Establishment and Characterization of a Novel Gill Cell Line, LG-1, from Atlantic Lumpfish (Cyclopterus lumpus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Husbandry and Ethical Considerations
2.2. Development of Primary Cells
2.3. Routine Maintenance
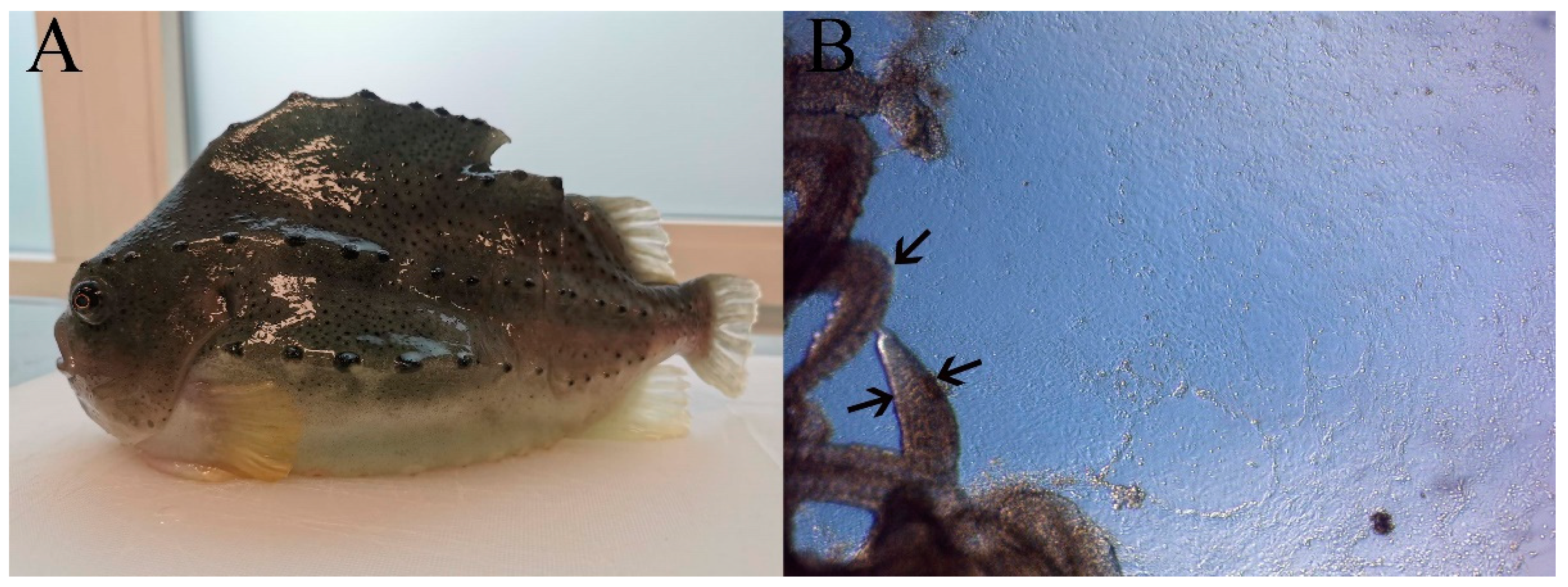
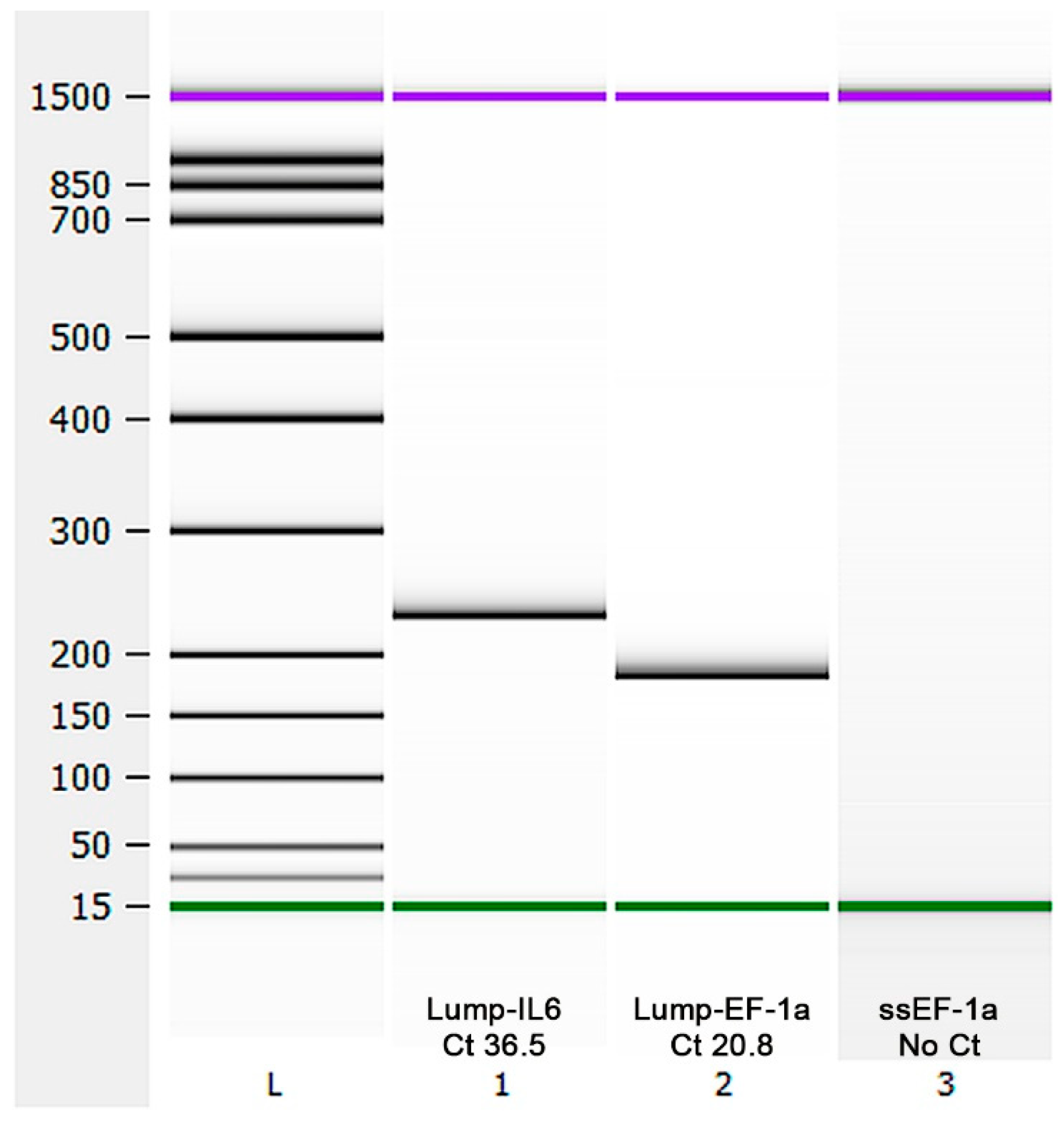
