Enhancer RNA Expression in Response to Glucocorticoid Treatment in Murine Macrophages
Abstract
:1. Introduction
2. Material and Methods
2.1. Animals
2.2. Isolation of Bone Marrow-Derived Macrophages
2.3. Treatment and 4sU Labelling
2.4. RNA Isolation and 4sU-Seq
2.5. RNA Isolation and Quantitative Real-Time (qRT-) PCR
2.6. NGS Data Analysis
2.6.1. Data Sources
2.6.2. RNA Sequencing (RNA-Seq)
2.6.3. sU-Seq
2.6.4. Defining the Reproducible GR Cistrome
2.6.5. Identification and Characterization of Enhancer RNAs
2.6.6. DNA Motif Enrichment
2.6.7. Gene Ontology (GO) Enrichment for Biological Processes
2.6.8. Analysis of ChIP Sequencing (ChIP-Seq) Data and Quantification
3. Results
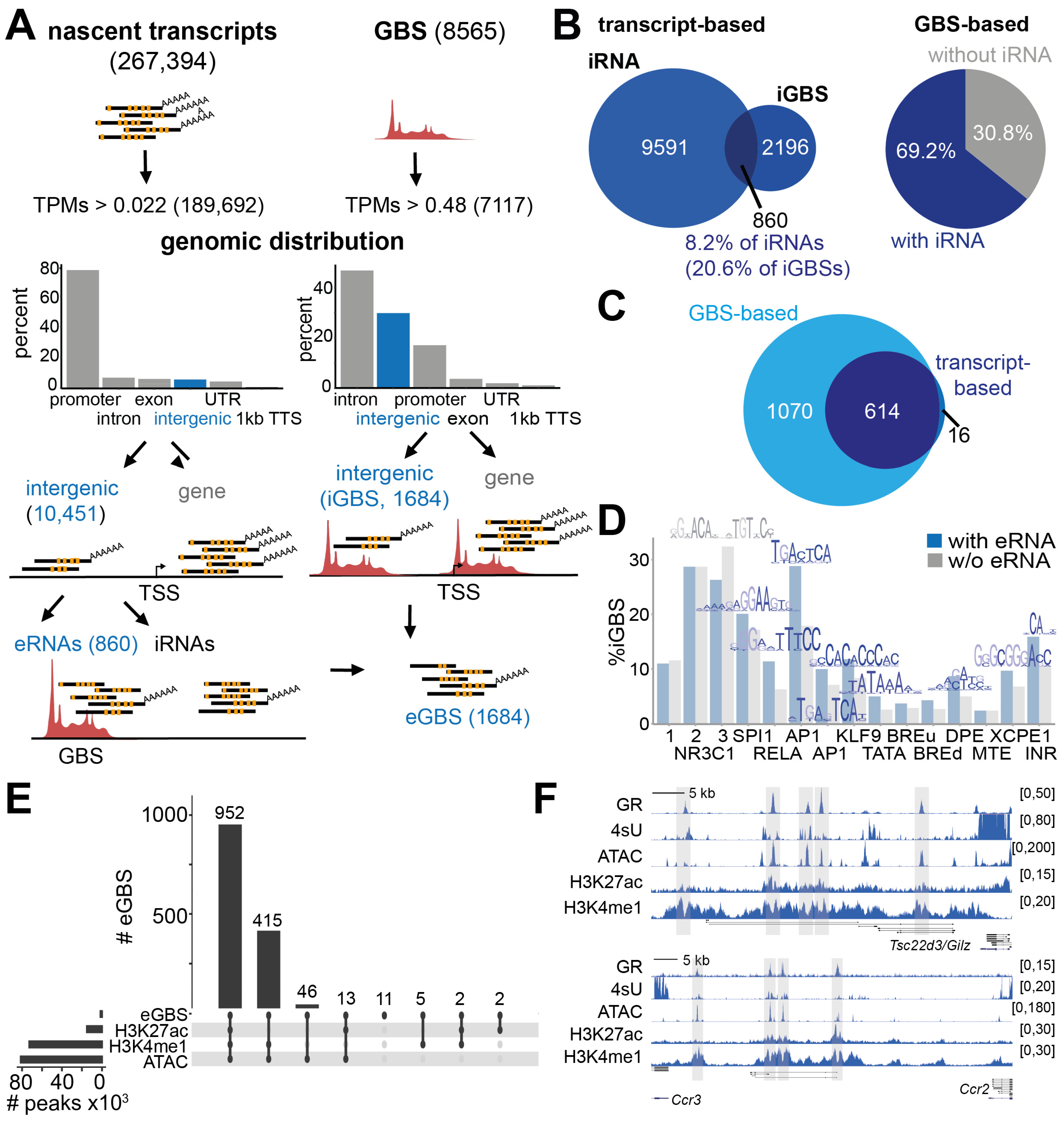
3.1. Intergenic Glucocorticoid Receptor Binding Coincides with eRNA Synthesis in Macrophages
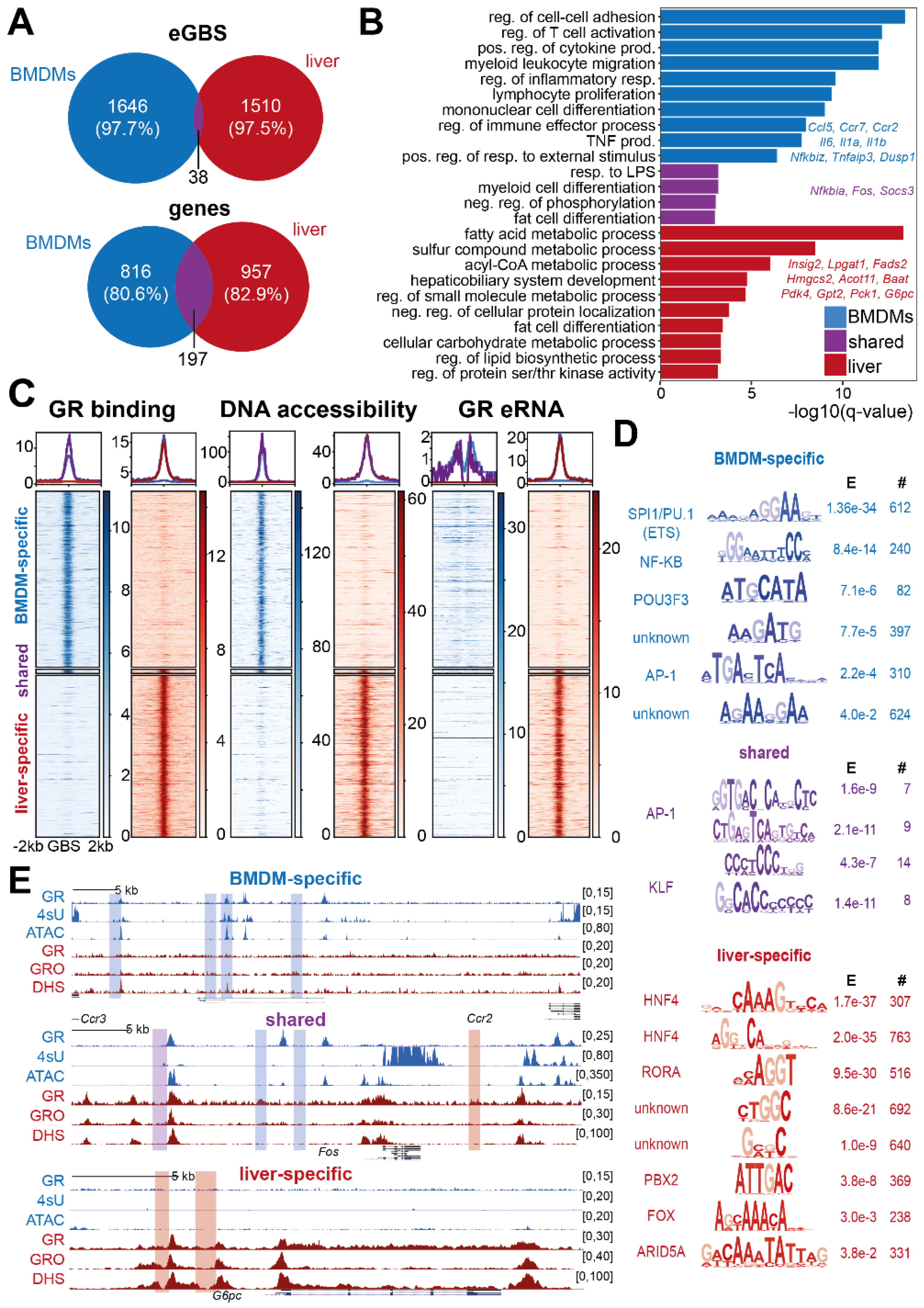
3.2. Enhancer RNA Synthesis at GR Binding Sites Is a Tissue-Specific Feature of the Glucocorticoid Response
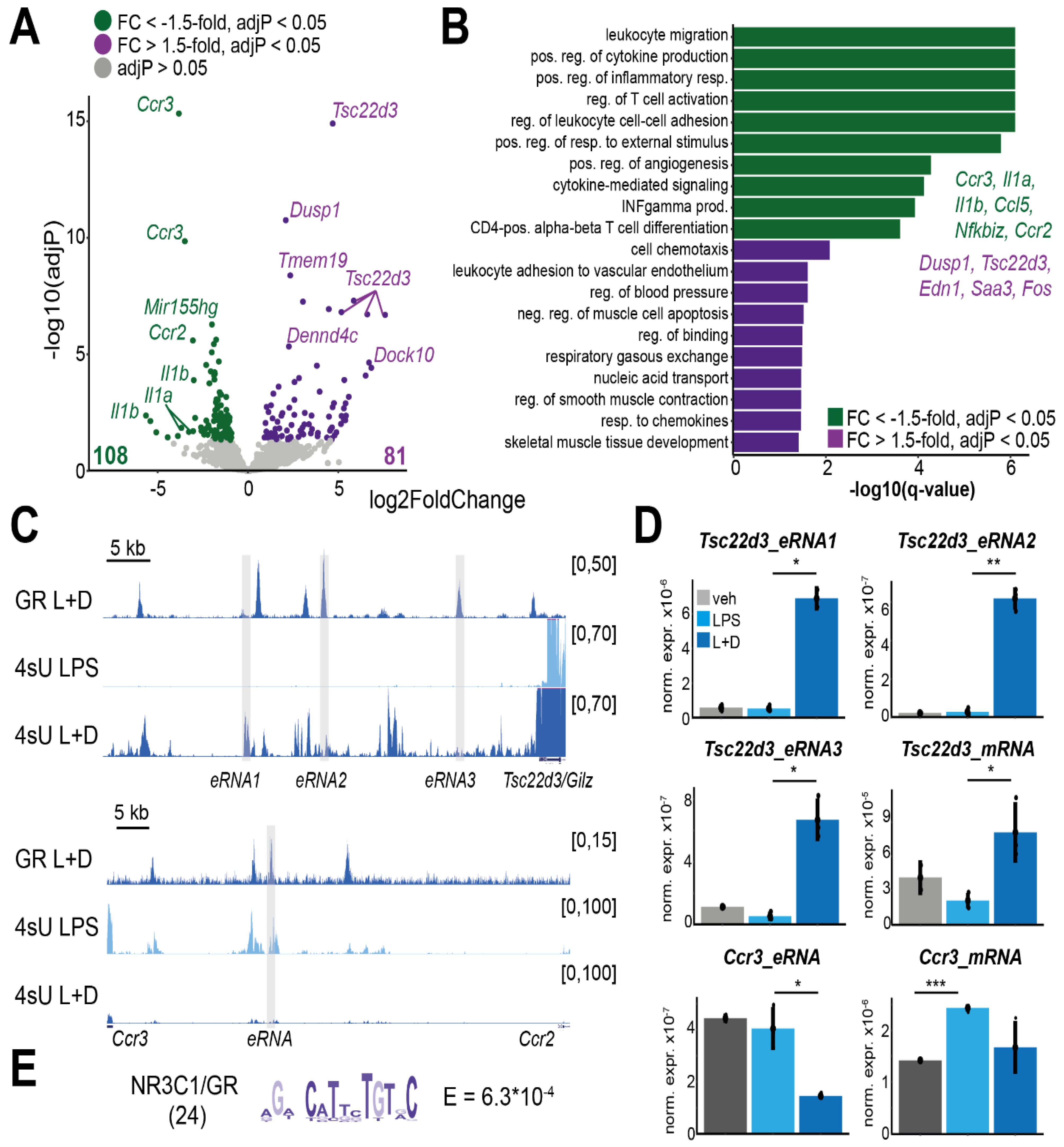
3.3. Differential Expression of eRNAs upon Glucocorticoid Treatment in Macrophages
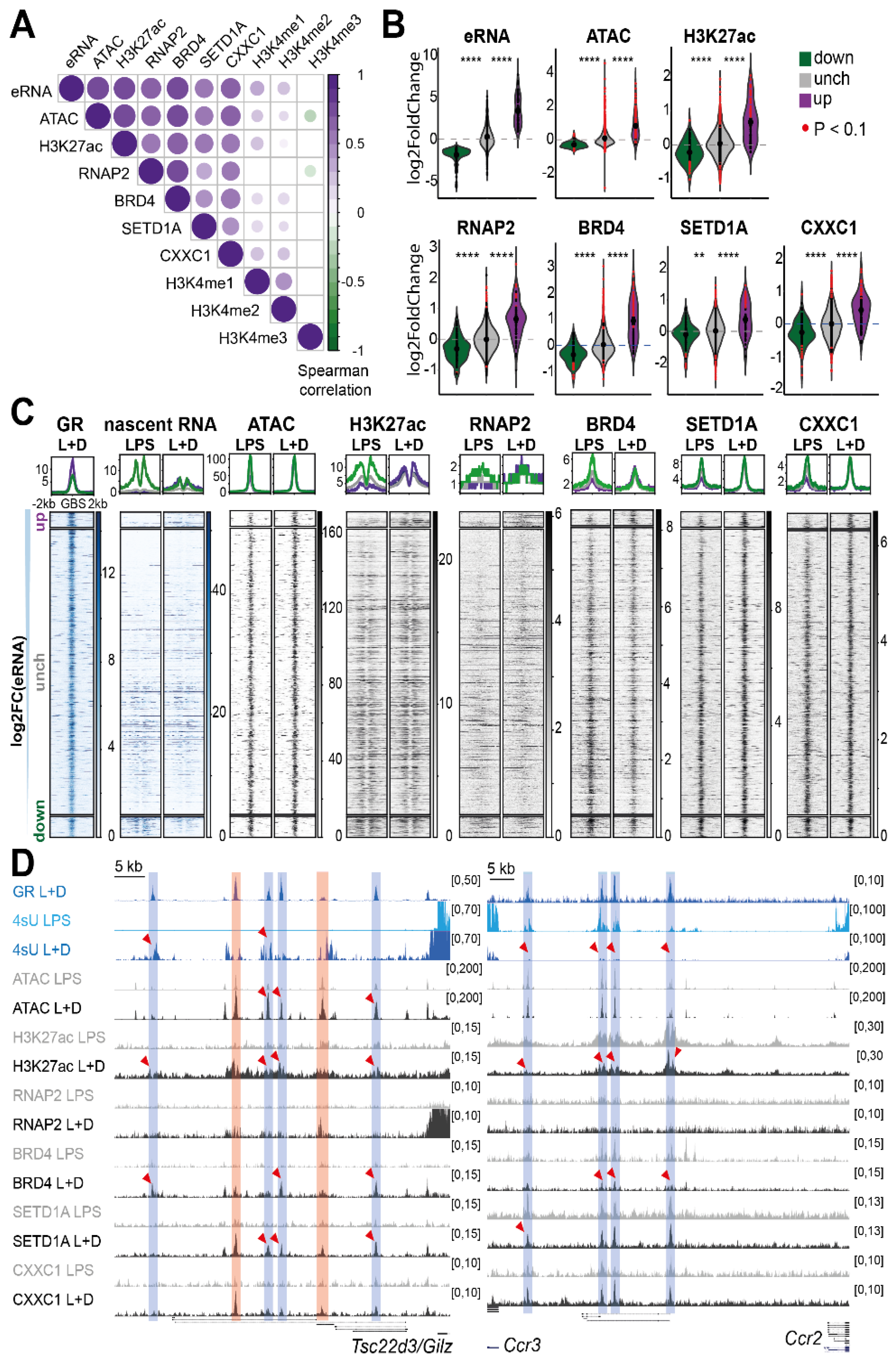
3.4. Enhancer RNA Expression Correlates with BRD4 and H3K27ac at GR Enhancer in Macrophages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheifetz, P.N. The daily rhythm of the secretion of corticotrophin and corticosterone in rats and mice. J. Endocrinol. 1971, 49, xi–xii. [Google Scholar] [PubMed]
- Lightman, S.; Wiles, C.; Atkinson, H.; Henley, D.; Russell, G.; Leendertz, J.; McKenna, M.; Spiga, F.; Wood, S.; Conway-Campbell, B. The significance of glucocorticoid pulsatility. Eur. J. Pharmacol. 2008, 583, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Quagliarini, F.; Mir, A.A.; Balazs, K.; Wierer, M.; Dyar, K.A.; Jouffe, C.; Makris, K.; Hawe, J.; Heinig, M.; Filipp, F.V.; et al. Cistromic Reprogramming of the Diurnal Glucocorticoid Hormone Response by High-Fat Diet. Mol. Cell 2019, 76, 531–545.e535. [Google Scholar] [CrossRef] [PubMed]
