Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss
Abstract
1. Introduction
2. The Diaphanous Family
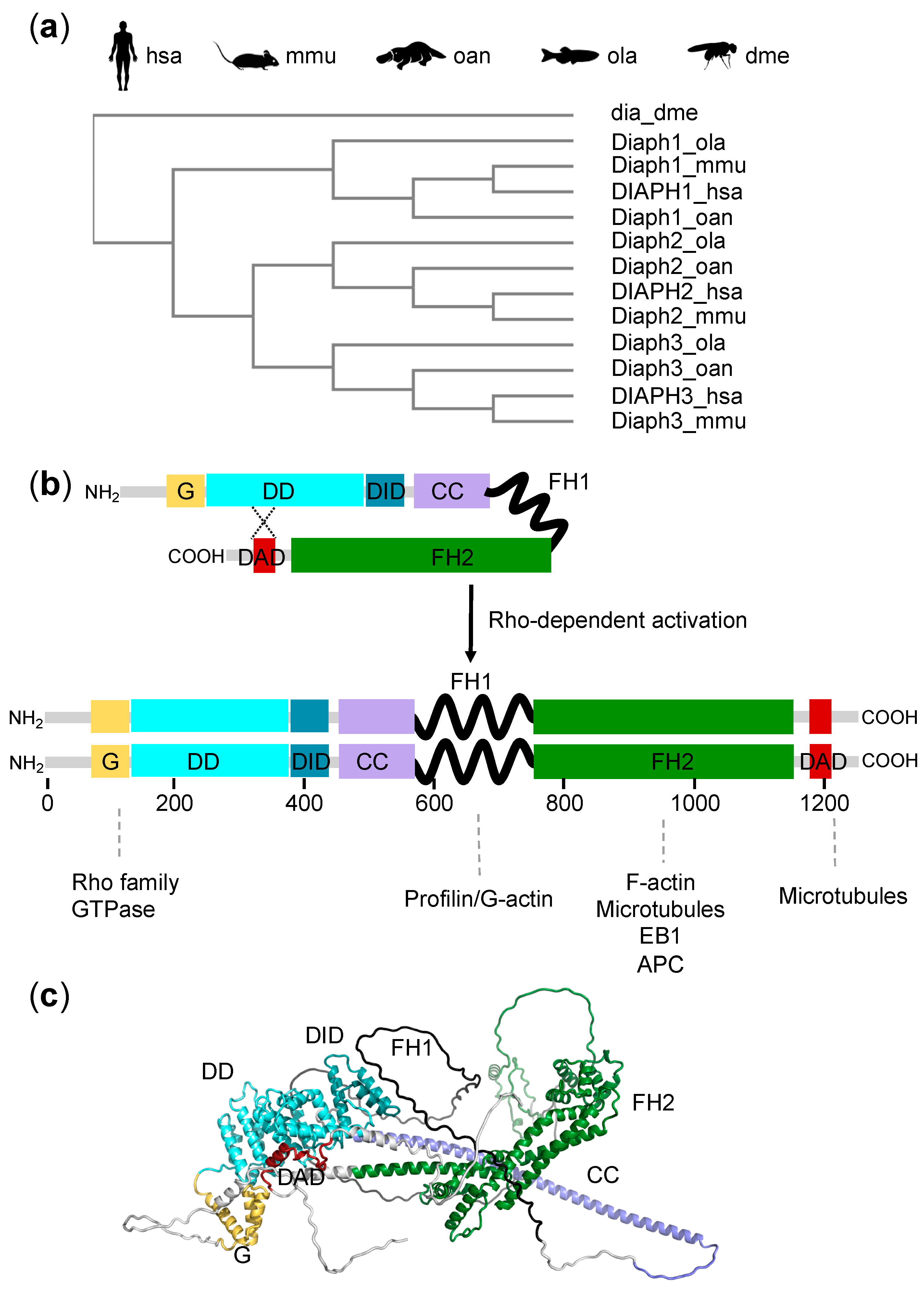
2.1. Evolution
2.2. Structure
2.3. Actin and Microtubule-Modulating Activity of Diaphanous Proteins
2.4. Expression of Diaphanous Proteins in the Inner Ear
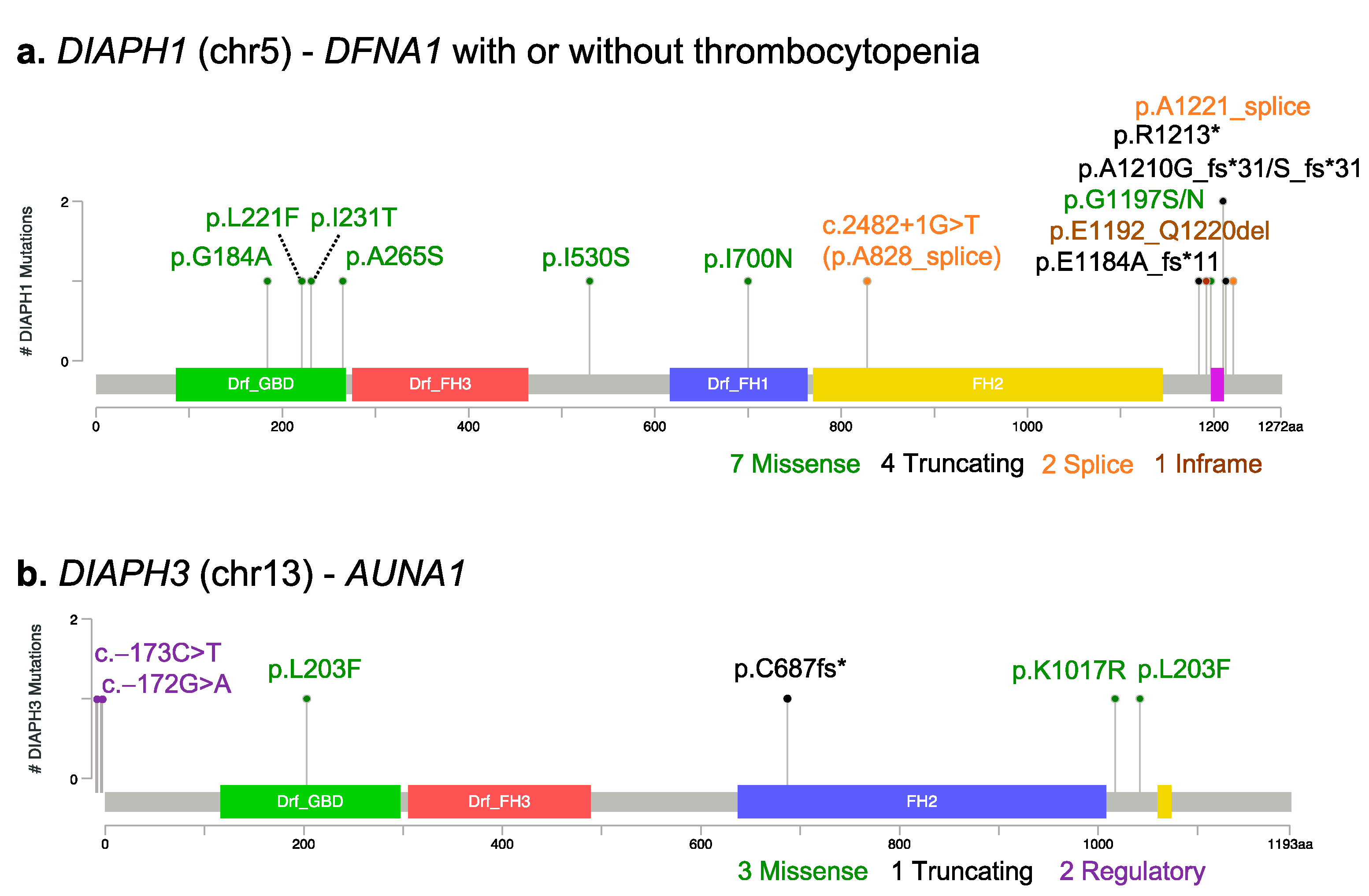
3. DRF Mutations and Hearing Loss
3.1. DIAPH1
3.2. DIAPH2
3.3. DIAPH3
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drummond, M.C.; Belyantseva, I.A.; Friderici, K.H.; Friedman, T.B. Actin in hair cells and hearing loss. Hear. Res. 2012, 288, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, S.; Schultz, J.; Grosshans, J. Formin’ cellular structures: Physiological roles of Diaphanous (Dia) in actin dynamics. Commun. Integr. Biol. 2013, 6, e27634. [Google Scholar] [CrossRef] [PubMed]
- Campellone, K.G.; Welch, M.D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Labat-De-Hoz, L.; Alonso, M.A. Formins in Human Disease. Cells 2021, 10, 2554. [Google Scholar] [CrossRef] [PubMed]
- Lammers, M.; Meyer, S.; Kühlmann, D.; Wittinghofer, A. Specificity of Interactions between mDia Isoforms and Rho Proteins. J. Biol. Chem. 2008, 283, 35236–35246. [Google Scholar] [CrossRef] [PubMed]
- Chesarone, M.A.; DuPage, A.G.; Goode, B.L. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 2009, 11, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, J.; Ramabhadran, V.; Neumanne, E.; Gurel, P.; Blanchoin, L.; Vantard, M.; Higgs, H.N. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol. Biol. Cell 2011, 22, 4575–4587. [Google Scholar] [CrossRef]
- Castrillon, D.; Wasserman, S. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 1994, 120, 3367–3377. [Google Scholar] [CrossRef]
- Grunt, M.; Žárský, V.; Cvrčková, F. Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 2008, 8, 115. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Palander, O.; Trimble, W.S. DIAPH1 regulates ciliogenesis and trafficking in primary cilia. FASEB J. 2020, 34, 16516–16535. [Google Scholar] [CrossRef] [PubMed]
- Palander, O.; Lam, A.; Collins, R.F.; Moraes, T.J.; Trimble, W.S. Nonredundant roles of DIAPHs in primary ciliogenesis. J. Biol. Chem. 2021, 296, 100680. [Google Scholar] [CrossRef] [PubMed]