2.4. Species Identification
2.5. Proliferation
2.6. Morphology
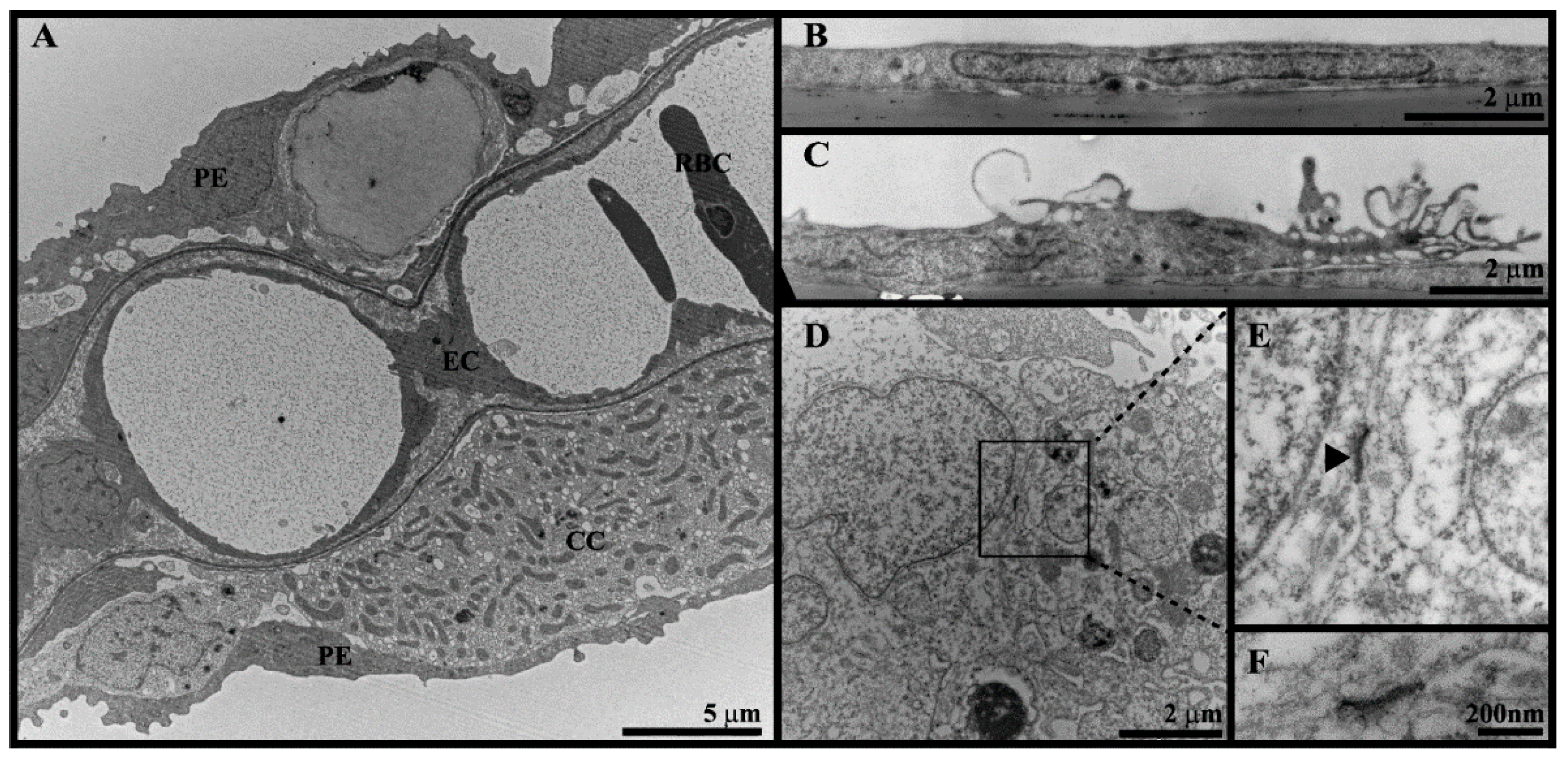
2.7. Transmission Electron Microscopy (TEM)
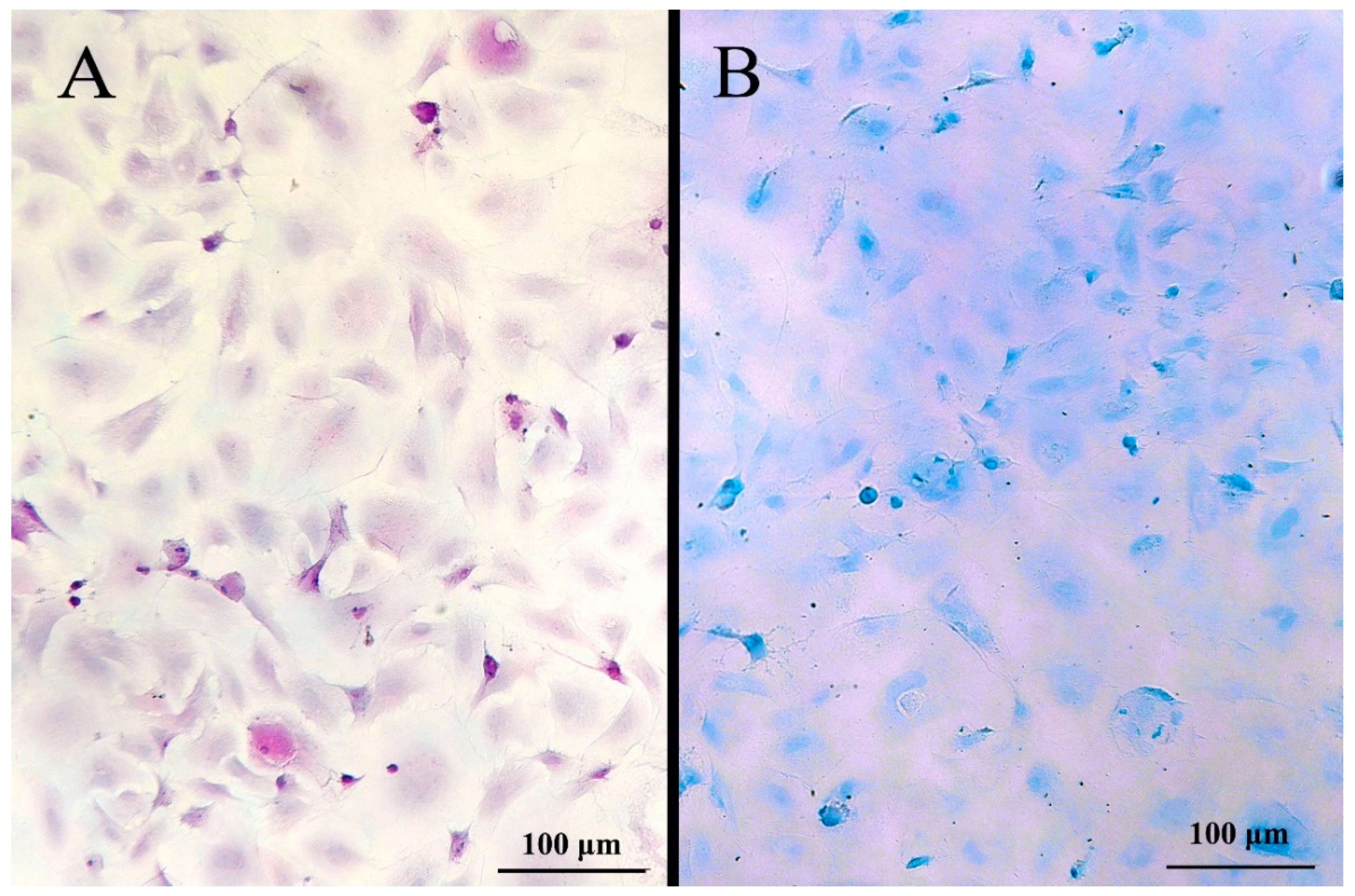
2.8. Periodic Acid-Schiff (PAS), Alcian Blue pH 1 Staining and Immunostaining for Chloride Cells
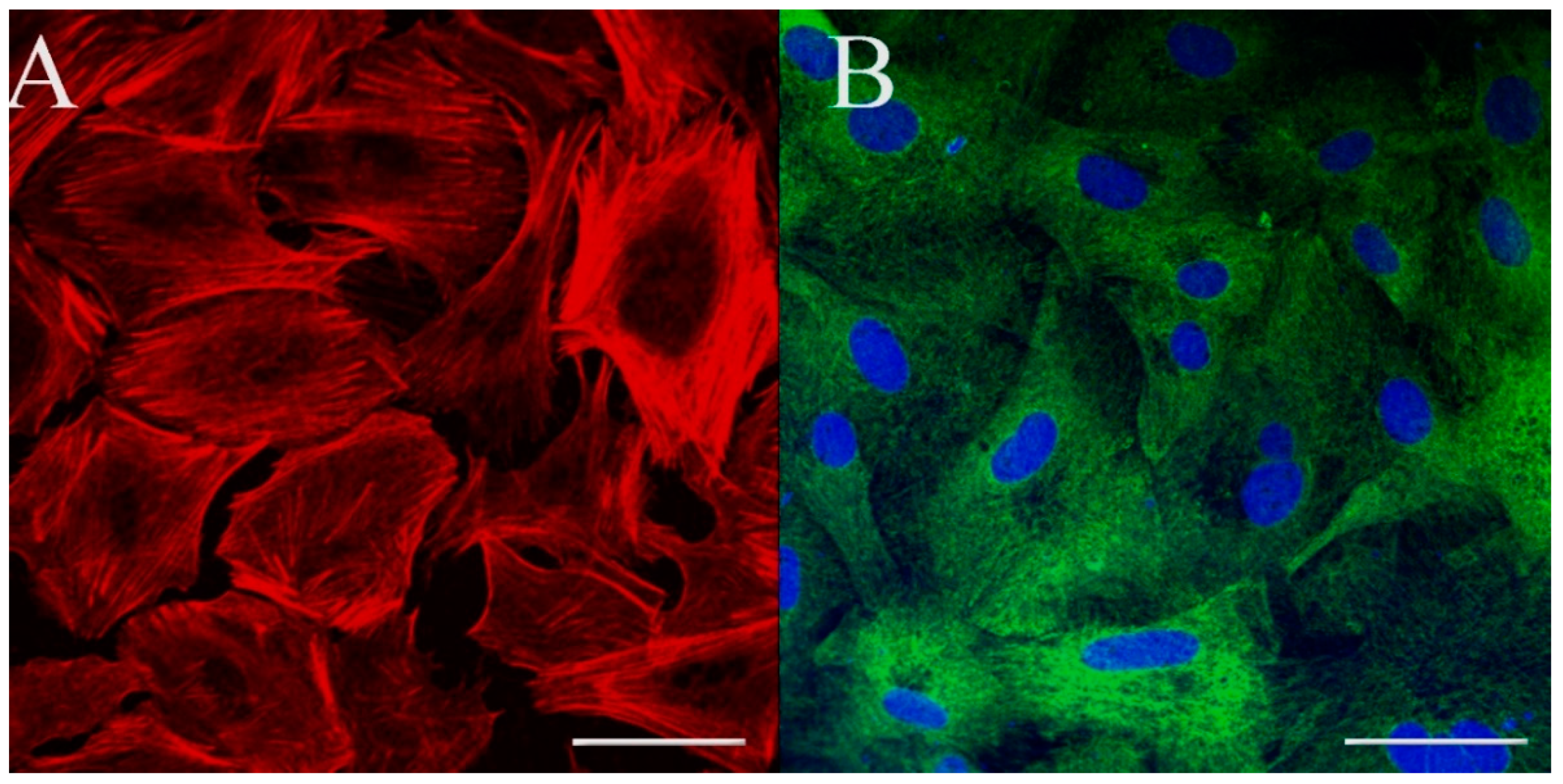