- Uhlenhaut, N.H.; Barish, G.D.; Yu, R.T.; Downes, M.; Karunasiri, M.; Liddle, C.; Schwalie, P.; Hubner, N.; Evans, R.M. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 2013, 49, 158–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vegiopoulos, A.; Herzig, S. Glucocorticoids, metabolism and metabolic diseases. Mol. Cell Endocrinol. 2007, 275, 43–61. [Google Scholar] [CrossRef] [Green Version]
- Oakley, R.H.; Cidlowski, J.A. Cellular processing of the glucocorticoid receptor gene and protein: New mechanisms for generating tissue-specific actions of glucocorticoids. J. Biol. Chem. 2011, 286, 3177–3184. [Google Scholar] [CrossRef] [Green Version]
- Barnes, P.J. Anti-inflammatory actions of glucocorticoids: Molecular mechanisms. Clin. Sci. 1998, 94, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Group, R.C.; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Kadmiel, M.; Cidlowski, J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013, 34, 518–530. [Google Scholar] [CrossRef] [Green Version]
- Schacke, H.; Docke, W.D.; Asadullah, K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 2002, 96, 23–43. [Google Scholar] [CrossRef]
- Cole, T.J.; Blendy, J.A.; Monaghan, A.P.; Krieglstein, K.; Schmid, W.; Aguzzi, A.; Fantuzzi, G.; Hummler, E.; Unsicker, K.; Schutz, G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995, 9, 1608–1621. [Google Scholar] [CrossRef] [Green Version]
- Escoter-Torres, L.; Greulich, F.; Quagliarini, F.; Wierer, M.; Uhlenhaut, N.H. Anti-inflammatory functions of the glucocorticoid receptor require DNA binding. Nucleic Acids Res. 2020, 48, 8393–8407. