Abstract
Traumatic brain injury (TBI) represents one of the leading causes of mortality and morbidity worldwide, placing an enormous socioeconomic burden on healthcare services and communities around the world. Survivors of TBI can experience complications ranging from temporary neurological and psychosocial problems to long-term, severe disability and neurodegenerative disease. The current lack of therapeutic agents able to mitigate the effects of secondary brain injury highlights the urgent need for novel target discovery. This study comprises two independent systematic reviews, investigating both microRNA (miRNA) and proteomic expression in rat models of severe TBI (sTBI). The results were combined to perform integrated miRNA-protein co-expression analyses with the aim of uncovering the potential roles of miRNAs in sTBI and to ultimately identify new targets for therapy. Thirty-four studies were included in total. Bioinformatic analysis was performed to identify any miRNA–protein associations. Endocytosis and TNF signalling pathways were highlighted as common pathways involving both miRNAs and proteins found to be differentially expressed in rat brain tissue following sTBI, suggesting efforts to find novel therapeutic targets that should be focused here. Further high-quality investigations are required to ascertain the involvement of these pathways and their miRNAs in the pathogenesis of TBI and other CNS diseases and to therefore uncover those targets with the greatest therapeutic potential.
1. Introduction
Traumatic brain injury (TBI) has been described as a problem of epidemic magnitude, with an estimated annual incidence of over 10 million and rising [1,2,3,4,5,6]. As such, TBI represents one of the leading causes of mortality and morbidity worldwide, and the principal cause of both in Western countries for individuals under the age of 45 [7,8,9,10]. This public health issue therefore places an enormous physical, psychological and socioeconomic burden on healthcare services, individuals and communities around the world [11,12,13]. For instance, in the United States, an annual economic burden of USD 76.5 billion has been estimated for this type of injury, encompassing total lifetime medical costs and productivity losses due to TBI [1,14].
Brain injury results from an external mechanical force to the head, face or neck which leads to a series of complex pathological alterations in neural homeostasis [15]. The most common modes of TBI include falls, motor vehicle accidents, sport-related injuries, military injuries and assaults. Survivors of TBI can experience complications ranging from temporary neurological and psychosocial problems to long-term, severe disability and neurodegenerative disease [13,16,17]. Clinical features may include prolonged coma, seizures, headaches, nausea, aphasia and amnesia, as well as behavioural abnormalities including increased aggression or anxiety. Symptoms can manifest within minutes of injury and may persist for months or years post-TBI [13,18,19]. Currently, there are no known effective treatments for TBI and no FDA-approved therapies to prevent or limit the deficits caused by brain injuries. This is a likely consequence of the heterogeneity of the disease and the many complex biochemical and pathophysiological events which occur at multiple time points following injury, the underlying mechanisms of which are yet to be fully elucidated [7,13,20].
MicroRNAs (miRNAs, miRs) are a class of small, endogenous, non-coding RNA molecules which regulate protein synthesis at the post transcriptional level by binding to the 3′-untranslated region (UTR) of target genes, resulting in translational suppression and mRNA degradation [21,22]. The human genome is believed to encode over 2000 miRNAs, which are estimated to play a role in targeting and regulating the expression of 30–60% of all genes, with single miRNAs possessing the ability to regulate up to several hundred target mRNAs [10,22,23,24]. These molecules therefore control a wide range of biological functions and processes including development, differentiation, apoptosis, immune responses and cell metabolism [20]. In addition, miRNAs are free circulating in the bloodstream as well as in other biofluids (CSF, saliva, urine, etc.), acting as effectors of cell-to-cell communication. They may also be passively released into biofluids as result of cell injury, thereby potentially serving as biomarkers of injury and/or toxicity. Several studies have already demonstrated the utility of microRNAs as potential circulating biomarkers in the context of TBI [1,10,22].
The ability of a single miRNA to regulate up to several hundred target genes makes miRNA particularly suitable for therapeutic purpose. In addition, tissue-specific viral injection, which allows precise spatially targeted therapies and avoids the pitfalls of systematically delivered oligonucleotides, are already in development.
Therefore, in this research, two independent systematic reviews were conducted, investigating both miRNA and proteomic expression in brain tissues of sTBI rat models with the aim to identify novel in situ-targets able to mitigate the devastating effects of secondary brain injury. By combining the two individual reviews, we hoped to establish associations between miRNA and protein expression following sTBI in order to better understand the pathways and underlying mechanisms involved in this disease pathology and to highlight any potential novel targets for the condition.
2. Methods
2.1. Literature Search
Throughout the conduction of this systematic review, the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement for systematic reviews was adhered to [25]. Two independent searches of the literature were conducted, the results of which were combined to perform integrated miRNA-protein co-expression analyses. To ensure that the literature was reviewed sufficiently and that all appropriate articles for inclusion were found, 3 independent databases were searched: PubMed, Web of Science and Scopus. The search terms applied to each database for the miRNA and protein searches were ‘traumatic brain injury’, ‘microRNA’ and ‘rat model’, and ‘traumatic brain injury’, ‘proteins’, ‘rat model’ and ‘biomarker’, respectively. Search terms were applied to all fields rather than simply to the title or abstract to ensure all suitable papers were captured in the search. To structure the search, Boolean operators were used as follows: (1) ‘(traumatic brain injury) AND (microRNA) AND (rat model)’ and (2) ‘(traumatic brain injury) AND (proteins) AND (rat model) AND (biomarker)’. Results from each search were copied into separate Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA, USA, available at: https://office.microsoft.com/excel, accessed on 15 June 2021) spreadsheets, including information surrounding the authors, title, publication year, digital object identifier (DOI) and web address for each search result.
2.2. Selection Criteria
Two authors (C.O. and V.D.P.) independently scrutinised the titles and abstracts of each paper generated from the literature search to assess their eligibility according to our inclusion and exclusion criteria. The selection criteria for the searches were developed based on the following inclusion (Table 1) and exclusion criteria (Table 2):

Table 1.
Inclusion criteria.

Table 2.
Exclusion criteria.
2.3. Data Collection
The two researchers independently extracted the following study characteristics from the selected, eligible studies for each review: authors, title, year, web address, access to full text, primary paper, English language, animal model used (rat), sample size, injury model used, severity of TBI, methodology of miRNA and protein expression analysis, time point of analysis, results (miRNAs/proteins named). Any additional, useful information regarding study findings or limitations was also recorded.