- Daou, P.; Hasan, S.; Breitsprecher, D.; Baudelet, E.; Camoin, L.; Audebert, S.; Goode, B.L.; Badache, A. Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol. Biol. Cell 2014, 25, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.L.; Eck, M.J. Mechanism and Function of Formins in the Control of Actin Assembly. Annu. Rev. Biochem. 2007, 76, 593–627. [Google Scholar] [CrossRef]
- Otomo, T.; Otomo, C.; Tomchick, D.; Machius, M.; Rosen, M.K. Structural Basis of Rho GTPase-Mediated Activation of the Formin mDia1. Mol. Cell 2005, 18, 273–281. [Google Scholar] [CrossRef]
- Rose, R.; Weyand, M.; Lammers, M.; Ishizaki, T.; Ahmadian, M.R.; Wittinghofer, A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 2005, 435, 513–518. [Google Scholar] [CrossRef]
- Young, K.G.; Copeland, J.W. Formins in cell signaling. Biochim. Biophys. Acta 2010, 1803, 183–190. [Google Scholar] [CrossRef]
- Watanabe, N.; Kato, T.; Fujita, A.; Ishizaki, T.; Narumiya, S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1999, 1, 136–143. [Google Scholar] [CrossRef]
- Ueyama, T.; Ninoyu, Y.; Nishio, S.-Y.; Miyoshi, T.; Torii, H.; Nishimura, K.; Sugahara, K.; Sakata, H.; Thumkeo, D.; Sakaguchi, H.; et al. Constitutive activation of DIA 1 ( DIAPH 1) via C-terminal truncation causes human sensorineural hearing loss. EMBO Mol. Med. 2016, 8, 1310–1324. [Google Scholar] [CrossRef]
- Kim, B.J.; Ueyama, T.; Miyoshi, T.; Lee, S.; Han, J.H.; Park, H.-R.; Kim, A.R.; Oh, J.; Kim, M.Y.; Kang, Y.S.; et al. Differential disruption of autoinhibition and defect in assembly of cytoskeleton during cell division decide the fate of human DIAPH1-related cytoskeletopathy. J. Med. Genet. 2019, 56, 818–827. [Google Scholar] [CrossRef]
- Nezami, A.G.; Poy, F.; Eck, M.J. Structure of the Autoinhibitory Switch in Formin mDia1. Structure 2006, 14, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Nezami, A.; Poy, F.; Toms, A.; Zheng, W.; Eck, M.J. Crystal Structure of a Complex between Amino and Carboxy Terminal Fragments of mDia1: Insights into Autoinhibition of Diaphanous-Related Formins. PLoS ONE 2010, 5, e12992. [Google Scholar] [CrossRef] [PubMed]
- Otomo, T.; Tomchick, D.R.; Otomo, C.; Machius, M.; Rosen, M.K. Crystal Structure of the Formin mDia1 in Autoinhibited Conformation. PLoS ONE 2010, 5, e12896. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Maiti, S.; Michelot, A.; Gould, C.; Blanchoin, L.; Sokolova, O.; Goode, B.L. Structure and activity of full-length formin mDia1. Cytoskeleton 2012, 69, 393–405. [Google Scholar] [CrossRef]