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.; Ganti, K.P.; Chambon, P. Glucocorticoid-induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/NCoR1-HDAC3 repressing complex. Proc. Natl. Acad. Sci. USA 2016, 113, E635–E643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsova, T.; Wang, S.Y.; Rao, N.A.; Mandoli, A.; Martens, J.H.; Rother, N.; Aartse, A.; Groh, L.; Janssen-Megens, E.M.; Li, G.; et al. Glucocorticoid receptor and nuclear factor kappa-b affect three-dimensional chromatin organization. Genome Biol. 2015, 16, 264. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorthy, S.; Cidlowski, J.A. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol. Cell. Biol. 2013, 33, 1711–1722. [Google Scholar] [CrossRef] [Green Version]
- Sacta, M.A.; Tharmalingam, B.; Coppo, M.; Rollins, D.A.; Deochand, D.K.; Benjamin, B.; Yu, L.; Zhang, B.; Hu, X.; Li, R.; et al. Gene-specific mechanisms direct glucocorticoid-receptor-driven repression of inflammatory response genes in macrophages. Elife 2018, 7, e34864. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.F.; Anderson, C.M.; Ma, H.; Hong, H.; Uht, R.M.; Kushner, P.J.; Stallcup, M.R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): Multiple motifs with different binding specificities. Mol. Endocrinol. 1998, 12, 302–313. [Google Scholar] [CrossRef]
- Escoter-Torres, L.; Caratti, G.; Mechtidou, A.; Tuckermann, J.; Uhlenhaut, N.H.; Vettorazzi, S. Fighting the Fire: Mechanisms of Inflammatory Gene Regulation by the Glucocorticoid Receptor. Front. Immunol. 2019, 10, 1859. [Google Scholar] [CrossRef]
- Greulich, F.; Wierer, M.; Mechtidou, A.; Gonzalez-Garcia, O.; Uhlenhaut, N.H. The glucocorticoid receptor recruits the COMPASS complex to regulate inflammatory transcription at macrophage enhancers. Cell Rep. 2021, 34, 108742. [Google Scholar] [CrossRef]
