Heterogeneity of Hepatic Stellate Cells in Fibrogenesis of the Liver: Insights from Single-Cell Transcriptomic Analysis in Liver Injury
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Animals
2.2. HSC Cell Isolation and Purification
2.3. Single-Cell Library Preparation and Sequencing
2.4. Seurat Analysis and Linear Dimensional Reduction
2.5. Determine Statistically Significant Principal Components
2.6. Human Liver Samples
3. Results
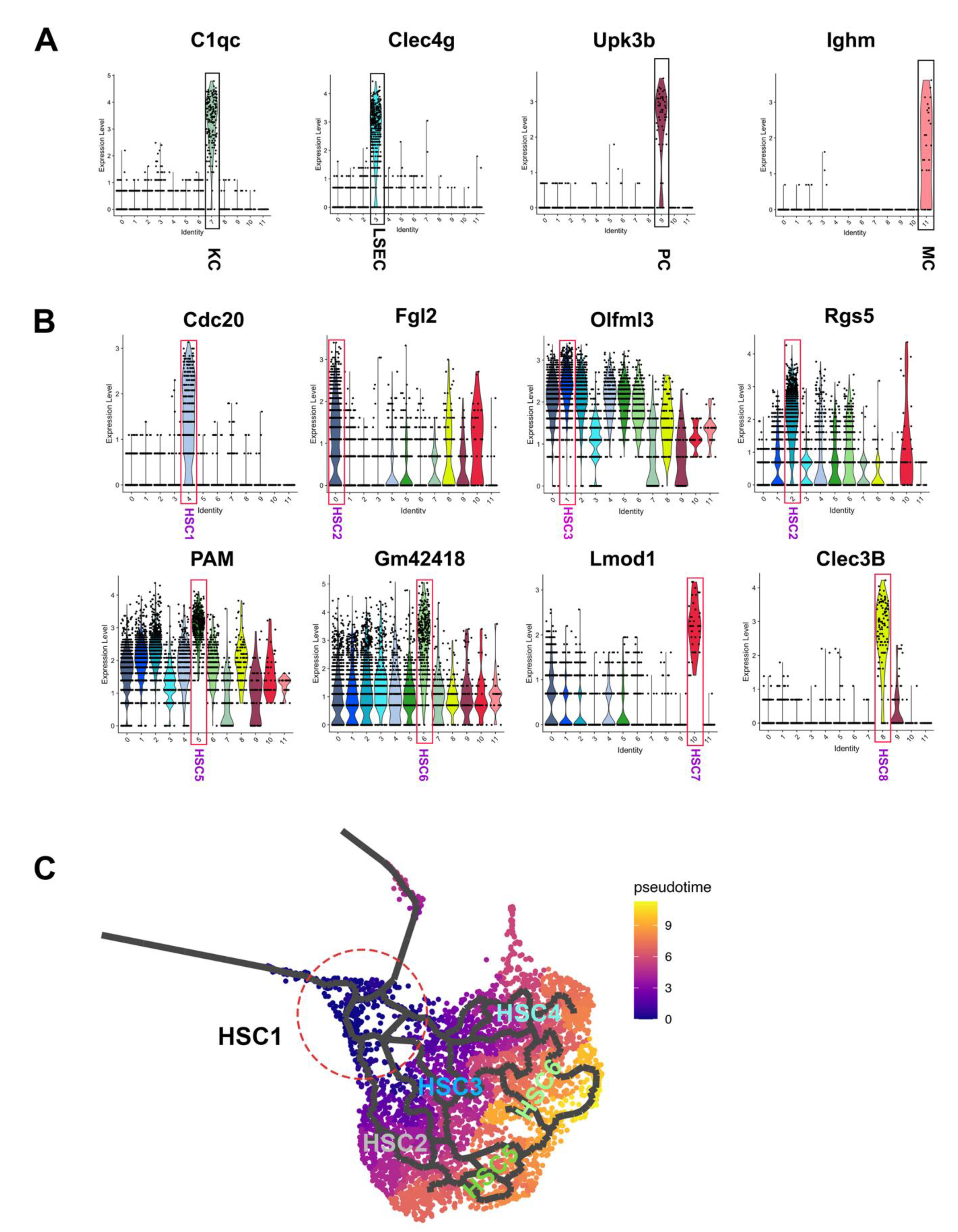
3.1. Single-Cell Transcriptomic Analysis of LRAT-Cre Reporter Traced Hepatic Cells in CCl4-Treated Mouse
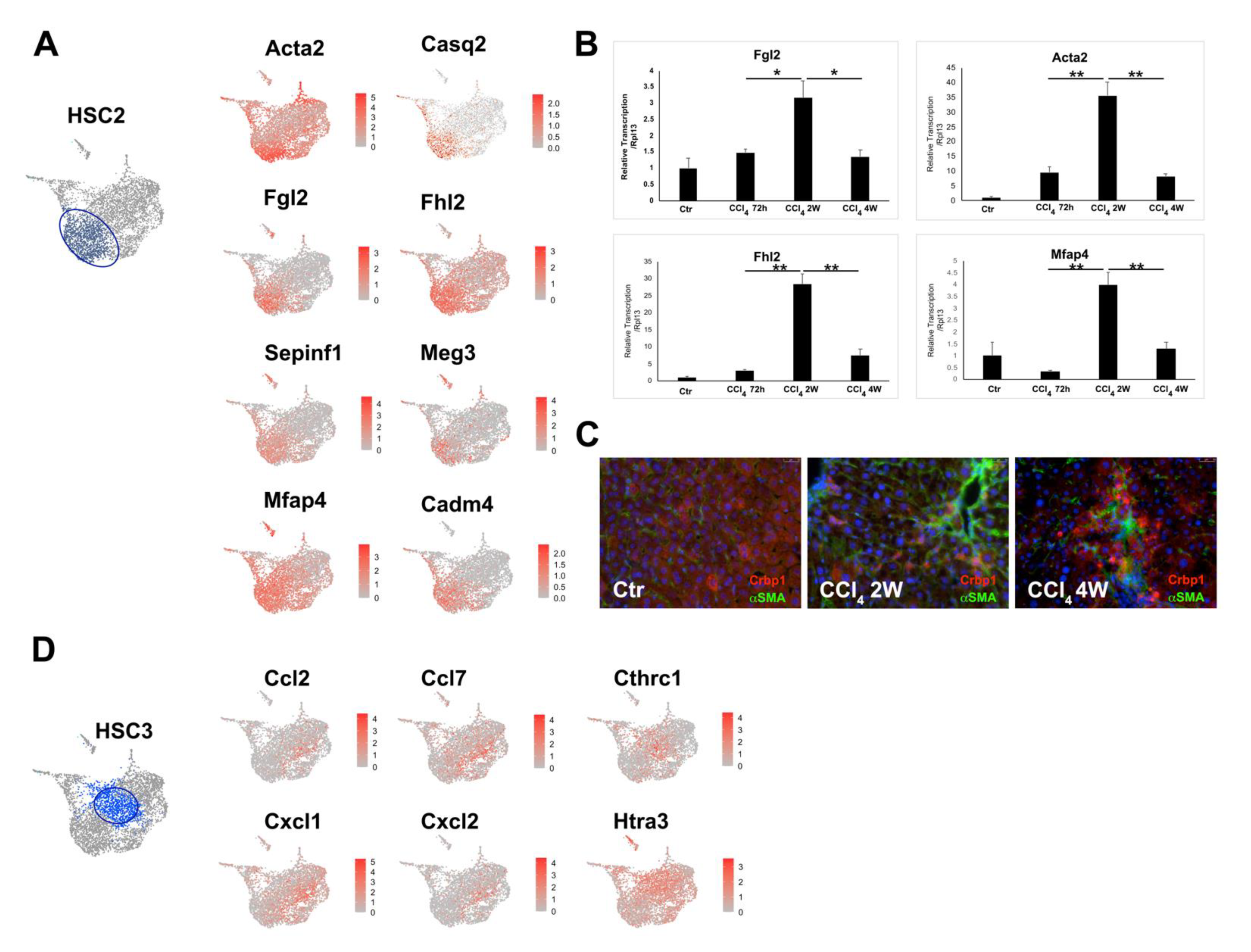
3.2. Heterogeneity of Activated HSCs in CCl4 Injured Mouse Liver
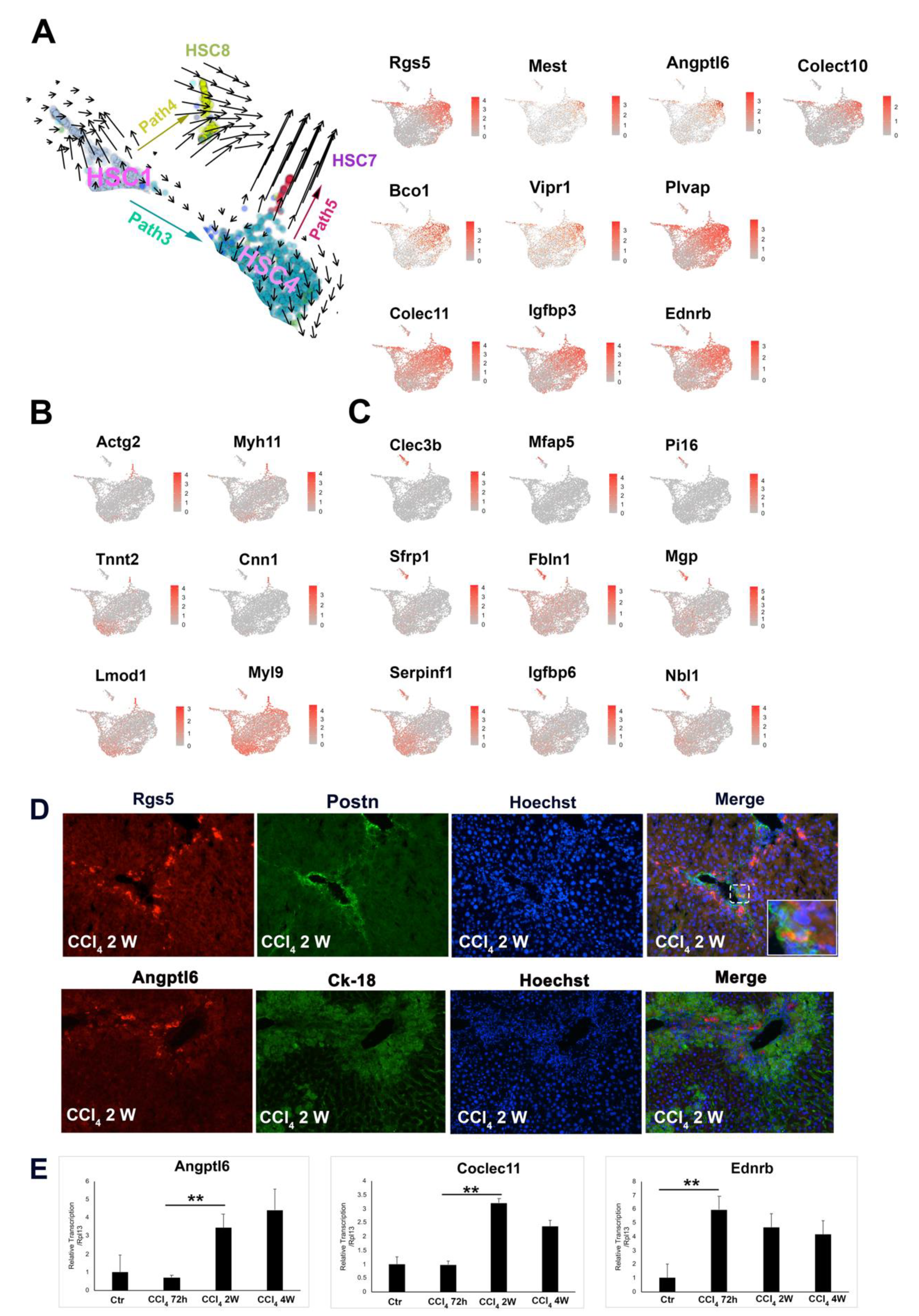
3.3. Two Major Differentiation Trajectory Paths Lead to Distinct Mature Myofibroblast Clusters
3.4. Unique HSC Differentiation Trajectory Path for Liver Repair and Vasoactive Modulation
3.5. Trajectory Paths That Contribute to the Obscure HSC Subtypes
3.6. The Expression of CV HSC Markers and PV HSC Markers in HSC Subtypes
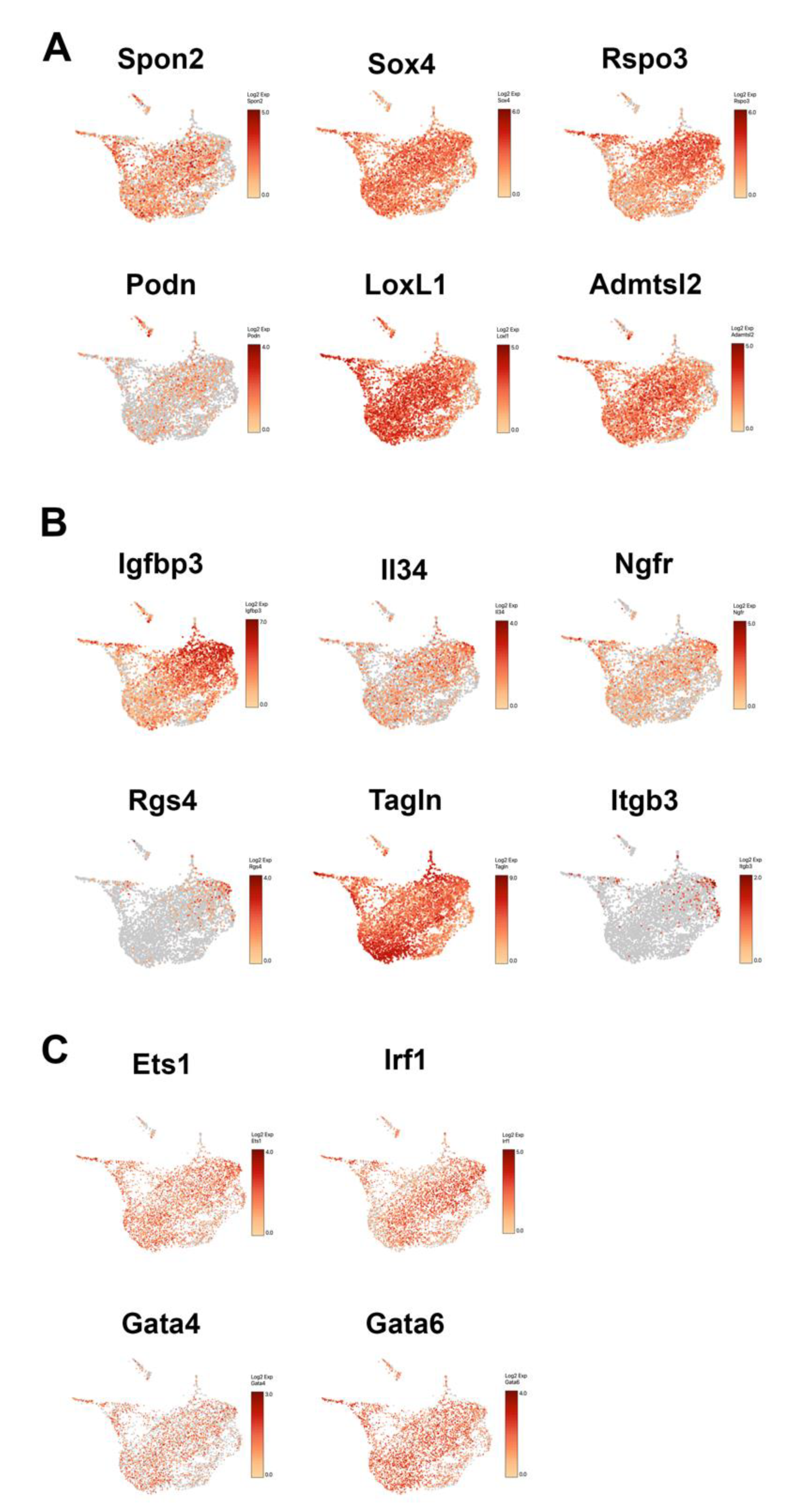
3.7. Heterogeneity of Activated Human HSC Population in Cirrhotic Liver
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar]