- Bartolini, F.; Gundersen, G. Formins and microtubules. Biochim. Biophys. Acta 2010, 1803, 164–173. [Google Scholar] [CrossRef]
- Schoen, C.J.; Burmeister, M.; Lesperance, M.M. Diaphanous homolog 3 (Diap3) Overexpression Causes Progressive Hearing Loss and Inner Hair Cell Defects in a Transgenic Mouse Model of Human Deafness. PLoS ONE 2013, 8, e56520. [Google Scholar] [CrossRef]
- Surel, C.; Guillet, M.; Lenoir, M.; Bourien, J.; Sendin, G.; Joly, W.; Delprat, B.; Lesperance, M.M.; Puel, J.-L.; Nouvian, R. Remodeling of the Inner Hair Cell Microtubule Meshwork in a Mouse Model of Auditory Neuropathy AUNA1. eNeuro 2016, 3, ENEURO.0295-16.2016. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2020, 49, D899–D907. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 23 February 2022).
- Neuhaus, C.; Lang-Roth, R.; Zimmermann, U.; Heller, R.; Eisenberger, T.; Weikert, M.; Markus, S.; Knipper, M.; Bolz, H.J. Extension of the clinical and molecular phenotype of DIAPH1-associated autosomal dominant hearing loss (DFNA1). Clin. Genet. 2016, 91, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Ninoyu, Y.; Sakaguchi, H.; Lin, C.; Suzuki, T.; Hirano, S.; Hisa, Y.; Saito, N.; Ueyama, T. The integrity of cochlear hair cells is established and maintained through the localization of Dia1 at apical junctional complexes and stereocilia. Cell Death Dis. 2020, 11, 536. [Google Scholar] [CrossRef] [PubMed]
- Hertzano, R.; Elkon, R.; Kurima, K.; Morrisson, A.; Chan, S.-L.; Sallin, M.; Biedlingmaier, A.; Darling, D.S.; Griffith, A.J.; Eisenman, D.J.; et al. Cell Type–Specific Transcriptome Analysis Reveals a Major Role for Zeb1 and miR-200b in Mouse Inner Ear Morphogenesis. PLoS Genet. 2011, 7, e1002309. [Google Scholar] [CrossRef] [PubMed]
- Boyer, O.; Nevo, F.; Plaisier, E.; Funalot, B.; Gribouval, O.; Benoit, G.; Cong, E.H.; Arrondel, C.; Tête, M.-J.; Montjean, R.; et al. INF2Mutations in Charcot–Marie–Tooth Disease with Glomerulopathy. N. Engl. J. Med. 2011, 365, 2377–2388. [Google Scholar] [CrossRef]
- Kim, T.B.; Isaacson, B.; Sivakumaran, T.A.; Starr, A.; Keats, B.J.B.; Lesperance, M.M. A gene responsible for autosomal dominant auditory neuropathy (AUNA1) maps to 13q14-21. J. Med. Genet. 2004, 41, 872–876. [Google Scholar] [CrossRef]
- Lynch, E.D.; Lee, M.K.; Morrow, J.E.; Welcsh, P.L.; León, P.E.; King, M.C. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science 1997, 278, 1315–1318. [Google Scholar] [CrossRef]