- Hemmer, M.C.; Wierer, M.; Schachtrup, K.; Downes, M.; Hubner, N.; Evans, R.M.; Uhlenhaut, N.H. E47 modulates hepatic glucocorticoid action. Nat. Commun. 2019, 10, 306. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Guo, C.; Li, W.; Li, J.; Wang, W.; Myatt, L.; Sun, K. Involvement of GR and p300 in the induction of H6PD by cortisol in human amnion fibroblasts. Endocrinology 2012, 153, 5993–6002. [Google Scholar] [CrossRef]
- Chen, W.; Roeder, R.G. The Mediator subunit MED1/TRAP220 is required for optimal glucocorticoid receptor-mediated transcription activation. Nucleic Acids Res. 2007, 35, 6161–6169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Drouin, J.; Trifiro, M.A.; Plante, R.K.; Nemer, M.; Eriksson, P.; Wrange, O. Glucocorticoid receptor binding to a specific DNA sequence is required for hormone-dependent repression of pro-opiomelanocortin gene transcription. Mol. Cell. Biol. 1989, 9, 5305–5314. [Google Scholar] [CrossRef] [PubMed]
- Hudson, W.H.; Vera, I.M.S.; Nwachukwu, J.C.; Weikum, E.R.; Herbst, A.G.; Yang, Q.; Bain, D.L.; Nettles, K.W.; Kojetin, D.J.; Ortlund, E.A. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappaB response elements. Nat. Commun. 2018, 9, 1337. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.; Paulen, L.; Chambon, P. GR SUMOylation and formation of an SUMO-SMRT/NCoR1-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc. Natl. Acad. Sci. USA 2016, 113, E626–E634. [Google Scholar] [CrossRef] [Green Version]
- Biddie, S.C.; John, S.; Sabo, P.J.; Thurman, R.E.; Johnson, T.A.; Schiltz, R.L.; Miranda, T.B.; Sung, M.H.; Trump, S.; Lightman, S.L.; et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 2011, 43, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Greulich, F.; Hemmer, M.C.; Rollins, D.A.; Rogatsky, I.; Uhlenhaut, N.H. There goes the neighborhood: Assembly of transcriptional complexes during the regulation of metabolism and inflammation by the glucocorticoid receptor. Steroids 2016, 114, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Grontved, L.; John, S.; Baek, S.; Liu, Y.; Buckley, J.R.; Vinson, C.; Aguilera, G.; Hager, G.L. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013, 32, 1568–1583. [Google Scholar] [CrossRef] [Green Version]