In order to be able to perform bioinformatic analysis, a separate table for both the miRNA literature search and the protein literature search were created, detailing the names of each miRNA or protein found to be differentially expressed (significantly upregulated, downregulated or both), along with the time point of analysis post-injury and the associated fold changes and p values. MiRNAs and proteins that were found to be differentially expressed across 3 or more of the selected papers were recorded. This additional information helped to determine whether it was possible to perform a meta-analysis.
2.4. Bioinformatic Analysis
To establish the target genes of the differentially expressed miRNAs found in the literature, DIANA Tools mirPath (DIANA-miRpath v.3, University of Thesselay, Greece, available on line: DIANA TOOLS—mirPath v.3 (grnet.gr), accessed on 15 June 2021) [26] was used and the resulting targets screened to determine any overlap with the proteins also found. An additional search was performed in order to validate miRNA-target interactions and using 2 separate and different tools: miRTargets, (miRDB, MO, USA, available online: http://mirdb.org/, accessed on 15 June 2021) and miRwalk (miRwalk, Heidelberg, DE, available online: http://mirwalk.umm.uni-heidelberg.de/, accessed on 1 Septemeber 2021). The direction of expression fold change for the associated miRNAs and proteins were evaluated, with any inverse relationships being highlighted—this was executed to account for the fact miRNAs inversely regulate protein expression through a negative feedback mechanism [3]. Additionally, these interactions were checked to observe whether they were experimentally validated, using TarBase (Tarbase v8.0, Greece, available online: https://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex, accessed on 1 September 2021), an online database for functional annotations. For those miRNAs and proteins with an inverse expression relationship, KEGG pathway analysis was performed to identify the biological pathways in which they are implicated. For the miRNAs, analysis was conducted using DIANA Tools mirPath [26], while for the proteins, DAVID tool (DAVID v6.8, MD, USA, available online: (https://david.ncifcrf.gov/content.jsp?file=Acknowledgements.html, accessed on 15 June 2021) [27,28] software was used. Pathways in common for both the miRNAs and proteins were then documented before DIANA Tools mirPath, and the KEGG Pathway Database (KEEG, Kyoto Encyclopedia of Genes and Genomes, Japan, available online: https://www.genome.jp/kegg/pathway.html, accessed on 15 July 2021) was used to reveal the miRNAs and proteins involved in these common pathways.
2.5. Risk of Bias
Risk of bias was assessed using the SYRCLE risk of bias (RoB) tool for animal studies [29]. Risk for each study was assessed across 10 domains by two independent reviewers (C.O. and Z.A.), with any disagreements being settled through discussion. The domains used to assess the potential risk of bias across the studies were: random sequence generation, baseline characteristics described, correct timing of randomization, allocation concealment, random housing, blinding, random outcome assessment, incomplete data, sample size calculation and primary outcome specified. Algorithms were then used to determine whether each study had either a low, some, or high risk of bias.
3. Results
3.1. Study Selection
The miRNA search yielded 161 results across the three databases. Of these, 64 duplicate studies were identified and excluded using the reference management software, EndNote (EndNote v.20, PA, USA) [30], along with nine papers which were excluded once filters for free full text/open access were applied within the databases themselves. The remaining 88 records were then manually screened and assessed for eligibility against the selection criteria previously described (Section 2.2). Seventy-eight results were excluded during this process, leaving 10 miRNA studies for inclusion in the systematic review [31,32,33,34,35,36,37,38,39,40]. Figure 1 provides an overview of the miRNA literature search and study selection process.
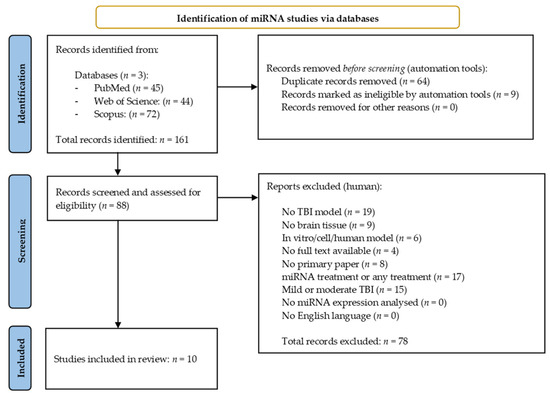
Figure 1.
Preferred reporting items for systematic reviews and meta-analysis (PRISMA) flow chart detailing the literature search process for miRNA studies.
A total of 530 records were identified across the three databases from the protein search. The same search filters were applied, leading to 295 results being excluded while a further 71 duplicates were removed using EndNote 20 [30]. A total of 140 papers were manually excluded following assessment against the selection criteria, leaving a total of 24 papers eligible for inclusion in the review [36,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63]. Figure 2 provides an overview of the protein literature search and study selection process.
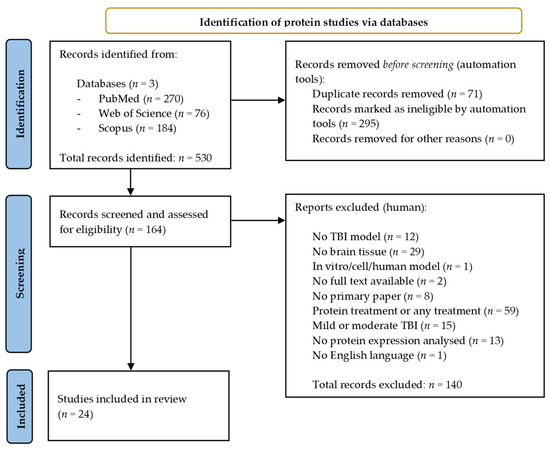
Figure 2.
Preferred reporting items for systematic reviews and meta-analysis (PRISMA) flow chart detailing the literature search process for protein studies.
3.2. Study Characteristics
All studies included are primary research papers investigating changes in either miRNA or protein expression following severe TBI in rats. The injury model, time point of analysis post-TBI and analysis method differed across the studies. Supplementary Materials Tables S1 and S2 provide an overview of the study characteristics for the included papers [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63]. The data collected here allowed us to establish whether performance of a meta-analysis was possible. In concurrence with a qualified statistician, the conclusion was reached that, due to the heterogeneous nature of data presentation, study techniques and analysis methods across the miRNA and protein studies, completion of a meta-analysis was not possible.