2.9. Immunostaining for Cell Markers
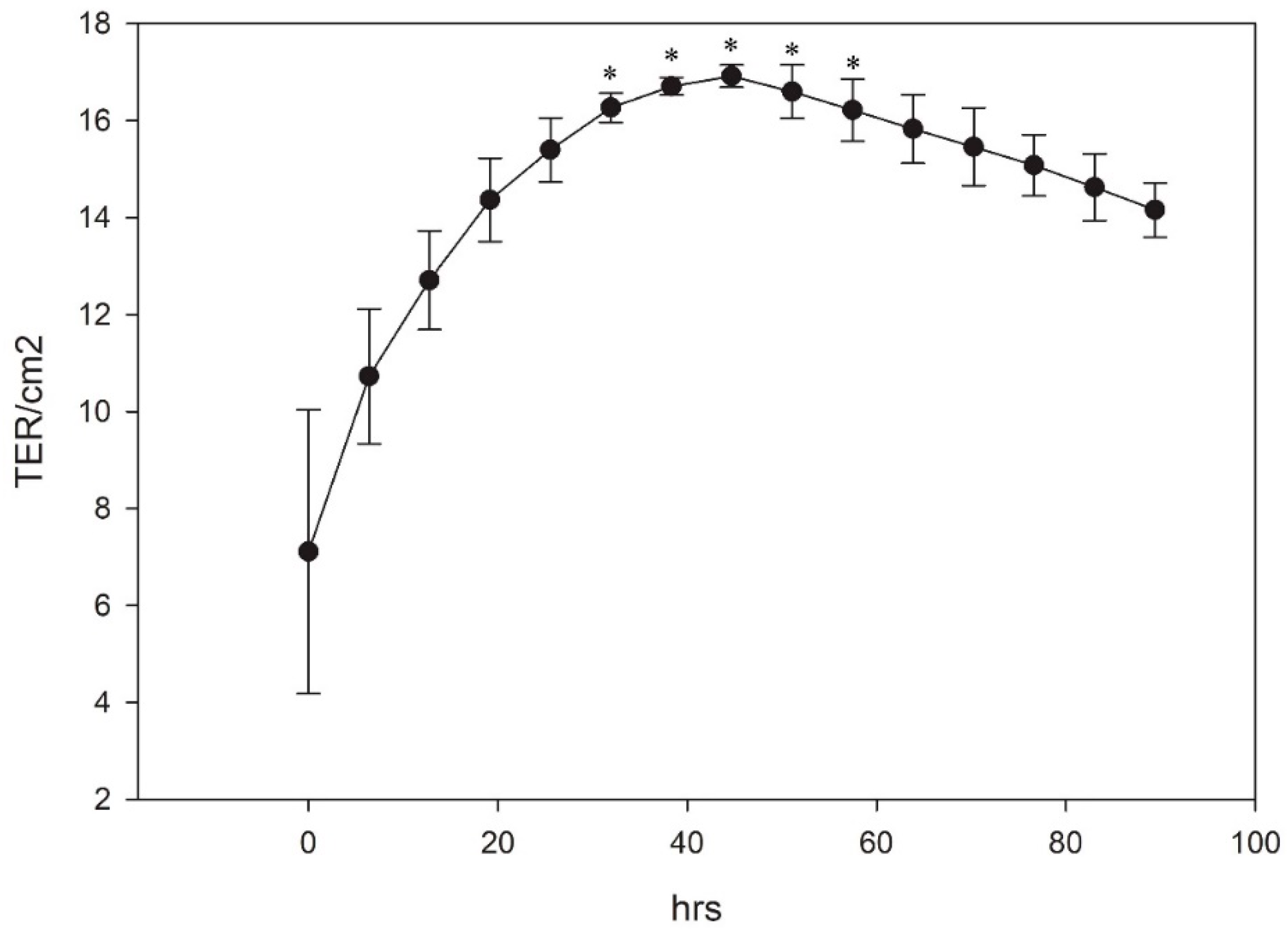
2.10. Transepithelial/Transendothelial Electrical Resistance (TER)
2.11. CYP1A Activity
2.12. Testing for Susceptibility to Fish Viruses
2.13. Statistical Analysis
3. Results
3.1. Development and Maintenance of the LG-1 Cell Line