- John, S.; Sabo, P.J.; Johnson, T.A.; Sung, M.H.; Biddie, S.C.; Lightman, S.L.; Voss, T.C.; Davis, S.R.; Meltzer, P.S.; Stamatoyannopoulos, J.A.; et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 2008, 29, 611–624. [Google Scholar] [CrossRef]
- John, S.; Sabo, P.J.; Thurman, R.E.; Sung, M.H.; Biddie, S.C.; Johnson, T.A.; Hager, G.L.; Stamatoyannopoulos, J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011, 43, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Whirledge, S.; DeFranco, D.B. Glucocorticoid Signaling in Health and Disease: Insights From Tissue-Specific GR Knockout Mice. Endocrinology 2018, 159, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Radle, B.; Rutkowski, A.J.; Ruzsics, Z.; Friedel, C.C.; Koszinowski, U.H.; Dolken, L. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J. Vis. Exp. 2013, 78, 50195. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Mechtidou, A.; Greulich, F.; Strickland, B.A.; Jouffe, C.; Cernilogar, F.M.; Schotta, G.; Uhlenhaut, N.H. BRG1 defines a genomic subset of inflammatory genes transcriptionally controlled by the glucocorticoid receptor. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sobel, J.A.; Krier, I.; Andersin, T.; Raghav, S.; Canella, D.; Gilardi, F.; Kalantzi, A.S.; Rey, G.; Weger, B.; Gachon, F.; et al. Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol. 2017, 15, e2001069. [Google Scholar] [CrossRef] [Green Version]