3.3. miRNA and Protein Expression Changes Post-TBI
The names of each miRNA and protein found to be differentially expressed in rat brain tissue following severe TBI were extracted from each of the included papers, along with whether expression was significantly up- or downregulated at each time point following injury. A comprehensive list of each differentially expressed miRNA or protein and the direction of change can be found in Tables S1 and S2 in Supplementary Materials, respectively.
3.4. Bioinformatic Analysis
To determine the gene targets of each differentially expressed miRNA, DIANA Tools mirPath v.3 software was used. The targets for each were copied and Microsoft Excel 2016 was used to identify any overlaps between those miRNAs targets and the proteins also found to be differentially expressed in the literature. In total, 57 miRNA target genes and proteins were found to overlap (Table S3 in Supplementary Materials). Data previously collected regarding the direction of expression fold change was consulted to uncover which of the overlapping miRNAs and proteins had an inverse expression relationship. Our results showed that expression of 58 miRNAs was inversely correlated to at least one of the proteins, while expression of 46 proteins was inversely correlated to at least one of the miRNAs. This data is illustrated in Table 3. In Table 3, we also show whether these interactions were detected using different prediction tools (miTargets and miRwalk) and whether they were experimentally validated.

Table 3.
miRNA-protein interactions predicted by DIANA tools and confirmed by miRtaregts and miRwalk tools. The table also reports the inverse expression relationships between miRNAs and proteins and whether the interaction was experimentally validated.
3.5. KEGG Pathways Analysis to Identify Potential miRNA Processes and Targets for TBI
For those miRNAs and proteins with an inverse expression relationship, KEGG pathway analysis was performed to identify the biological pathways in which they are implicated. For miRNAs, analysis was conducted using DIANA Tools mirPath v.3 [26], and for proteins, DAVID v6.8 [27,28] software was used. A full list of the KEGG pathways identified for the miRNAs and proteins with inverse expression relationships can be found in the Supplementary Materials (Table S4 in Supplementary Materials). Two KEGG pathways were found to be in common between the miRNAs and proteins: endocytosis and TNF signalling. The miRNAs and proteins/genes implicated in these pathways are shown in Table 4.

Table 4.
miRNAs and proteins implicated in the Endocytosis and TNF signalling KEGG pathways.
3.6. Risk of Bias (RoB)
RoB analysis found that all included articles specified the primary outcomes and described baseline characteristics. Nearly all studies (97%) had full data availability with no missing data. However, no studies calculated sample size, randomly housed animals or concealed treatment allocations. Incidences of random sequence generation, correct timing of randomisation, blinding and random outcome assessment were also low, with 85–100% of studies reporting no information concerning these domains. As a result, overall risk of bias was judged to be high and hence may invalidate certain findings of the included papers. RoB analysis results are depicted in a stacked bar chart in Figure 3.
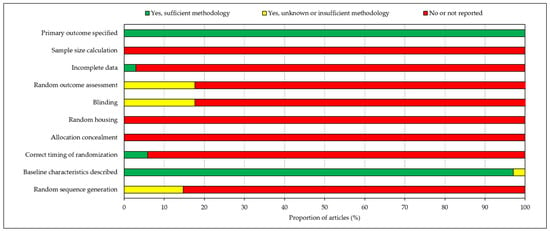
Figure 3.
Risk of bias assessment according to the SYRCLE risk of bias (RoB) tool for animal studies. Risk of bias was judged to be high.
4. Discussion
This systematic review identified 34 studies (10 miRNA and 14 protein studies) that met our inclusion and exclusion criteria in relation to changes in miRNA and protein expression following TBI in the brain tissue of rats. Due to the heterogeneous nature of parameters including data presentation, study technique and analysis method across those included, meta-analysis was not deemed possible, hence studies were analysed qualitatively. Our results demonstrated that 57 gene targets of the differentially expressed miRNAs identified in the literature overlapped with the proteins also found to be significantly up- or downregulated following sTBI. By studying those overlapping miRNAs and proteins which had inverse expression relationships, we were able to uncover the common KEGG pathways and highlight potential novel targets for therapeutic intervention.
Two KEGG pathways were found to be in common: the endocytosis pathway and TNF signalling pathway. Upon review of the literature, little was found to associate the endocytosis pathway with TBI or any other CNS pathologies. Endocytosis involves the internalisation of cell surface receptors, ligands, nutrients, plasma membrane proteins and lipids, bringing them to the interior of the cell [82]. This process occurs via pinocytosis, phagocytosis or receptor-mediated endocytosis, with cargo progressing through several stages of sorting and routing before being degraded by lysosomes and recycled back to the cell surface or secreted [82,83]. Despite the fact that several endocytic proteins are known to be dysregulated in certain disease states and that functionally active miRs, such as miR-29a and miR-125a, are secreted in the synaptosomes via exocytosis and endocytosis pathways, the involvement of the endocytosis pathway in TBI does not appear to be well documented in the literature [84].
On the contrary, the implication of the TNF signalling pathway in TBI has previously been reported. Several miRs have been already described as regulators of TNF [85,86,87]. TNF is a critical cytokine able to induce a number of important intracellular signalling pathways, including both inflammation and cell death (apoptosis), with dysregulation of its downstream NF-Κb signalling pathway, giving rise to chronic inflammation responsible for several human pathologies [88]. TNFR1 is the principal receptor for TNF and is expressed in almost all cells, while the second receptor, TNFR2, is expressed in a limited range of cells including microglia, oligodendrocytes and specific neuron subtypes [89]. The previously documented involvement of this pathway in inflammation and apoptosis processes, as well as the CNS-specific expression of its receptors, allows us to be confident when speculating the involvement of this pathway in the pathogenesis of TBI and other CNS diseases as our results would suggest.