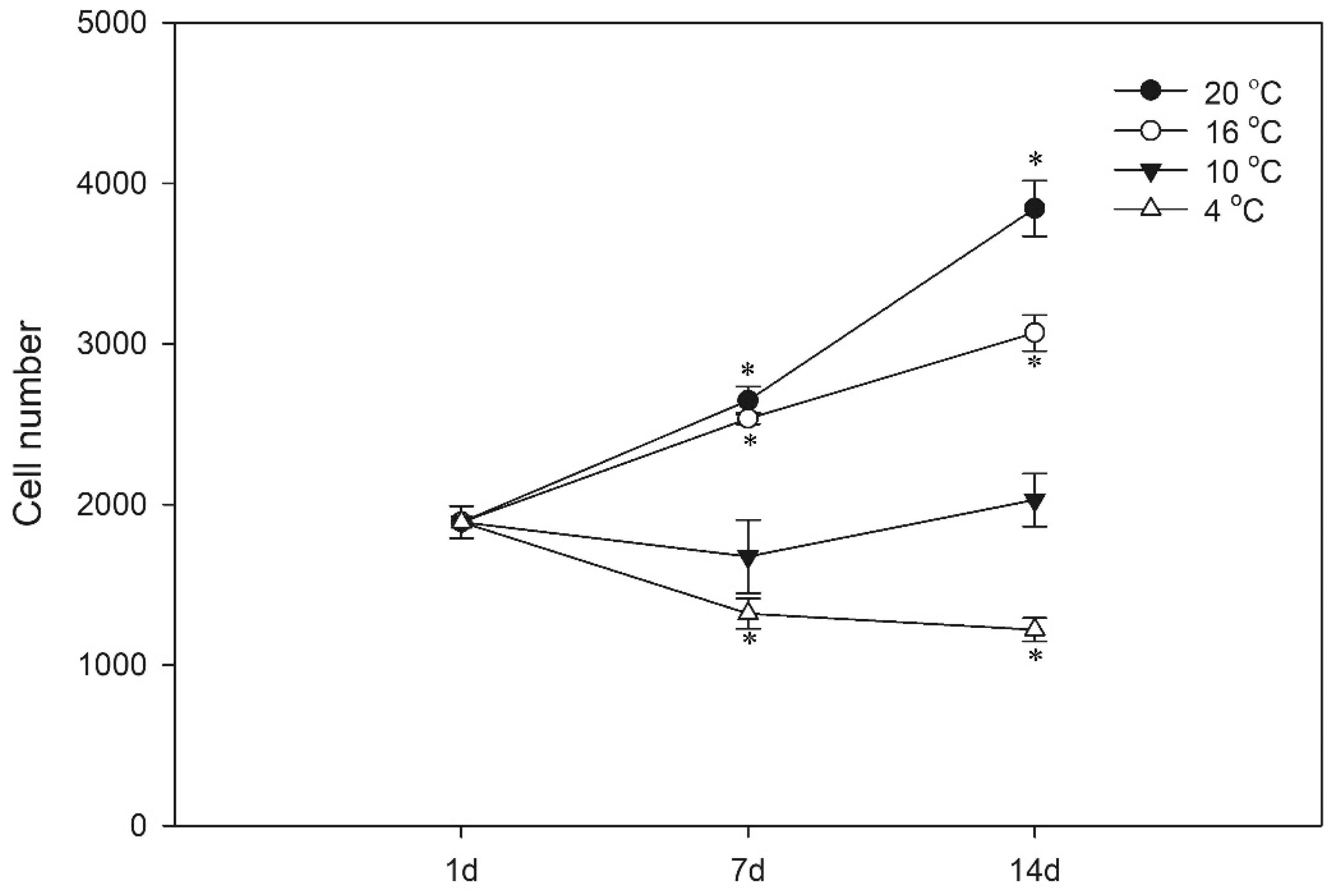
3.2. Proliferation
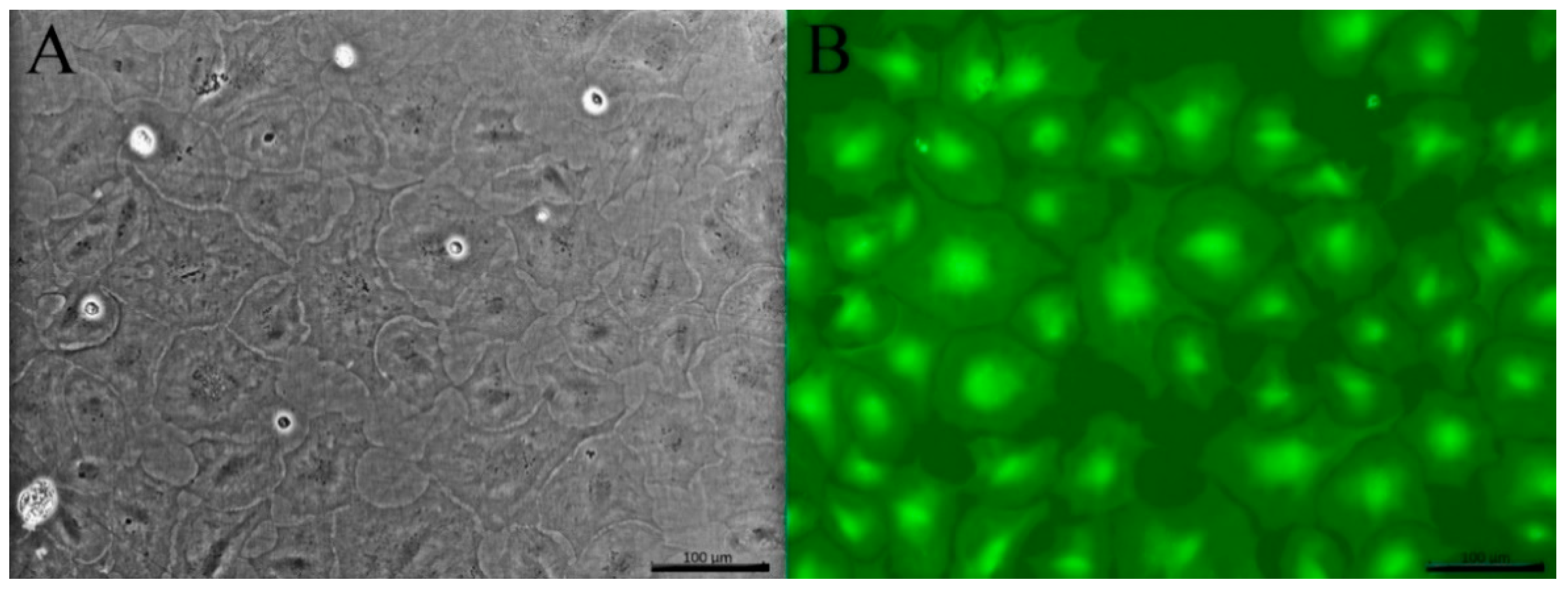
3.3. Cell Morphology
3.4. Generation of TER
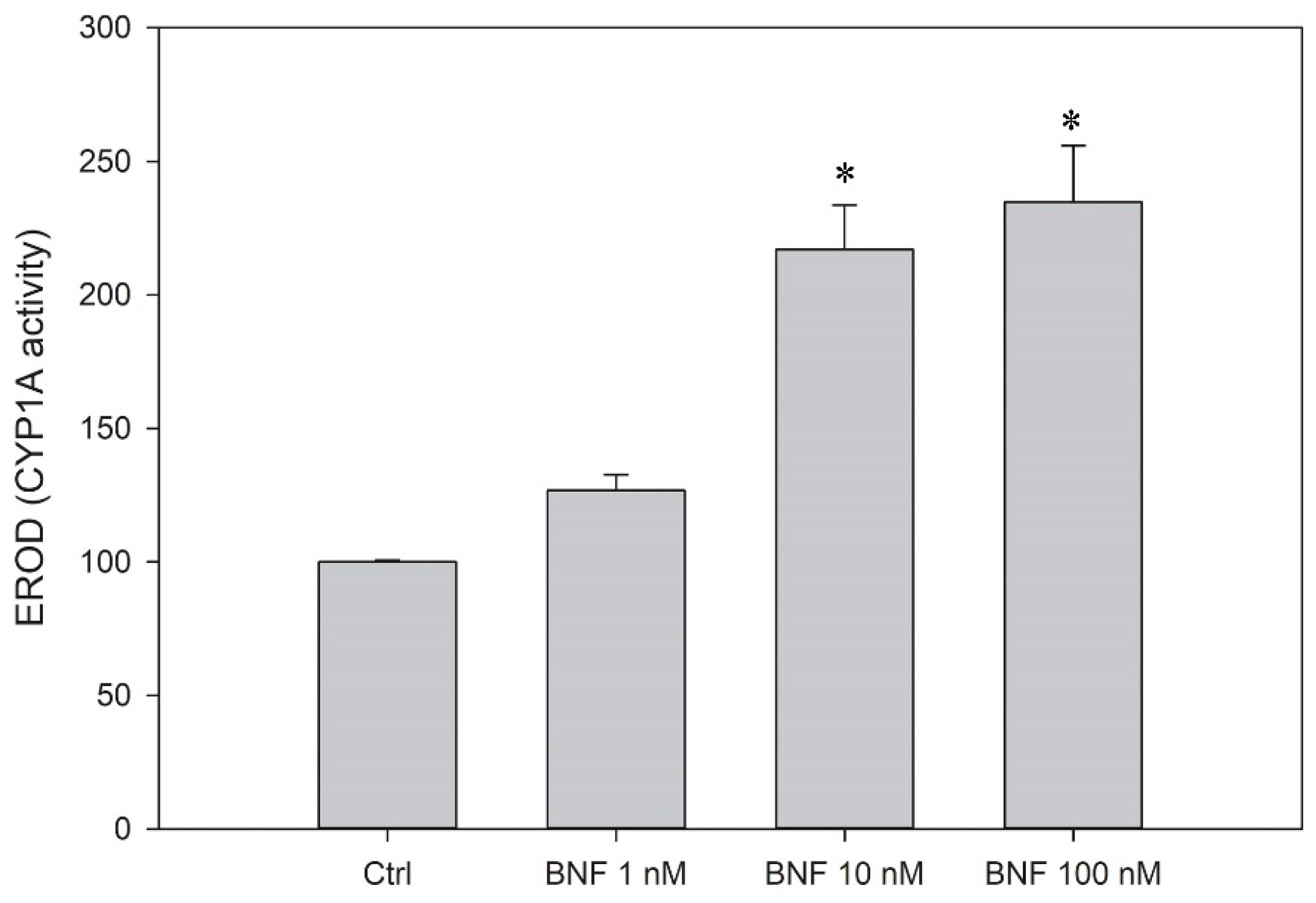
3.5. CYP1A Induction
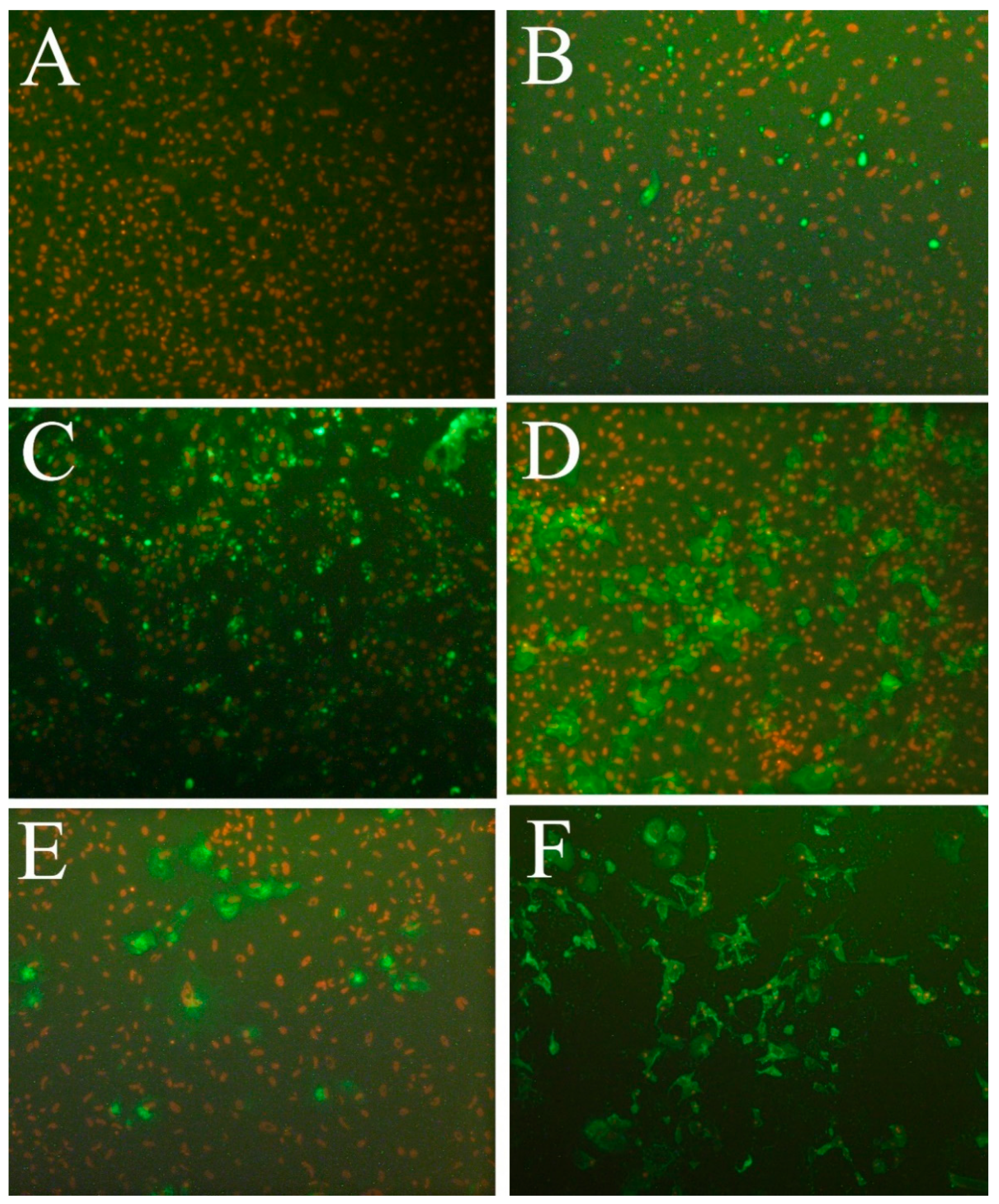
3.6. Susceptibility to Fish Viruses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Directorate_of_Fisheries. Cleanerfish (Lumpfish and Wrasse). Available online: https://www.fiskeridir.no/English/Aquaculture/Statistics/Cleanerfish-Lumpfish-and-Wrasse/_/attachment/download/d70cefea-4c9a-4537-8250-6f1ef7e6827e:ac417b3f12fdb0f0a04792c4fc2078384e6cd9ef/sta-laks-mat-10-rensefisk.xlsx (accessed on 13 August 2021).