- Shearer, A.E.; Black-Ziegelbein, E.A.; Hildebrand, M.; Eppsteiner, R.W.; Ravi, H.; Joshi, S.; Guiffre, A.C.; Sloan, C.M.; Happe, S.; Howard, S.D.; et al. Advancing genetic testing for deafness with genomic technology. J. Med. Genet. 2013, 50, 627–634. [Google Scholar] [CrossRef]
- Iwasa, Y.-I.; Nishio, S.-Y.; Usami, S.-I. Comprehensive Genetic Analysis of Japanese Autosomal Dominant Sensorineural Hearing Loss Patients. PLoS ONE 2016, 11, e0166781. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef]
- Kang, T.-H.; Baek, J.-I.; Sagong, B.; Park, H.-J.; Park, C.I.; Lee, K.-Y.; Kim, U.-K. A novel missense variant in the DIAPH1 gene in a Korean family with autosomal dominant nonsyndromic hearing loss. Genes Genet. Syst. 2016, 91, 289–292. [Google Scholar] [CrossRef]
- Brozkova, D.S.; Marková, S.P.; Mészárosová, A.U.; Jenčík, J.; Čejnová, V.; Čada, Z.; Laštůvková, J.; Rašková, D.; Seeman, P. Spectrum and frequencies of non GJB2 gene mutations in Czech patients with early non-syndromic hearing loss detected by gene panel NGS and whole-exome sequencing. Clin. Genet. 2020, 98, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, T.; Belyantseva, I.A.; Kitajiri, S.-I.; Miyajima, H.; Nishio, S.-Y.; Usami, S.-I.; Kim, B.J.; Choi, B.Y.; Omori, K.; Shroff, H.; et al. Human deafness-associated variants alter the dynamics of key molecules in hair cell stereocilia F-actin cores. Qual. Life Res. 2021, 141, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Stritt, S.; Nurden, P.; Turro, E.; Greene, D.; Jansen, S.B.; Westbury, S.; Petersen, R.; Astle, W.; Marlin, S.; Bariana, T.K.; et al. A gain-of-function variant in DIAPH1 causes dominant macrothrombocytopenia and hearing loss. Blood 2016, 127, 2903–2914. [Google Scholar] [CrossRef] [PubMed]
- Ganaha, A.; Kaname, T.; Shinjou, A.; Chinen, Y.; Yanagi, K.; Higa, T.; Kondo, S.; Suzuki, M. Progressive macrothrombocytopenia and hearing loss in a large family with DIAPH1 related disease. Am. J. Med. Genet. Part A 2017, 173, 2826–2830. [Google Scholar] [CrossRef]
- Karki, N.R.; Ajebo, G.; Savage, N.; Kutlar, A. DIAPH1 Mutation as a Novel Cause of Autosomal Dominant Macrothrombocytopenia and Hearing Loss. Acta Haematol. 2020, 144, 91–94. [Google Scholar] [CrossRef]
- Westbury, S.K.; Downes, K.; Burney, C.; Lozano, M.L.; Obaji, S.G.; Toh, C.H.; Sevivas, T.; Morgan, N.V.; Erber, W.; Kempster, C.; et al. Phenotype description and response to thrombopoietin receptor agonist in DIAPH1-related disorder. Blood Adv. 2018, 2, 2341–2346. [Google Scholar] [CrossRef]