Evidence exists which indicates that TNF signalling plays an important role in exacerbating both Amyloid-β (Aβ) and tau pathologies in vivo [90]. APP (amyloid precursor protein) and MAPT (microtubule associated protein tau) were both found to exhibit an inverse expression relationship with multiple miRNAs in this systematic review. In our results, APP expression was shown to be inversely correlated with that of miR-185-5p and miR-144-3p, and MAPT with miR-671-5p and miR-298-5p. Wu et al. have reported that, through a series of signalling cascades, TBI results in the cleavage of both APP and tau (at APP N585a and Tau N368 sites, respectively), mediating Alzheimer’s disease pathogenesis through the promotion of Aβ production and tau hyperphosphorylation, processes which ultimately induce neuroinflammation and neurotoxicity [91]. Furthermore, the presence of extracellular Aβ plaques has been recorded both acutely and chronically following severe TBI, and similar aggregates are also found in elderly people of normal cognitive status [92,93,94]. The involvement of APP and MAPT in certain neurodegenerative diseases, ageing, and most importantly, in TBI is well documented and supportive of our findings. Therefore, targeting miRNAs which negatively regulate TNF signalling or APP expression directly may be used as strategy to prevent accumulation of this protein and subsequent neurological disease.
Our current knowledge of miRNAs and their roles in several physiological and pathological processes have led to them serving as important emerging candidates for novel therapeutic targets in CNS injury [95]. These molecules possess numerous characteristics which make them extremely attractive tools for therapeutic intervention; in addition to the previously described knowledge that manipulation of one miRNA can affect many targets, certain other desirable features have contributed to the increasing interest surrounding these molecules in terms of drug development [95,96]. For example, not only does their short length (~22 nucleotides) allow miRNA-based drugs to be easily designed; they are also frequently conserved between species, and several drug delivery systems have been approved for human use which allow in vivo delivery of these emerging therapeutics [95,97,98,99]. Examples of miRNA-based therapeutics are miRNA mimics (agomir) and miRNA inhibitors (antagomiRs), which decrease and increase target gene expression, respectively [95]. As the name suggests, miRNA mimics and can act as compensatory agents in the event of a loss of miRNA functional activity due to downregulation of expression in correlation with disease progression. Conversely, in a situation where specific miRNAs are upregulated in response to injury and appear to be contributing towards disease pathogenesis, miRNA inhibitors can be used with the aim of suppressing this overexpression [95].
It is possible to reliably predict potential gene targets based on the partially complementary sequences between mature miRNAs and mRNA candidate targets. As a result, several pharmaceutical companies have been investigating the application of miRNAs as therapeutics in recent years, with various drugs advancing to human trial stages, such as Miravirsen, RG-101, RG-125/AZD4076, MRX34, and TagomiRs [95,99,100,101,102]. However, despite certain miRNA drug candidates displaying promising effects and an ability to improve neurological deficits post-injury in preclinical studies, phase I and phase II trials, of the more than 30 clinical trials which have taken place over the past 3 decades, all candidates have failed to demonstrate therapeutic efficacy in humans with TBI [7,13,103]. This widespread failure may be attributed to the heterogeneity of this form of injury, as well as the variability in treatment protocols between trial sites [7]. Regardless of the reason, this lack of success has highlighted the urgent need for novel target discovery for this increasingly devastating disease.
Limitations
One of the main limitations of this study is the variation in time point of expression analysis from the initial time of injury (ranging from 2 h to 12 months post-TBI). Given that biomarker signatures are constantly changing and evolving following a brain injury, this variation in analysis time point makes comparisons between studies extremely difficult. Another significant limitation of this study is that included studies used different models of TBI, including the fluid-percussion, cortical impact, penetrating ballistic-like brain injury models, which may affect different microRNAs due to the different mechanisms of injury. Certain studies also used laser capture microdissection, whilst the majority of the studies used specific regions of the brain to harvest mRNA and protein, potentially introducing greater variety. Other limitations include the heterogeneity between studies in terms of data presentation, study technique and analysis method, which rendered the performance of a meta-analysis impossible. Furthermore, studies failed to report sample sizes; this along with a number of other shortcomings meant that overall risk of bias was judged to be high. High risk of bias should be avoided going forward through adherence to guidelines outlining standardised techniques for animal studies such as the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines [104]. In the future, investigators should focus efforts towards identifying clear miRNA and/or protein biomarker signatures across a set of reasonable time points, reducing variation in study techniques and analysis methods, all while ensuring adequate sample sizes are used and appropriate guidelines are adhered to.
5. Conclusions
In part, our results support the existing literature surrounding the involvement of the TNF signalling pathway, APP, MAPT and their associated miRNAs in the pathogenesis of TBI and other neuropathological conditions, and hence their potential as therapeutic targets. However, the involvement of the endocytosis pathway in this field of research has yet to be widely discussed. Although we can speculate involvement of both biological pathways in TBI, the limitations discussed above as well as the high risk of bias and lack of meta-analysis are considerably limiting factors in this review, hence caution is advised when interpreting this data and further high-quality studies are required. In recent years, miRNAs have shown immense potential as diagnostic biomarkers and therapeutic targets in a number of pathologies, including TBI. Further studies are needed in both animal and human models of TBI to better understand the mechanisms underlying the pathogenesis of this increasingly devastating disease, the miRNA-protein interactions which occur at different time points following brain injury, and to ultimately uncover the most effective targets for treatment.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cells10092425/s1, Table S1: Study characteristics and differentially regulated miRNAs post-sTBI, Table S2: Study characteristics and differentially regulated proteins post-sTBI, Table S3: Common miRNA targets and differentially expressed proteins, Table S4: KEGG pathways identified for miRNAs and proteins with inverse expression relationships and their significance (p-value). Pathways highlighted in blue represent common pathways.
Author Contributions
Conceptualization, C.O. and V.D.P.; methodology, C.O., V.D.P., and Z.A.; formal analysis, C.O. and V.D.P.; data curation, C.O. and V.D.P.; writing—original draft preparation, C.O. and V.D.P.; writing—review and editing, C.O., V.D.P., and Z.A.; supervision, V.D.P. and Z.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived for this study as per advice from the NHS Health Research Authority (UK) decision tool since it is a systematic review of published literature.
Informed Consent Statement
Patient consent was waived because no patients or members of the public were involved in the design, conduct of this study, or reporting of this research.