- Fang, B.; Everett, L.J.; Jager, J.; Briggs, E.; Armour, S.M.; Feng, D.; Roy, A.; Gerhart-Hines, Z.; Sun, Z.; Lazar, M.A. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 2014, 159, 1140–1152. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Res 2020, 9. [Google Scholar] [CrossRef]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [Green Version]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 9354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.J.; Gazin, C.; Lawson, N.D.; Pages, H.; Lin, S.M.; Lapointe, D.S.; Green, M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010, 11, 237. [Google Scholar] [CrossRef] [Green Version]
- Bioconductor Core Team; Bioconductor Package Maintainer. TxDb.Mmusculus.UCSC.mm10.knowngene: Annotation Package for TxDb Object(s). 2019. Available online: https://bioconductor.org/packages/release/data/annotation/html/TxDb.Mmusculus.UCSC.mm10.knownGene.html (accessed on 29 October 2021).
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [Green Version]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: https://www.repeatmasker.org/ (accessed on 29 October 2021).
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D1284. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newburger, D.E.; Bulyk, M.L. UniPROBE: An online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009, 37, D77–D82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pachkov, M.; Balwierz, P.J.; Arnold, P.; Ozonov, E.; van Nimwegen, E. SwissRegulon, a database of genome-wide annotations of regulatory sites: Recent updates. Nucleic Acids Res. 2013, 41, D214–D220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, C.E.; Bailey, T.L. XSTREME: Comprehensive motif analysis of biological sequence datasets. bioRxiv 2021. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wei, T.; Simko, S. R Package ‘Corrplot’: Visualization of a Correlation Matrix. 2021. Available online: https://cran.r-project.org/web/packages/corrplot/corrplot.pdf (accessed on 29 October 2021).