- Turro, E.; Astle, W.J.; Megy, K.; Gräf, S.; Greene, D.; Shamardina, O.; Allen, H.L.; Sanchis-Juan, A.; Frontini, M.; Thys, C.; et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 2020, 583, 96–102. [Google Scholar] [CrossRef]
- Lakha, R.; Montero, A.M.; Jabeen, T.; Costeas, C.C.; Ma, J.; Vizcarra, C.L. Variable Autoinhibition among Deafness-Associated Variants of Diaphanous 1 (DIAPH1). Biochemistry 2021, 60, 2320–2329. [Google Scholar] [CrossRef]
- Wu, K.; Wang, H.; Guan, J.; Lan, L.; Zhao, C.; Zhang, M.; Wang, D.; Wang, Q. A novel variant in diaphanous homolog 1 (DIAPH1) as the cause of auditory neuropathy in a Chinese family. Int. J. Pediatr. Otorhinolaryngol. 2020, 133, 109947. [Google Scholar] [CrossRef]
- Ercan-Sencicek, A.G.; Jambi, S.; Franjic, D.; Nishimura, S.; Li, M.; El-Fishawy, P.; Morgan, T.M.; Sanders, S.J.; Bilguvar, K.; Suri, M.; et al. Homozygous loss of DIAPH1 is a novel cause of microcephaly in humans. Eur. J. Hum. Genet. 2014, 23, 165–172. [Google Scholar] [CrossRef]
- Al-Maawali, A.; Barry, B.J.; Rajab, A.; El-Quessny, M.; Seman, A.; Coury, S.N.; Barkovich, A.J.; Yang, E.; Walsh, C.A.; Mochida, G.H.; et al. Novel loss-of-function variants in DIAPH1 associated with syndromic microcephaly, blindness, and early onset seizures. Am. J. Med. Genet. Part A 2015, 170, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Kaustio, M.; Nayebzadeh, N.; Hinttala, R.; Tapiainen, T.; Åström, P.; Mamia, K.; Pernaa, N.; Lehtonen, J.; Glumoff, V.; Rahikkala, E.; et al. Loss of DIAPH1 causes SCBMS, combined immunodeficiency, and mitochondrial dysfunction. J. Allergy Clin. Immunol. 2021, 148, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Kitchen, S.M.; West, R.A.; Sigler, R.; Eisenmann, K.M.; Alberts, A.S. Myeloproliferative Defects following Targeting of the Drf1 Gene Encoding the Mammalian Diaphanous–Related Formin mDia1. Cancer Res. 2007, 67, 7565–7571. [Google Scholar] [CrossRef] [PubMed]
- Eisenmann, K.M.; West, R.A.; Hildebrand, D.; Kitchen, S.M.; Peng, J.; Sigler, R.; Zhang, J.; Siminovitch, K.A.; Alberts, A.S. T Cell Responses in Mammalian Diaphanous-related Formin mDia1 Knock-out Mice. J. Biol. Chem. 2007, 282, 25152–25158. [Google Scholar] [CrossRef] [PubMed]