Data Availability Statement
All data generated as part of this study are included in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bhomia, M.; Balakathiresan, N.S.; Wang, K.K.; Papa, L.; Maheshwari, R.K. A Panel of Serum MiRNA Biomarkers for the Diagnosis of Severe to Mild Traumatic Brain Injury in Humans. Sci. Rep. 2016, 6, 28148. [Google Scholar] [CrossRef]
- Faul, M.; Coronado, V. Epidemiology of traumatic brain injury. Handb. Clin. Neurol. 2015, 127, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Capizzi, A.; Woo, J.; Verduzco-Gutierres, M. Traumatic Brain Injury: An Overview of Epidemiology, Pathophysiology, and Medical Management. Med. Clin. N. Am. 2020, 104, 213–238. [Google Scholar] [CrossRef] [PubMed]
- Hyder, A.A.; Wunderlich, C.A.; Puvanachandra, P.; Gururaj, G.; Kobusingye, O.C. The impact of traumatic brain injuries: A global perspective. NeuroRehabilitation 2007, 22, 341–353. [Google Scholar] [CrossRef]
- Corrigan, J.D.; Selassie, A.W.; Orman, J.A. The epidemiology of traumatic brain injury. J. Head Trauma Rehabil. 2010, 25, 72–80. [Google Scholar] [CrossRef]
- Feigin, V.L.; Theadom, A.; Barker-Collo, S.; Starkey, N.J.; McPherson, K.; Kahan, M.; Dowell, A.; Brown, P.; Parag, V.; Kydd, R.; et al. Incidence of traumatic brain injury in New Zealand: A population-based study. Lancet Neurol. 2013, 12, 53–64. [Google Scholar] [CrossRef]
- Toffolo, K.; Osei, J.; Kelly, W.; Poulsen, A.; Donahue, K.; Wang, J.; Hunter, M.; Bard, J.; Wang, J.; Poulsen, D. Circulating microRNAs as biomarkers in traumatic brain injury. Neuropharmacology 2019, 145, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Langlois, J.A.; Rutland-Brown, W.; Wald, M.M. The epidemiology and impact of traumatic brain injury: A brief overview. J. Head Trauma Rehabil. 2006, 21, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Andelic, N.; Howe, E.I.; Hellstrøm, T.; Sanchez, M.F.; Lu, J.; Løvstad, M.; Røe, C. Disability and quality of life 20 years after traumatic brain injury. Brain Behav. 2018, 8, e01018. [Google Scholar] [CrossRef]
- Di Pietro, V.; Ragusa, M.; Davies, D.; Su, Z.; Hazeldine, J.; Lazzarino, G.; Hill, L.J.; Crombie, N.; Foster, M.; Purrello, M.; et al. MicroRNAs as Novel Biomarkers for the Diagnosis and Prognosis of Mild and Severe Traumatic Brain Injury. J. Neurotrauma 2017, 34, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.A.; Bell, J.M.; Breiding, M.J.; Xu, L. Traumatic Brain Injury-Related Emergency Department Visits, Hospitalizations, and Deaths—United States, 2007 and 2013. MMWR Surveill. Summ. 2017, 66, 1–16. [Google Scholar] [CrossRef]
- De Guzman, E.; Ament, A. Neurobehavioral Management of Traumatic Brain Injury in the Critical Care Setting: An Update. Crit. Care Clin. 2017, 33, 423–440. [Google Scholar] [CrossRef]
- Ng, S.Y.; Lee, A.Y.W. Traumatic Brain Injuries: Pathophysiology and Potential Therapeutic Targets. Front. Cell. Neurosci. 2019, 13, 528. [Google Scholar] [CrossRef] [PubMed]
- Rockhill, C.M.; Jaffe, K.; Zhou, C.; Fan, M.Y.; Katon, W.; Fann, J.R. Health care costs associated with traumatic brain injury and psychiatric illness in adults. J. Neurotrauma 2012, 29, 1038–1046. [Google Scholar] [CrossRef]
- Menon, D.K.; Schwab, K.; Wright, D.W.; Maas, A.I. Position statement: Definition of traumatic brain injury. Arch. Phys. Med. Rehabil. 2010, 91, 1637–1640. [Google Scholar] [CrossRef]
- Zaloshnja, E.; Miller, T.; Langlois, J.A.; Selassie, A.W. Prevalence of long-term disability from traumatic brain injury in the civilian population of the United States, 2005. J. Head Trauma Rehabil. 2008, 23, 394–400. [Google Scholar] [CrossRef]
- Bazarian, J.J.; Cernak, I.; Noble-Haeusslein, L.; Potolicchio, S.; Temkin, N. Long-term neurologic outcomes after traumatic brain injury. J. Head Trauma Rehabil. 2009, 24, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Bruns, J., Jr.; Hauser, W.A. The epidemiology of traumatic brain injury: A review. Epilepsia 2003, 44, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Andriessen, T.M.; Jacobs, B.; Vos, P.E. Clinical characteristics and pathophysiological mechanisms of focal and diffuse traumatic brain injury. J. Cell. Mol. Med. 2010, 14, 2381–2392. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Nam, J.W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Redell, J.B.; Moore, A.N.; Ward, N.H.; Hergenroeder, G.W.; Dash, P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 2010, 27, 2147–2156. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, 460–466. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Hooijmans, C.R.; Rovers, M.M.; de Vries, R.B.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- The EndNote Team. EndNote; EndNote 20: Clarivate: Philadelphia, PA, USA, 2013. [Google Scholar]
- Hu, Z.; Yu, D.; Almeida-Suhett, C.; Tu, K.; Marini, A.M.; Eiden, L.; Braga, M.F.; Zhu, J.; Li, Z. Expression of miRNAs and their cooperative regulation of the pathophysiology in traumatic brain injury. PLoS ONE 2012, 7, e39357. [Google Scholar] [CrossRef]
- Jadhav, S.P.; Kamath, S.P.; Choolani, M.; Lu, J.; Dheen, S.T. microRNA-200b modulates microglia-mediated neuroinflammation via the cJun/MAPK pathway. J. Neurochem. 2014, 130, 388–401. [Google Scholar] [CrossRef]