- Friedman, S.L. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar] [CrossRef]
- Miyahara, T.; Schrum, L.; Rippe, R.; Xiong, S.; Yee, H.F.; Motomura, K., Jr.; Anania, F.A.; Willson, T.M.; Tsukamoto, H. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J. Biol. Chem. 2000, 275, 35715–35722. [Google Scholar] [CrossRef] [Green Version]
- MacParland, S.A.; Liu, J.C.; Ma, X.Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Kuang, H.; Ansari, S.; Liu, T.; Gong, J.; Wang, S.; Zhao, X.Y.; Ji, Y.; Li, C.; Guo, L.; et al. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol. Cell 2019, 75, 644–660.e645. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shi, Y.; Zou, X.; Lu, Z.N.; Xie, G.; Yang, J.Y.; Wu, C.C.; Cui, X.F.; He, K.Y.; Luo, Q.; et al. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genom. 2017, 18, 946. [Google Scholar] [CrossRef]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Krenkel, O.; Hundertmark, J.; Ritz, T.P.; Weiskirchen, R.; Tacke, F. Single Cell RNA Sequencing Identifies Subsets of Hepatic Stellate Cells and Myofibroblasts in Liver Fibrosis. Cells 2019, 8, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenthal, S.B.; Liu, X.; Ganguly, S.; Dhar, D.; Pasillas, M.P.; Ricciardelli, E.; Li, R.Z.; Troutman, T.D.; Kisseleva, T.; Glass, C.K.; et