- Helland, S.; Dahle, S.V.W.; Hough, C.; Borthen, J. Production of ballan wrasse (Labrus bergylta). Science and Practice. Available online: https://www.fhf.no/prosjekter/prosjektbasen/900554/ (accessed on 13 August 2021).
- Nytrø, A.V.; Vikingstad, E.; Foss, A.; Hangstad, T.A.; Reynolds, P.; Eliassen, G.; Elvegård, T.A.; Falk-Petersen, I.-B.; Imsland, A.K. The effect of temperature and fish size on growth of juvenile lumpfish (Cyclopterus lumpus L.). Aquaculture 2014, 434, 296–302. [Google Scholar] [CrossRef] [Green Version]
- Rimstad, E.; Basic, D.; Gulla, S.; Hjeltnes, B.; Mortensen, S. Risk assessment of fish health associated with the use of cleaner fish in aquaculture. Opinion of the Panel on Animal Health and Welfare of the Norwegian Scientific Committee for Food and Environment. VKM Report 2017. Available online: https://vkm.no/download/18.40cd61cf160507e9844ca6c/1513852770245/Risk%20assessment%20of%20fish%20health%20associated%20with%20the%20use%20of%20cleaner%20fish%20in%20aquaculture.pdf (accessed on 13 August 2021).
- Sommerset, I.; Bang Jensen, B.; Bornø, G.; Haukaas, A.; Brun, E. The Health Situation in Norwegian Aquaculture 2020. Available online: https://www.vetinst.no/rapporter-og-publikasjoner/rapporter/2021/fish-health-report-2020/_/attachment/download/d2c81a9f-2126-488c-828b-10285c988997:187b69c8c0e3ff3065bb49c49462de2e9d0c1cae/rl2230-fiskehelse-eng-3.pdf (accessed on 13 August 2021).
- Parrish, C.R.; Holmes, E.C.; Morens, D.M.; Park, E.-C.; Burke, D.S.; Calisher, C.H.; Laughlin, C.A.; Saif, L.J.; Daszak, P. Cross-Species Virus Transmission and the Emergence of New Epidemic Diseases. Microbiol. Mol. Biol. Rev. 2008, 72, 457–470. [Google Scholar] [CrossRef] [Green Version]
- Erkinharju, T.; Dalmo, R.A.; Hansen, M.; Seternes, T. Cleaner fish in aquaculture: Review on diseases and vaccination. Rev. Aquac. 2021, 13, 189–237. [Google Scholar] [CrossRef]
- Guðmundsdóttir, S.; Vendramin, N.; Cuenca, A.; Sigurðardóttir, H.; Kristmundsson, A.; Iburg, T.M.; Olesen, N.J. Outbreak of viral haemorrhagic septicaemia (VHS) in lumpfish (Cyclopterus lumpus) in Iceland caused by VHS virus genotype IV. J. Fish Dis. 2019, 42, 47–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stagg, H.E.B.; Guðmundsdóttir, S.; Vendramin, N.; Ruane, N.M.; Sigurðardóttir, H.; Christiansen, D.H.; Cuenca, A.; Petersen, P.E.; Munro, E.S.; Popov, V.L.; et al. Characterization of ranaviruses isolated from lumpfish Cyclopterus lumpus L. in the North Atlantic area: Proposal for a new ranavirus species (European North Atlantic Ranavirus). J. Gen. Virol. 2020, 101, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Skoge, R.H.; Brattespe, J.; Økland, A.L.; Plarre, H.; Nylund, A. New virus of the family Flaviviridae detected in lumpfish (Cyclopterus lumpus). Arch. Virol. 2018, 163, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Haugland, G.T.; Olsen, A.-B.; Rønneseth, A.; Andersen, L. Lumpfish (Cyclopterus lumpus L.) develop amoebic gill disease (AGD) after experimental challenge with Paramoeba perurans and can transfer amoebae to Atlantic salmon (Salmo salar L.). Aquaculture 2017, 478, 48–55. [Google Scholar] [CrossRef]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co. Ltd.: London, UK, 1959. [Google Scholar]
- Tannenbaum, J.; Bennett, B.T. Russell and Burch’s 3Rs then and now: The need for clarity in definition and purpose. J. Am. Assoc. Lab. Anim. Sci. 2015, 54, 120–132. [Google Scholar] [PubMed]
- Villena, A.J. Applications and needs of fish and shellfish cell culture for disease control in aquaculture. Rev. Fish Biol. Fish. 2003, 13, 111–140. [Google Scholar] [CrossRef]
- Fischer, M.; Belanger, S.E.; Berckmans, P.; Bernhard, M.J.; Bláha, L.; Coman Schmid, D.E.; Dyer, S.D.; Haupt, T.; Hermens, J.L.M.; Hultman, M.T.; et al. Repeatability and Reproducibility of the RTgill-W1 Cell Line Assay for Predicting Fish Acute Toxicity. Toxicol. Sci. 2019, 169, 353–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Schirmer, K. Cell culture-based biosensing techniques for detecting toxicity in water. Curr. Opin. Biotechnol. 2017, 45, 59–68. [Google Scholar] [CrossRef]
- Eichbaum, K.; Brinkmann, M.; Buchinger, S.; Reifferscheid, G.; Hecker, M.; Giesy, J.P.; Engwall, M.; van Bavel, B.; Hollert, H. In vitro bioassays for detecting dioxin-like activity—Application potentials and limits of detection, a review. Sci. Total Environ. 2014, 487, 37–48. [Google Scholar] [CrossRef]
- Herrero, A.; Thompson, K.D.; Ashby, A.; Rodger, H.D.; Dagleish, M.P. Complex Gill Disease: An Emerging Syndrome in Farmed Atlantic Salmon (Salmo salar L.). J. Comp. Pathol. 2018, 163, 23–28. [Google Scholar] [CrossRef]
- Adelmann, M.; Kollner, B.; Bergmann, S.M.; Fischer, U.; Lange, B.; Weitschies, W.; Enzmann, P.J.; Fichtner, D. Development of an oral vaccine for immunisation of rainbow trout (Oncorhynchus mykiss) against viral haemorrhagic septicaemia. Vaccine 2008, 26, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Gjessing, M.C.; Aamelfot, M.; Batts, W.N.; Benestad, S.L.; Dale, O.B.; Thoen, E.; Weli, S.C.; Winton, J.R. Development and characterization of two cell lines from gills of Atlantic salmon. PLoS ONE 2018, 13, e0191792. [Google Scholar] [CrossRef] [Green Version]
- Volff, J.N. Genome evolution and biodiversity in teleost fish. Heredity 2005, 94, 280–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.F.; Clyburne, S. New Cell Line from the Marine Lumpfish, Cyclopterus lumpus. J. Fish. Res. Board Can. 1977, 34, 134–139. [Google Scholar] [CrossRef]
- NIH. Primer-BLAST Software. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 13 August 2021).