- Korotkov, A.; Puhakka, N.; Gupta, S.D.; Vuokila, N.; Broekaart, D.W.M.; Anink, J.J.; Heiskanen, M.; Karttunen, J.; van Scheppingen, J.; Huitinga, I.; et al. Increased expression of miR142 and miR155 in glial and immune cells after traumatic brain injury may contribute to neuroinflammation via astrocyte activation. Brain Pathol. 2020, 30, 897–912. [Google Scholar] [CrossRef]
- Puhakka, N.; Bot, A.M.; Vuokila, N.; Debski, K.J.; Lukasiuk, K.; Pitkänen, A. Chronically dysregulated NOTCH1 interactome in the dentate gyrus after traumatic brain injury. PLoS ONE 2017, 12, e0172521. [Google Scholar] [CrossRef]
- Redell, J.B.; Liu, Y.; Dash, P.K. Traumatic brain injury alters expression of hippocampal microRNAs: Potential regulators of multiple pathophysiological processes. J. Neurosci. Res. 2009, 87, 1435–1448. [Google Scholar] [CrossRef]
- Thangavelu, B.; Wilfred, B.S.; Johnson, D.; Gilsdorf, J.S.; Shear, D.A.; Boutte, A.M. Penetrating Ballistic-Like Brain Injury Leads to MicroRNA Dysregulation, BACE1 Upregulation, and Amyloid Precursor Protein Loss in Lesioned Rat Brain Tissues. Front. Neurosci. 2020, 14, 915. [Google Scholar] [CrossRef]
- Vuokila, N.; Aronica, E.; Korotkov, A.; van Vliet, E.A.; Nuzhat, S.; Puhakka, N.; Pitkänen, A. Chronic regulation of miR-124-3p in the perilesional cortex after experimental and human TBI. Int. J. Mol. Sci. 2020, 21, 2418. [Google Scholar] [CrossRef]
- Vuokila, N.; Lukasiuk, K.; Bot, A.M.; van Vliet, E.A.; Aronica, E.; Pitkänen, A.; Puhakka, N. miR-124-3p is a chronic regulator of gene expression after brain injury. Cell Mol. Life Sci. 2018, 75, 4557–4581. [Google Scholar] [CrossRef]
- Wang, W.X.; Prajapati, P.; Vekaria, H.; Spry, M.; Cloud, A.; Sullivan, P.; Springer, J. Temporal changes in inflammatory mitochondria-enriched microRNAs following traumatic brain injury and effects of miR-146a nanoparticle delivery. Neural Regen. Res. 2021, 16, 514–522. [Google Scholar] [CrossRef]
- Xiao, X.; Bai, P.; Cao, S.; Jiang, Y.; Liang, W.; Wang, T.; Luo, X.; Guan, Q.; Gao, L.; Zhang, L. Integrated Bioinformatics Analysis for the Identification of Key Molecules and Pathways in the Hippocampus of Rats After Traumatic Brain Injury. Neurochem. Res. 2020, 45, 928–939. [Google Scholar] [CrossRef]
- Anderson, K.J.; Scheff, S.W.; Miller, K.M.; Roberts, K.N.; Gilmer, L.K.; Yang, C.; Shaw, G. The phosphorylated axonal form of the neurofilament subunit NF-H (pNF-H) as a blood biomarker of traumatic brain injury. J. Neurotrauma 2008, 25, 1079–1085. [Google Scholar] [CrossRef]
- Anwer, M.; Lara-Valderrabano, L.; Karttunen, J.; Ndode-Ekane, X.E.; Puhakka, N.; Pitkänen, A. Acute Downregulation of Novel Hypothalamic Protein Sushi Repeat-Containing Protein X-Linked 2 after Experimental Traumatic Brain Injury. J. Neurotrauma 2020, 37, 924–938. [Google Scholar] [CrossRef]
- Bonneh-Barkay, D.; Zagadailov, P.; Zou, H.; Niyonkuru, C.; Figley, M.; Starkey, A.; Wang, G.; Bissel, S.J.; Wiley, C.A.; Wagner, A.K. YKL-40 expression in traumatic brain injury: An initial analysis. J. Neurotrauma 2010, 27, 1215–1223. [Google Scholar] [CrossRef]
- Cui, G.; Yu, Z.; Li, Z.; Wang, W.; Lu, T.; Qian, C.; Li, J.; Ding, Y. Increased expression of Foxj1 after traumatic brain injury. J. Mol. Neurosci. 2011, 45, 145–153. [Google Scholar] [CrossRef][Green Version]
- Dalgard, C.L.; Cole, J.T.; Kean, W.S.; Lucky, J.J.; Sukumar, G.; McMullen, D.C.; Pollard, H.B.; Watson, W.D. The cytokine temporal profile in rat cortex after controlled cortical impact. Front. Mol. Neurosci. 2012. [Google Scholar] [CrossRef]
- Das Gupta, S.; Lipponen, A.; Paldanius, K.M.A.; Puhakka, N.; Pitkanen, A. Dynamics of clusterin protein expression in the brain and plasma following experimental traumatic brain injury. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Dawish, H.; Mahmood, A.; Schallert, T.; Chopp, M.; Therrien, B. Mild traumatic brain injury (MTBI) leads to spatial learning deficits. Brain Inj. 2012, 26, 151–165. [Google Scholar] [CrossRef]
- DeDominicis, K.E.; Hwang, H.; Cartagena, C.M.; Shear, D.A.; Boutte, A.M. Cerebrospinal Fluid Biomarkers Are Associated with Glial Fibrillary Acidic Protein and alpha II-spectrin Breakdown Products in Brain Tissues Following Penetrating Ballistic-Like Brain Injury in Rats. Front. Neurol. 2018, 9, 490. [Google Scholar] [CrossRef]
- Kilbourne, M.; Kuehn, R.; Tosun, C.; Caridi, J.; Keledjian, K.; Bochicchio, G.; Scalea, T.; Gerzanich, V.; Simard, J.M. Novel model of frontal impact closed head injury in the rat. J. Neurotrauma 2009, 26, 2233–2243. [Google Scholar] [CrossRef]
- Kobeissy, F.H.; Guingab-Cagmat, J.D.; Zhang, Z.; Moghieb, A.; Glushakova, O.Y.; Mondello, S.; Boutté, A.M.; Anagli, J.; Rubenstein, R.; Bahmad, H.; et al. Neuroproteomics and Systems Biology Approach to Identify Temporal Biomarker Changes Post Experimental Traumatic Brain Injury in Rats. Front. Neurol. 2016, 7, 198. [Google Scholar] [CrossRef]
- Kobeissy, F.H.; Liu, M.C.; Yang, Z.; Zhang, Z.; Zheng, W.; Glushakova, O.; Mondello, S.; Anagli, J.; Hayes, R.L.; Wang, K.K. Degradation of βII-Spectrin Protein by Calpain-2 and Caspase-3 Under Neurotoxic and Traumatic Brain Injury Conditions. Mol. Neurobiol. 2015, 52, 696–709. [Google Scholar] [CrossRef]