- Sakata, D.; Taniguchi, H.; Yasuda, S.; Adachi-Morishima, A.; Hamazaki, Y.; Nakayama, R.; Miki, T.; Minato, N.; Narumiya, S. Impaired T lymphocyte trafficking in mice deficient in an actin-nucleating protein, mDia1. J. Exp. Med. 2007, 204, 2031–2038. [Google Scholar] [CrossRef] [PubMed]
- Salaa, C.; Arrigoa, G.; Torri, G.P.; Martinazzia, F.; Riva, P.; Larizzab, L.; Philippec, C.; Jonveauxc, P.; Sloanc, F.; Labellad, T.; et al. Eleven X Chromosome Breakpoints Associated with Premature Ovarian Failure (POF) Map to a 15-Mb YAC Contig Spanning Xq21. Genomics 1997, 40, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Bione, S.; Sala, C.; Manzini, M.C.; Arrigo, G.; Zuffardi, O.; Banfi, S.; Borsani, G.; Jonveaux, P.; Philippe, C.; Zuccotti, M.; et al. A Human Homologue of the Drosophila melanogaster diaphanous Gene Is Disrupted in a Patient with Premature Ovarian Failure: Evidence for Conserved Function in Oogenesis and Implications for Human Sterility. Am. J. Hum. Genet. 1998, 62, 533–541. [Google Scholar] [CrossRef]
- Marozzi, A.; Manfredini, E.; Tibiletti, M.; Furlan, D.; Villa, N.; Vegetti, W.; Crosignani, P.; Ginelli, E.; Meneveri, R.; Dalprà, L. Molecular definition of Xq common-deleted region in patients affected by premature ovarian failure. Qual. Life Res. 2000, 107, 304–311. [Google Scholar] [CrossRef]
- Genesio, R.; Mormile, A.; Licenziati, M.R.; De Brasi, D.; Leone, G.; Balzano, S.; Izzo, A.; Bonfiglio, F.; Conti, A.; Fioretti, G.; et al. Short stature and primary ovarian insufficiency possibly due to chromosomal position effect in a balanced X;1 translocation. Mol. Cytogenet. 2015, 8, 50. [Google Scholar] [CrossRef][Green Version]
- Bestetti, I.; Castronovo, C.; Sironi, A.; Caslini, C.; Sala, C.; Rossetti, R.; Crippa, M.; Ferrari, I.; Pistocchi, A.; Toniolo, D.; et al. High-resolution array-CGH analysis on 46,XX patients affected by early onset primary ovarian insufficiency discloses new genes involved in ovarian function. Hum. Reprod. 2019, 34, 574–583. [Google Scholar] [CrossRef]
- Schoen, C.J.; Emery, S.B.; Thorne, M.C.; Ammana, H.R.; Śliwerska, E.; Arnett, J.; Hortsch, M.; Hannan, F.; Burmeister, M.; Lesperance, M.M. Increased activity of Diaphanous homolog 3 (DIAPH3)/diaphanous causes hearing defects in humans with auditory neuropathy and in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 13396–13401. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martínez, A.; Benito-Orejas, J.I.; Tellería-Orriols, J.J.; Alonso-Ramos, M.J. Neuropatía auditiva autosómica dominante y variante DIAPH3 (c.