- Malik, M.S.; Teige, L.H.; Braaen, S.; Olsen, A.B.; Nordberg, M.; Amundsen, M.M.; Dhamotharan, K.; Svenning, S.; Edholm, E.S.; Takano, T.; et al. Piscine Orthoreovirus (PRV)-3, but Not PRV-2, Cross-Protects against PRV-1 and Heart and Skeletal Muscle Inflammation in Atlantic Salmon. Vaccines 2021, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Erkinharju, T.; Strandskog, G.; Vågnes, Ø.; Hordvik, I.; Dalmo, R.A.; Seternes, T. Intramuscular vaccination of Atlantic lumpfish (Cyclopterus lumpus L.) induces inflammatory reactions and local immunoglobulin M production at the vaccine administration site. J. Fish Dis. 2019, 42, 1731–1743. [Google Scholar] [CrossRef] [Green Version]
- Suvarna, S.K.; Layton, C.; Bancroft, J.D. Bancroft’s Theory and Practice of Histological Techniques; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; p. 557. [Google Scholar]
- Heinrich, P.; Diehl, U.; Förster, F.; Braunbeck, T. Improving the in vitro ethoxyresorufin-O-deethylase (EROD) assay with RTL-W1 by metabolic normalization and use of β-naphthoflavone as the reference substance. Comp. Biochem. Physiol. C Toxicol. Pharm. 2014, 164, 27–34. [Google Scholar] [CrossRef]
- Dale, O.B.; Orpetveit, I.; Lyngstad, T.M.; Kahns, S.; Skall, H.F.; Olesen, N.J.; Dannevig, B.H. Outbreak of viral haemorrhagic septicaemia (VHS) in seawater-farmed rainbow trout in Norway caused by VHS virus Genotype III. Dis. Aquat. Organ. 2009, 85, 93–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Totland, G.K.; Grotmol, S.; Morita, Y.; Nishioka, T.; Nakai, T. Pathogenicity of nodavirus strains from striped jack Pseudocaranx dentex and Atlantic halibut Hippoglossus hippoglossus, studied by waterborne challenge of yolk-sac larvae of both teleost species. Dis. Aquat. Organ. 1999, 38, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dannevig, B.H.; Nilsen, R.; Modahl, I.; Jankowska, M.; Taksdal, T.; Press, C.M. Isolation in cell culture of nodavirus from farmed Atlantic halibut Hippoglossus hippoglossus in Norway. Dis. Aquat. Organ. 2000, 43, 183–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriette, C.; LeBerre, M.; Boscher, S.K.; Castric, J.; Bremont, M. Characterization and mapping of monoclonal antibodies against the Sleeping disease virus, an aquatic alphavirus. J. Gen. Virol. 2005, 86, 3119–3127. [Google Scholar] [CrossRef]
- Grove, S.; Johansen, R.; Dannevig, B.H.; Reitan, L.J.; Ranheim, T. Experimental infection of Atlantic halibut Hippoglossus hippoglossus with nodavirus: Tissue distribution and immune response. Dis. Aquat. Organ. 2003, 53, 211–221. [Google Scholar] [CrossRef]
- Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Arch. Für Exp. Pathol. Und Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]
- Krueger, J.; Ray, A.; Tamm, I.; Sehgal, P.B. Expression and function of interleukin-6 in epithelial cells. J. Cell Biochem. 1991, 45, 327–334. [Google Scholar] [CrossRef]
- May, L.T.; Torcia, G.; Cozzolino, F.; Ray, A.; Tatter, S.B.; Santhanam, U.; Sehgal, P.B.; Stern, D. Interleukin-6 gene expression in human endothelial cells: RNA start sites, multiple IL-6 proteins and inhibition of proliferation. Biochem. Biophys. Res. Commun. 1989, 159, 991–998. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [Green Version]
- Tanneberger, K.; Knöbel, M.; Busser, F.J.; Sinnige, T.L.; Hermens, J.L.; Schirmer, K. Predicting fish acute toxicity using a fish gill cell line-based toxicity assay. Environ. Sci. Technol. 2013, 47, 1110–1119. [Google Scholar] [CrossRef]
- Mdegela, R.; Myburgh, J.; Correia, D.; Braathen, M.; Ejobi, F.; Botha, C.; Sandvik, M.; Skaare, J.U. Evaluation of the gill filament-based EROD assay in African sharptooth catfish (Clarias gariepinus) as a monitoring tool for waterborne PAH-type contaminants. Ecotoxicology 2006, 15, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Gravato, C.; Guilhermino, L.; Galindo-Riaño, M.D.; Perales, J.A. EROD activity and cytochrome P4501A induction in liver and gills of Senegal sole Solea senegalensis from a polluted Huelva estuary (SW Spain). Comp. Biochem. Physiol. C Toxicol. Pharm. 2014, 166, 134–144. [Google Scholar] [CrossRef]
- Leguen, I.I.; Carlsson, C.; Perdu-Durand, E.; Prunet, P.; Pärt, P.; Cravedi, J.P. Xenobiotic and steroid biotransformation activities in rainbow trout gill epithelial cells in culture. Aquat. Toxicol. 2000, 48, 165–176. [Google Scholar] [CrossRef]
- Clemons, J.H.; Heuvel, M.R.v.d.; Stegeman, J.J.; Dixon, D.G.; Bols, N.C. Comparison of Toxic Equivalent Factors for Selected Dioxin and Furan Congeners Derived Using Fish and Mammalian Liver Cell Lines. Can. J. Fish. Aquat. Sci. 1994, 51, 1577–1584. [Google Scholar] [CrossRef]
- OIE. Aquatic Manual, Chapter 2.3.8. Infection with Salmonid Alphavirus. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/aquatic-manual-online-access/ (accessed on 13 August 2021).
