1 (cleaved into μ1C and μ1N) | M2 | Forms pores in endosomes | Induces apoptosis | outer capsid | [56,57] |
| μNS + μNSC | M3 | Forms viral factories Provides scaffolding for progeny core assembly | Imhibits IRF3 signaling | non-structural | [26,55] |
| σ1 + σ1s | S1 | σ1 binds to host cell receptor such as JAM-A Glycosidase | σ1 binds host cell σ1s can induce cell cycle arrest | σ1 = outer capsid σ1s = non-structural | [55,58,59,60] |
| σ2 | S2 | Interacts with λ1 to form the viral core dsRNA binding | Unknown | inner capsid | [28,45,61] |
| σNS | S4 | RNA binding Viral factory formation May be involved in genome packaging | Unknown | non-structrual | [31,62,63] |
| σ3 | S3 | dsRNA binding May mediate binding to NgR1 | Blocks PKR Blocks RLR signaling Binds µ1 to attenuate apoptosis | outer capsid | [37,64,65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeAntoneo, C.; Danthi, P.; Balachandran, S. Reovirus Activated Cell Death Pathways. Cells 2022, 11, 1757. https://doi.org/10.3390/cells11111757
DeAntoneo C, Danthi P, Balachandran S. Reovirus Activated Cell Death Pathways. Cells. 2022; 11(11):1757. https://doi.org/10.3390/cells11111757
Chicago/Turabian StyleDeAntoneo, Carly, Pranav Danthi, and Siddharth Balachandran. 2022. "Reovirus Activated Cell Death Pathways" Cells 11, no. 11: 1757. https://doi.org/10.3390/cells11111757
APA StyleDeAntoneo, C., Danthi, P., & Balachandran, S. (2022). Reovirus Activated Cell Death Pathways. Cells, 11(11), 1757. https://doi.org/10.3390/cells11111757