- Kobeissy, F.H.; Ottens, A.K.; Zhang, Z.; Liu, M.C.; Denslow, N.D.; Dave, J.R.; Tortella, F.C.; Hayes, R.L.; Wang, K.K. Novel differential neuroproteomics analysis of traumatic brain injury in rats. Mol. Cell. Proteom. 2006, 5, 1887–1898. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, C.; Liu, W.; Luo, P.; Zhang, L.; Wang, Y.; Wang, Z.; Fei, Z. A novel rat model of blast-induced traumatic brain injury simulating different damage degree: Implications for morphological, neurological, and biomarker changes. Front. Cell. Neurosci. 2015, 9, 168. [Google Scholar] [CrossRef]
- Liu, M.C.; Akle, V.; Zheng, W.; Kitlen, J.; O’Steen, B.; Larner, S.F.; Dave, J.R.; Tortella, F.C.; Hayes, R.L.; Wang, K.K. Extensive degradation of myelin basic protein isoforms by calpain following traumatic brain injury. J. Neurochem. 2006, 98, 700–712. [Google Scholar] [CrossRef]
- McDonald, W.S.; Jones, E.E.; Wojciak, J.M.; Drake, R.R.; Sabbadini, R.A.; Harris, N.G. Matrix-Assisted Laser Desorption Ionization Mapping of Lysophosphatidic Acid Changes after Traumatic Brain Injury and the Relationship to Cellular Pathology. Am. J. Pathol. 2018, 188, 1779–1793. [Google Scholar] [CrossRef]
- Mrozek, S.; Delamarre, L.; Capilla, F.; Al-Saati, T.; Fourcade, O.; Constantin, J.M.; Geeraerts, T. Cerebral Expression of Glial Fibrillary Acidic Protein, Ubiquitin Carboxy-Terminal Hydrolase-L1, and Matrix Metalloproteinase 9 After Traumatic Brain Injury and Secondary Brain Insults in Rats. Biomark Insights 2019, 14, 1177271919851515. [Google Scholar] [CrossRef]
- Ottens, A.K.; Bustamante, L.; Golden, E.C.; Yao, C.; Hayes, R.L.; Wang, K.K.; Tortella, F.C.; Dave, J.R. Neuroproteomics: A biochemical means to discriminate the extent and modality of brain injury. J. Neurotrauma 2010, 27, 1837–1852. [Google Scholar] [CrossRef]
- Ottens, A.K.; Golden, E.C.; Bustamante, L.; Hayes, R.L.; Denslow, N.D.; Wang, K.K. Proteolysis of multiple myelin basic protein isoforms after neurotrauma: Characterization by mass spectrometry. J. Neurochem. 2008, 104, 1404–1414. [Google Scholar] [CrossRef]
- Pabón, M.M.; Acosta, S.; Guedes, V.A.; Tajiri, N.; Kaneko, Y.; Borlongan, C.V. Brain Region-Specific Histopathological Effects of Varying Trajectories of Controlled Cortical Impact Injury Model of Traumatic Brain Injury. CNS Neurosci. Ther. 2016, 22, 200–211. [Google Scholar] [CrossRef]
- Rubenstein, R.; Chang, B.; Davies, P.; Wagner, A.K.; Robertson, C.S.; Wang, K.K. A novel, ultrasensitive assay for tau: Potential for assessing traumatic brain injury in tissues and biofluids. J. Neurotrauma 2015, 32, 342–352. [Google Scholar] [CrossRef]
- Schober, M.E.; Requena, D.F.; Davis, L.J.; Metzger, R.R.; Bennett, K.S.; Morita, D.; Niedzwecki, C.; Yang, Z.; Wang, K.K. Alpha II Spectrin breakdown products in immature Sprague Dawley rat hippocampus and cortex after traumatic brain injury. Brain Res. 2014, 1574, 105–112. [Google Scholar] [CrossRef]
- Yao, C.; Williams, A.J.; Ottens, A.K.; Lu, X.C.; Liu, M.C.; Hayes, R.L.; Wang, K.K.; Tortella, F.C.; Dave, J.R. P43/pro-EMAPII: A potential biomarker for discriminating traumatic versus ischemic brain injury. J. Neurotrauma 2009, 26, 1295–1305. [Google Scholar] [CrossRef]
- Zheng, F.; Zhou, Y.T.; Feng, D.D.; Li, P.F.; Tang, T.; Luo, J.K.; Wang, Y. Metabolomics analysis of the hippocampus in a rat model of traumatic brain injury during the acute phase. Brain Behav. 2020, 10, e01520. [Google Scholar] [CrossRef]
- Boudreau, R.L.; Jiang, P.; Gilmore, B.L.; Spengler, R.M.; Tirabassi, R.; Nelson, J.A.; Ross, C.A.; Xing, X.; Davidson, B.L. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron 2014, 81, 294–305. [Google Scholar] [CrossRef]
- Pellegrino, L.; Stebbing, J.; Braga, V.M.; Frampton, A.E.; Jacob, J.; Buluwela, L.; Jiao, L.R.; Periyasamy, M.; Madsen, C.D.; Caley, M.P.; et al. miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013, 41, 5400–5412. [Google Scholar] [CrossRef]
- Pillai, M.M.; Gillen, A.E.; Yamamoto, T.M.; Kline, E.; Brown, J.; Flory, K.; Hesselberth, J.R.; Kabos, P. HITS-CLIP reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res. Treat. 2014, 8, 559–564. [Google Scholar] [CrossRef]
- Kishore, S.; Jaskiewicz, L.; Burger, L.; Hausser, J.; Khorshid, M.; Zavolan, M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 2011, 8, 559–564. [Google Scholar] [CrossRef]
- Karginov, F.D.; Hannon, G.J. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013, 27, 1624–1632. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef]
- Haecker, I.; Gay, L.A.; Yang, Y.; Hu, J.; Morse, A.M.; McIntyre, L.M.; Renne, R. Ago HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012, 8, e1002884. [Google Scholar] [CrossRef]
- Kameswaran, V.; Bramswig, N.C.; McKenna, L.B.; Penn, M.; Schug, J.; Hand, N.J.; Chen, J.; Choi, I.; Vourekas, A.; Won, K.J.; et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic isle Cell. Cell. Metab. 2014, 19, 135–145. [Google Scholar] [CrossRef]