- Simons, J.; Bruno, D.W.; Ho, Y.M.; Murray, W.; Matejusova, I. Common dab, Limanda limanda (L.), as a natural carrier of salmonid alphavirus (SAV) from waters off north-west Ireland. J. Fish. Dis. 2015. [Google Scholar] [CrossRef]
- McCleary, S.; Giltrap, M.; Henshilwood, K.; Ruane, N.M. Detection of salmonid alphavirus RNA in Celtic and Irish Sea flatfish. Dis. Aquat. Organ. 2014, 109, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLoughlin, M.F.; Graham, D.A. Alphavirus infections in salmonids--a review. J. Fish Dis. 2007, 30, 511–531. [Google Scholar] [CrossRef]
- Hellberg, H.; Kvellestad, A.; Dannevig, B.; Borno, G.; Modahl, I.; Haldorsen, R.N.; Vik-Mo, F.; Ottesen, K.; Saetre, E.M.; Sindre, H. Outbreaks of viral nervous necrosis in juvenile and adult farmed Atlantic cod, Gadus morhua L., in Norway. J. Fish Dis. 2010, 33, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Korsnes, K.; Bergh, Ø.; Vik-Mo, F.; Pedersen, J.; Nerland, A.H. Nodavirus in farmed Atlantic cod Gadus morhua in Norway. Dis. Aquat. Organ. 2007, 77, 169–173. [Google Scholar] [CrossRef] [Green Version]
- OIE. Aquatic Manual—Archived version of Chapter 2.3.12. Viral Encephalopathy and Retinopathy 14/11/2019. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/chapitre_viral_encephalopathy_retinopathy.pdf (accessed on 11 August 2021).
- Korsnes, K.; Devold, M.; Nerland, A.H.; Nylund, A. Viral encephalopathy and retinopathy (VER) in Atlantic salmon Salmo salar after intraperitoneal challenge with a nodavirus from Atlantic halibut Hippoglossus hippoglossus. Dis. Aquat. Organ. 2005, 68, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Munro, E.S.; McIntosh, R.E.; Weir, S.J.; Noguera, P.A.; Sandilands, J.M.; Matejusova, I.; Mayes, A.S.; Smith, R. A mortality event in wrasse species (Labridae) associated with the presence of viral haemorrhagic septicaemia virus. J. Fish Dis. 2015, 38, 335–341. [Google Scholar] [CrossRef]
- Johansen, R.; Bergh, Ø.; Modahl, I.; Dahle, G.; Gjerset, B.; Holst, J.C.; Sandlund, N. High prevalence of viral haemorrhagic septicaemia virus (VHSV) in Norwegian spring-spawning herring. Mar. Ecol. Prog. Ser. 2013, 478, 223–230. [Google Scholar] [CrossRef]
- Mortensen, H.F.; Heuer, O.E.; Lorenzen, N.; Otte, L.; Olesen, N.J. Isolation of viral haemorrhagic septicaemia virus (VHSV) from wild marine fish species in the Baltic Sea, Kattegat, Skagerrak and the North Sea. Virus Res. 1999, 63, 95–106. [Google Scholar] [CrossRef]
- Smail, D.A. Isolation and identification of Viral Haemorrhagic Septicaemia (VHS) viruses from cod Gadus morhua with the ulcus syndrome and from haddock Melanogrammus aeglefinus having skin haemorrhages in the North Sea. Dis. Aquat. Organ. 2000, 41, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Graham, R.L.; Baric, R.S. Jumping species-a mechanism for coronavirus persistence and survival. Curr. Opin. Virol. 2017, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wu, Y.; Zhang, W.; Qi, J.; Gao, G.F. Enabling the ‘host jump’: Structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 2014, 12, 822–831. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primers | Amplicon Length | Accession | References |
|---|---|---|---|---|
| Cyclopterus lumpus IL-6 | Fwd: 5′-CAC CAT CAA CCA CAG ACG GA-3′ Rev: 5′-AAC GGC GCT TAC TGA GTT GA-3′ | 224 | MN093127.1 | Designed in house |
| Cyclopterus lumpus EF1a | Fwd: 5′-GGC CAG ATC AAT GCC GGA TA-3′ Rev: 5′-CTC CAC AAC CAT GGG CTT CT-3′ | 189 | XM_034545962.1 | [24] |
| Salmo salar EF1a | Fwd: 5′-TGC CCC TCC AGG ATG TCT AC-3′ Rev: 5′-TCA CCA GGC ATA GCC GAT TC-3′ | 175 | XM_014141923.1 | [25] |
| Virus Isolate | Primary Antibody |
|---|---|
| ISAV Glesvaer AF404340 | anti-ISAV P10, Aquatic Diagnostics, Glasgow, UK |
| SAV1 140699 SPDV WSV | Anti-SAV 17H23, [31] |
| SAV2 MR-R5-2011, HE863664 | Anti-SAV 17H23, [31] |
| SAV3 R-1_2007, LT630447 | Anti-SAV 17H23, [31] |
| VHSV genogroup III [28] | Anti-VHS, IP5B11, BIO-X Diagnostics, Rochefort, Belgium |
| IHNV genogroup M, LR-80, AY442514 | 050690 Mab G Protein, gift Oregon State University, USA |
| Betanodavirus, AH95NorA (BFNNV genotype), [29]) “fish nodavirus” | Anti-VER K67 [32] |
| IPNV Sp, SK1433/09 field isolate Norway | Anti-IPN BIO 345, BIO-X Diagnostics |
| lumpfish flavivirus CLuV, field outbreak | na |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sindre, H.; Gjessing, M.C.; Fosse, J.H.; Hermansen, L.C.; Böckerman, I.; Amundsen, M.M.; Dahle, M.K.; Solhaug, A. Establishment and Characterization of a Novel Gill Cell Line, LG-1, from Atlantic Lumpfish (Cyclopterus lumpus L.). Cells 2021, 10, 2442. https://doi.org/10.3390/cells10092442
Sindre H, Gjessing MC, Fosse JH, Hermansen LC, Böckerman I, Amundsen MM, Dahle MK, Solhaug A. Establishment and Characterization of a Novel Gill Cell Line, LG-1, from Atlantic Lumpfish (Cyclopterus lumpus L.). Cells. 2021; 10(9):2442. https://doi.org/10.3390/cells10092442
Chicago/Turabian StyleSindre, Hilde, Mona C. Gjessing, Johanna Hol Fosse, Lene C. Hermansen, Inger Böckerman, Marit M. Amundsen, Maria K. Dahle, and Anita Solhaug. 2021. "Establishment and Characterization of a Novel Gill Cell Line, LG-1, from Atlantic Lumpfish (Cyclopterus lumpus L.)" Cells 10, no. 9: 2442. https://doi.org/10.3390/cells10092442
APA StyleSindre, H., Gjessing, M. C., Fosse, J. H., Hermansen, L. C., Böckerman, I., Amundsen, M. M., Dahle, M. K., & Solhaug, A. (2021). Establishment and Characterization of a Novel Gill Cell Line, LG-1, from Atlantic Lumpfish (Cyclopterus lumpus L.). Cells, 10(9), 2442. https://doi.org/10.3390/cells10092442