-173C>T). Acta Otorrinolaringol. Esp. 2017, 68, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Sommen, M.; Schrauwen, I.; Vandeweyer, G.; Boeckx, N.; Corneveaux, J.J.; Ende, J.V.D.; Boudewyns, A.; De Leenheer, E.; Janssens, S.; Claes, K.; et al. DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum. Mutat. 2016, 37, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.-H.; Baek, J.-I.; Lee, J.D.; Song, M.H.; Kwon, T.-J.; Oh, S.-K.; Jeong, J.Y.; Choi, J.Y.; Lee, K.-Y.; Kim, U.-K. Genetic analysis of auditory neuropathy spectrum disorder in the Korean population. Gene 2013, 522, 65–69. [Google Scholar] [CrossRef]
- Lai, S.-L.; Chan, T.-H.; Lin, M.-J.; Huang, W.-P.; Lou, S.-W.; Lee, S.-J. Diaphanous-Related Formin 2 and Profilin I Are Required for Gastrulation Cell Movements. PLoS ONE 2008, 3, e3439. [Google Scholar] [CrossRef] [PubMed]
- Damiani, D.; Goffinet, A.M.; Alberts, A.; Tissir, F. Lack of Diaph3 relaxes the spindle checkpoint causing the loss of neural progenitors. Nat. Commun. 2016, 7, 13509. [Google Scholar] [CrossRef]
- Lau, E.O.-C.; Damiani, D.; Chehade, G.; Ruiz-Reig, N.; Saade, R.; Jossin, Y.; Aittaleb, M.; Schakman, O.; Tajeddine, N.; Gailly, P.; et al. DIAPH3 deficiency links microtubules to mitotic errors, defective neurogenesis, and brain dysfunction. eLife 2021, 10, e61974. [Google Scholar] [CrossRef]
- Vorstman, J.A.S.; Van Daalen, E.; Jalali, G.R.; Schmidt, E.R.; Pasterkamp, R.J.; De Jonge, M.; Hennekam, E.A.M.; Janson, E.; Staal, W.G.; Van Der Zwaag, B.; et al. A double hit implicates DIAPH3 as an autism risk gene. Mol. Psychiatry 2010, 16, 442–451. [Google Scholar] [CrossRef]
- Xie, J.; Li, H.; Zhu, H.; Huang, L.; Li, H.; Zhang, X.; Zhou, Y.; Zhou, Q.; Xu, W. Analysis of DIAPH3 gene mutation in a boy with autism spectrum disorder. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2016, 33, 481–484. (In Chinese) [Google Scholar] [CrossRef]
- Azaiez, H.; Booth, K.T.; Ephraim, S.S.; Crone, B.; Black-Ziegelbein, E.A.; Marini, R.J.; Shearer, A.E.; Sloan-Heggen, C.M.; Kolbe, D.; Casavant, T.; et al. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am. J. Hum. Genet. 2018, 103, 484–497. [Google Scholar] [CrossRef]