- Xue, Y.; Ouyang, K.; Huang, J.; Zhou, Y.; Ouyang, H.; Li, H.; Wang, G.; Wu, Q.; Wei, C.; Bi, Y.; et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013, 152, 82–96. [Google Scholar] [CrossRef]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, P.G.; Bartel, P.B. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Whisnant, A.D.; Bogerd, H.P.; Flores, O.; Ho, P.; Powers, J.G.; Sharova, N.; Stevenson, M.; Chen, C.H.; Cullen, B.R. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 2013, 4, e000193. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, I.; Yang, X.; Brown, J.; Ramakrishnan, A.; Torok-Storb, B.; Kabos, P.; Hesselberth, J.R.; Pillai, M.M. Genome-wide analysis of miRNA-mRNA interactions in marrow stromal cells. Stem. Cells 2014, 32, 662–673. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tavazoie, S.F.; Alarcón, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massagué, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Kai-How Farh, K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef]
- Luo, D.; Wilson, J.M.; Harvel, N.; Liu, J.; Pei, L.; Huang, S.; Hawthorn, L.A.; Shi, H. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J. Transl. Med. 2013, 11, 57. [Google Scholar] [CrossRef]
- Mandemakers, W.; Abuhatzira, L.; Xu, H.; Caromile, L.A.; Hébert, S.S.; Snellinx, A.; Morais, V.A.; Matta, S.; Cai, T.; Notkins, A.L.; et al. Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2 β: Identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia 2013, 56, 1547–1556. [Google Scholar] [CrossRef]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased. BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef]
- Khan, I.; Steeg, P.S. Endocytosis: A pivotal pathway for regulating metastasis. Br. J. Cancer 2021, 124, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.J.; Steinberg, F. To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2018, 19, 679–696. [Google Scholar] [CrossRef]
- Jie, X.; Qun, C.; Ke, Z.; Chenyu, Z.; Qipeng, Z. Synaptosomes secrete and uptake functionally active microRNAs via exocytosis and endocytosis pathways. JNC 2012, 124, 15–25. [Google Scholar]
- Xing, L.; Wanwen, L.; Yuanyuan, Z.; Chaoquan, P. MiR-125b overexpression in EPCs ameliorated TNF-α-induced activation of the NF-κB pathway. Cardiol. Res. Pract. 2020, 7, 6210847. [Google Scholar]
- Kim, S.W.; Ramasamy, K.; Bouamar, H.; Lin, A.P.; Jiang, D.; Aguiar, R.C. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc. Natl. Acad. Sci. USA 2012, 109, 7865–7870. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, I.; Lee, S.G.; Kim, Y.J.; Silwal, P.; Kim, J.Y.; Kim, J.K.; Seo, W.; Chung, C.; Cho, H.K.; et al. MiR-144-3p is associated with pathological inflammation in patients infected with Mycobacteroides abscessus. Exp. Mol. Med. 2021, 53, 136–149. [Google Scholar] [CrossRef]
- Van Quickelberghe, E.; De Sutter, D.; van Loo, G.; Eyckerman, S.; Gevaert, K. A protein-protein interaction map of the TNF-induced NF-κB signal transduction pathway. Sci. Data 2018, 5, 180289. [Google Scholar] [CrossRef]
- Medler, J.; Wajant, H. Tumor necrosis factor receptor-2 (TNFR2): An overview of an emerging drug target. Expert Opin. Targets 2019, 23, 295–307. [Google Scholar] [CrossRef]
- Decourt, B.; Lahiri, D.K.; Sabbagh, M.N. Targeting Tumor Necrosis Factor Alpha for Alzheimer’s Disease. Curr. Alzheimer Res. 2017, 14, 412–425. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Z.H.; Liu, X.; Zhang, Z.; Gu, X.; Yu, S.P.; Keene, C.D.; Cheng, L.; Ye, K. Traumatic brain injury triggers APP and Tau cleavage by delta-secretase, mediating Alzheimer’s disease pathology. Prog. Neurobiol. 2020, 185, 101730. [Google Scholar] [CrossRef] [PubMed]
- Mufson, E.J.; Ikonomovic, M.D.; Counts, S.E.; Perez, S.E.; Malek-Ahmadi, M.; Scheff, S.W.; Ginsberg, S.D. Molecular and cellular pathophysiology of preclinical Alzheimer’s disease. Behav. Brain Res. 2016, 311, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Ikonomovic, M.D.; Uryu, K.; Abrahamson, E.E.; Ciallella, J.R.; Trojanowski, J.Q.; Lee, V.M.; Clark, R.S.; Marion, D.W.; Wisniewski, S.R.; DeKosky, S.T. Alzheimer’s pathology in human temporal cortex surgically excised after severe brain injury. Exp. Neurol. 2004, 190, 192–203. [Google Scholar] [CrossRef]
- Johnson, V.E.; Stewart, W.; Smith, D.H. Widespread τ and amyloid-β pathology many years after a single traumatic brain injury in humans. Brain Pathol. 2012, 22, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Liu, D.Z.; Jickling, G.C.; Sharp, F.R.; Yin, K.J. MicroRNA-based therapeutics in central nervous system injuries. J. Cereb. Blood Flow Metab. 2018, 38, 1125–1148. [Google Scholar] [CrossRef]
- Madathil, S.K.; Nelson, P.T.; Saatman, K.E.; Wilfred, B.R. MicroRNAs in CNS injury: Potential roles and therapeutic implications. Bioessays 2011, 33, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar] [CrossRef]
- Chang, H.I.; Yeh, M.K. Clinical development of liposome-based drugs: Formulation, characterization, and therapeutic efficacy. Int. J. Nanomed. 2012, 7, 49–60. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Schmidt, M.F. Drug target miRNAs: Chances and challenges. Trends Biotechnol. 2014, 32, 578–585. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.G. Embracing failure: What the Phase III progesterone studies can teach about TBI clinical trials. Brain Inj. 2015, 29, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Percie du Sert, N.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; Emerson, M.; et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020, 18, e3000411. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).