al. Heterogeneity of hepatic stellate cells in a mouse model of non-alcoholic steatohepatitis (NASH). Hepatology 2021. [Google Scholar] [CrossRef]
- Dobie, R.; Wilson-Kanamori, J.R.; Henderson, B.E.; Smith, J.R.; Matchett, K.P.; Portman, J.R.; Wallenborg, K.; Picelli, S.; Zagorska, A.; Pendem, S.V.; et al. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep. 2019, 29, 1832–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mederacke, I.; Hsu, C.C.; Troeger, J.S.; Huebener, P.; Mu, X.; Dapito, D.H.; Pradere, J.P.; Schwabe, R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 2013, 4, 2823. [Google Scholar] [CrossRef] [Green Version]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L.A. global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef]
- Kumar, P.; Smith, T.; Raeman, R.; Chopyk, D.M.; Brink, H.; Liu, Y.; Sulchek, T.; Anania, F.A. Periostin promotes liver fibrogenesis by activating lysyl oxidase in hepatic stellate cells. J. Biol. Chem. 2018, 293, 12781–12792. [Google Scholar] [CrossRef] [Green Version]
- Mederacke, I.; Dapito, D.H.; Affo, S.; Uchinami, H.; Schwabe, R.F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 2015, 10, 305–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.J.; Campbell, K.R.; Lun, A.T.; Wills, Q.F. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017, 33, 1179–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.; Liao, J.; Lu, X.; Xue, R.; Ai, N.; Fan, X. scCATCH: Automatic Annotation on Cell Types of Clusters from Single-Cell RNA Sequencing Data. IScience 2020, 23, 100882. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Kato, K.; Goodman, D.S. Immunocytochemical studies on the localization of plasma and of cellular retinol-binding proteins and of transthyretin (prealbumin) in rat liver and kidney. J. Cell Biol. 1984, 98, 1696–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitkopf-Heinlein, K.; Meyer, C.; König, C.; Gaitantzi, H.; Addante, A.; Thomas, M.; Wiercinska, E.; Cai, C.; Li, Q.; Wan, F.; et al. BMP-9 interferes with liver regeneration and promotes liver fibrosis. Gut 2017, 66, 939–954. [Google Scholar] [CrossRef] [Green Version]
- Lua, I.; Li, Y.; Pappoe, L.S.; Asahina, K. Myofibroblastic Conversion and Regeneration of Mesothelial Cells in Peritoneal and Liver Fibrosis. Am. J. Pathol. 2015, 185, 3258–3273. [Google Scholar] [CrossRef]