- Ji, H.; Lu, J.; Wang, J.; Li, H.; Lin, X. Combined examination of sequence and copy number variations in human deafness genes improves diagnosis for cases of genetic deafness. BMC Ear Nose Throat Disord. 2014, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Pavlenkova, Z.; Varga, L.; Borecka, S.; Karhanek, M.; Huckova, M.; Skopkova, M.; Profant, M.; Gasperikova, D. Comprehensive molecular-genetic analysis of mid-frequency sensorineural hearing loss. Sci. Rep. 2021, 11, 22488. [Google Scholar] [CrossRef] [PubMed]
- Misceo, D.; Rødningen, O.; Barøy, T.; Sorte, H.; Mellembakken, J.; Strømme, P.; Fannemel, M.; Frengen, E. A translocation between Xq21.33 and 22q13.33 causes an intragenic SHANK3 deletion in a woman with Phelan-McDermid syndrome and hypergonadotropic hypogonadism. Am. J. Med. Genet. Part A 2011, 155, 403–408. [Google Scholar] [CrossRef] [PubMed]
- The Human Gene Mutation Database (HGMD). Available online: http://www.hgmd.cf.ac.uk/ac/index.php (accessed on 14 March 2022).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Kearney, H.M.; Thorland, E.C.; Brown, K.K.; Quintero-Rivera, F.; South, S.T. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011, 13, 680–685. [Google Scholar] [CrossRef]
| DIAPH1 | DIAPH3 | |
|---|---|---|
| Inheritance | AD | AD |
| HL onset | First decade | Second decade 1 |
| Affected frequencies | All | High frequencies more affected |
| Progression | Yes | Yes |
| ABR findings | Absent/abnormal | Absent/abnormal |
| Middle ear reflexes | Present | Absent |
| Affected structures | OHC | IHC, auditory nerve |
| Other symptoms | May be accompanied by thrombocytopenia | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiereghin, C.; Robusto, M.; Massa, V.; Castorina, P.; Ambrosetti, U.; Asselta, R.; Soldà, G. Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss. Cells 2022, 11, 1726. https://doi.org/10.3390/cells11111726
Chiereghin C, Robusto M, Massa V, Castorina P, Ambrosetti U, Asselta R, Soldà G. Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss. Cells. 2022; 11(11):1726. https://doi.org/10.3390/cells11111726
Chicago/Turabian StyleChiereghin, Chiara, Michela Robusto, Valentina Massa, Pierangela Castorina, Umberto Ambrosetti, Rosanna Asselta, and Giulia Soldà. 2022. "Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss" Cells 11, no. 11: 1726. https://doi.org/10.3390/cells11111726
APA StyleChiereghin, C., Robusto, M., Massa, V., Castorina, P., Ambrosetti, U., Asselta, R., & Soldà, G. (2022). Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss. Cells, 11(11), 1726. https://doi.org/10.3390/cells11111726