- Juven-Gershon, T.; Kadonaga, J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, M.; Wu, J.; Workman, J.L.; Li, B. Readers of histone modifications. Cell Res. 2011, 21, 564–578. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.; Shilatifard, A. The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 2010, 40, 689–701. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef]
- McDowell, I.C.; Barrera, A.; D’Ippolito, A.M.; Vockley, C.M.; Hong, L.K.; Leichter, S.M.; Bartelt, L.C.; Majoros, W.H.; Song, L.; Safi, A.; et al. Glucocorticoid receptor recruits to enhancers and drives activation by motif-directed binding. Genome Res. 2018, 28, 1272–1284. [Google Scholar] [CrossRef] [Green Version]
- Franco, L.M.; Gadkari, M.; Howe, K.N.; Sun, J.; Kardava, L.; Kumar, P.; Kumari, S.; Hu, Z.; Fraser, I.D.C.; Moir, S.; et al. Immune regulation by glucocorticoids can be linked to cell type-dependent transcriptional responses. J. Exp. Med. 2019, 216, 384–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, A.; Prefontaine, K.E. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 1994, 91, 752–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang-Yen, H.F.; Chambard, J.C.; Sun, Y.L.; Smeal, T.; Schmidt, T.J.; Drouin, J.; Karin, M. Transcriptional interference between c-Jun and the glucocorticoid receptor: Mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 1990, 62, 1205–1215. [Google Scholar] [CrossRef]
- Hunter, A.L.; Poolman, T.M.; Kim, D.; Gonzalez, F.J.; Bechtold, D.A.; Loudon, A.S.I.; Iqbal, M.; Ray, D.W. HNF4A is required to specify glucocorticoid action in the liver. bioRxiv 2021. [Google Scholar] [CrossRef]
- Iwafuchi-Doi, M.; Donahue, G.; Kakumanu, A.; Watts, J.A.; Mahony, S.; Pugh, B.F.; Lee, D.; Kaestner, K.H.; Zaret, K.S. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol. Cell 2016, 62, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Jubb, A.W.; Boyle, S.; Hume, D.A.; Bickmore, W.A. Glucocorticoid Receptor Binding Induces Rapid and Prolonged Large-Scale Chromatin Decompaction at Multiple Target Loci. Cell Rep. 2017, 21, 3022–3031. [Google Scholar] [CrossRef] [Green Version]
- So, A.Y.; Chaivorapol, C.; Bolton, E.C.; Li, H.; Yamamoto, K.R. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007, 3, e94. [Google Scholar] [CrossRef]
- Meijsing, S.H.; Pufall, M.A.; So, A.Y.; Bates, D.L.; Chen, L.; Yamamoto, K.R. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 2009, 324, 407–410. [Google Scholar] [CrossRef] [Green Version]
- Schone, S.; Jurk, M.; Helabad, M.B.; Dror, I.; Lebars, I.; Kieffer, B.; Imhof, P.; Rohs, R.; Vingron, M.; Thomas-Chollier, M.; et al. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nat. Commun. 2016, 7, 12621. [Google Scholar] [CrossRef]
- Dehe, P.M.; Geli, V. The multiple faces of Set1. Biochem. Cell Biol. 2006, 84, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chen, W.Y.; Shimada, M.; Nguyen, U.T.; Kim, J.; Sun, X.J.; Sengoku, T.; McGinty, R.K.; Fernandez, J.P.; Muir, T.W.; et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 2013, 154, 297–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itzen, F.; Greifenberg, A.K.; Bosken, C.A.; Geyer, M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014, 42, 7577–7590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.E.; Park, Y.K.; Park, S.; Jang, Y.; Waring, N.; Dey, A.; Ozato, K.; Lai, B.; Peng, W.; Ge, K. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat. Commun. 2017, 8, 2217. [Google Scholar] [CrossRef]
- Zhang, W.; Prakash, C.; Sum, C.; Gong, Y.; Li, Y.; Kwok, J.J.; Thiessen, N.; Pettersson, S.; Jones, S.J.; Knapp, S.; et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J. Biol. Chem. 2012, 287, 43137–43155. [Google Scholar] [CrossRef] [Green Version]
- Hargreaves, D.C.; Horng, T.; Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009, 138, 129–145. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [Green Version]
- De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.L.; Natoli, G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010, 8, e1000384. [Google Scholar] [CrossRef] [Green Version]
- Sasse, S.K.; Gruca, M.; Allen, M.A.; Kadiyala, V.; Song, T.; Gally, F.; Gupta, A.; Pufall, M.A.; Dowell, R.D.; Gerber, A.N. Nascent transcript analysis of glucocorticoid crosstalk with TNF defines primary and cooperative inflammatory repression. Genome Res. 2019, 29, 1753–1765. [Google Scholar] [CrossRef]
- Wissink, E.M.; Martinez, D.M.; Ehmsen, K.T.; Yamamoto, K.R.; Lis, J.T. Glucocorticoid receptor collaborates with pioneer factors and AP-1 to execute genome-wide regulation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Vockley, C.M.; D’Ippolito, A.M.; McDowell, I.C.; Majoros, W.H.; Safi, A.; Song, L.; Crawford, G.E.; Reddy, T.E. Direct GR Binding Sites Potentiate Clusters of TF Binding across the Human Genome. Cell 2016, 166, 1269–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liang, N.; Goni, S.; Damdimopoulos, A.; Wang, C.; Ballaire, R.; Jager, J.; Niskanen, H.; Han, H.; Jakobsson, T.; et al. The corepressors GPS2 and SMRT control enhancer and silencer remodeling via eRNA transcription during inflammatory activation of macrophages. Mol. Cell 2021, 81, 953–968.e9. [Google Scholar] [CrossRef] [PubMed]
- Ilott, N.E.; Heward, J.A.; Roux, B.; Tsitsiou, E.; Fenwick, P.S.; Lenzi, L.; Goodhead, I.; Hertz-Fowler, C.; Heger, A.; Hall, N.; et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014, 5, 3979. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Lee, J.H.; Zhang, Z.; De La Rosa, R.; Bi, M.; Tan, Y.; Liao, Y.; Hong, J.; Du, B.; Wu, Y.; et al. Enhancer RNAs Mediate Estrogen-Induced Decommissioning of Selective Enhancers by Recruiting ERalpha and Its Cofactor. Cell Rep. 2020, 31, 107803. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Spann, N.J.; Heinz, S.; Romanoski, C.E.; Allison, K.A.; Stender, J.D.; Chun, H.B.; Tough, D.F.; Prinjha, R.K.; Benner, C.; et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 2013, 51, 310–325. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, S.; Hossan, T.; Alawi, M.; Najafova, Z.; Indenbirken, D.; Bedi, U.; Taipaleenmaki, H.; Ben-Batalla, I.; Scheller, M.; Loges, S.; et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014, 8, 460–469. [Google Scholar] [CrossRef] [Green Version]
- Bose, D.A.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 2017, 168, 135–149.e122. [Google Scholar] [CrossRef] [Green Version]
- Rahnamoun, H.; Lee, J.; Sun, Z.; Lu, H.; Ramsey, K.M.; Komives, E.A.; Lauberth, S.M. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol. 2018, 25, 687–697. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [Google Scholar] [CrossRef] [Green Version]
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 2019, 76, 473–484.e7. [Google Scholar] [CrossRef] [Green Version]
- Crump, N.T.; Ballabio, E.; Godfrey, L.; Thorne, R.; Repapi, E.; Kerry, J.; Tapia, M.; Hua, P.; Lagerholm, C.; Filippakopoulos, P.; et al. BET inhibition disrupts transcription but retains enhancer-promoter contact. Nat. Commun. 2021, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Haberle, V.; Arnold, C.D.; Pagani, M.; Rath, M.; Schernhuber, K.; Stark, A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 2019, 570, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhang, B.; Deochand, D.; Sacta, M.A.; Coppo, M.; Shang, Y.; Guo, Z.; Zeng, X.; Rollins, D.A.; Tharmalingam, B.; et al. Negative elongation factor complex enables macrophage inflammatory responses by controlling anti-inflammatory gene expression. Nat. Commun. 2020, 11, 2286. [Google Scholar] [CrossRef]
- Davari, K.; Lichti, J.; Gallus, C.; Greulich, F.; Uhlenhaut, N.H.; Heinig, M.; Friedel, C.C.; Glasmacher, E. Rapid Genome-wide Recruitment of RNA Polymerase II Drives Transcription, Splicing, and Translation Events during T Cell Responses. Cell Rep. 2017, 19, 643–654. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greulich, F.; Bielefeld, K.A.; Scheundel, R.; Mechtidou, A.; Strickland, B.; Uhlenhaut, N.H. Enhancer RNA Expression in Response to Glucocorticoid Treatment in Murine Macrophages. Cells 2022, 11, 28. https://doi.org/10.3390/cells11010028
Greulich F, Bielefeld KA, Scheundel R, Mechtidou A, Strickland B, Uhlenhaut NH. Enhancer RNA Expression in Response to Glucocorticoid Treatment in Murine Macrophages. Cells. 2022; 11(1):28. https://doi.org/10.3390/cells11010028
Chicago/Turabian StyleGreulich, Franziska, Kirsten Adele Bielefeld, Ronny Scheundel, Aikaterini Mechtidou, Benjamin Strickland, and Nina Henriette Uhlenhaut. 2022. "Enhancer RNA Expression in Response to Glucocorticoid Treatment in Murine Macrophages" Cells 11, no. 1: 28. https://doi.org/10.3390/cells11010028
APA StyleGreulich, F., Bielefeld, K. A., Scheundel, R., Mechtidou, A., Strickland, B., & Uhlenhaut, N. H. (2022). Enhancer RNA Expression in Response to Glucocorticoid Treatment in Murine Macrophages. Cells, 11(1), 28. https://doi.org/10.3390/cells11010028