- Kanamori-Katayama, M.; Kaiho, A.; Ishizu, Y.; Okamura-Oho, Y.; Hino, O.; Abe, M.; Kishimoto, T.; Sekihara, H.; Nakamura, Y.; Suzuki, K.; et al. LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS ONE 2011, 6, e25391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, S.V.; Vergnolle, M.A.; Hussein, D.; Wozniak, M.J.; Allan, V.J.; Taylor, S.S. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J. Cell Sci. 2005, 118, 4889–4900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamers, F.; Van Der Ploeg, I.; Schild, L.; Ebus, M.E.; Koster, J.; Hansen, B.R.; Koch, T.; Versteeg, R.; Caron, H.N.; Molenaar, J.J. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocr. Relat. Cancer 2011, 18, 657–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, C.L.; Luster, A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillet, E.; Ouarné, M.; Desroches-Castan, A.; Mallet, C.; Subileau, M.; Didier, R.; Lioutsko, A.; Belthier, G.; Feige, J.J.; Bailly, S. A heterodimer formed by bone morphogenetic protein 9 (BMP9) and BMP10 provides most BMP biological activity in plasma. J. Biol. Chem. 2018, 293, 10963–10974. [Google Scholar] [CrossRef] [Green Version]
- Xie, T.; Wang, Y.; Deng, N.; Huang, G.; Taghavifar, F.; Geng, Y.; Liu, N.; Kulur, V.; Yao, C.; Chen, P.; et al. Single-Cell Deconvolution of Fibroblast Heterogeneity in Mouse Pulmonary Fibrosis. Cell Rep. 2018, 22, 3625–3640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhmar, P.; Trinh, R.T.P.; Ma, J.; Huo, X.; Peng, B.; Heijnen, C.J.; Kavelaars, A. The fibroblast-derived protein PI16 controls neuropathic pain. Proc. Natl. Acad. Sci. USA 2020, 117, 5463–5471. [Google Scholar] [CrossRef]
- Kraja, A.T.; Liu, C.; Fetterman, J.L.; Graff, M.; Have, C.T.; Gu, C.; Yanek, L.R.; Feitosa, M.F.; Arking, D.E.; Chasman, D.I.; et al. Associations of Mitochondrial and Nuclear Mitochondrial Variants and Genes with Seven Metabolic Traits. Am. J. Hum. Genet. 2019, 104, 112–138. [Google Scholar] [CrossRef] [Green Version]
- Sherry, T.; Handley, A.; Nicholas, H.R.; Pocock, R. Harmonization of L1CAM expression facilitates axon outgrowth and guidance of a motor neuron. Development 2020, 147, dev193805. [Google Scholar] [CrossRef]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Galli, A.; Bochaton-Piallat, M.L.; Gabbiani, G. The myofibroblast: One function, multiple origins. Am. J. Pathol. 2007, 170, 1807–1816. [Google Scholar] [CrossRef]
- Holobotovskyy, V.; Manzur, M.; Tare, M.; Burchell, J.; Bolitho, E.; Viola, H.; Hool, L.C.; Arnolda, L.F.; McKitrick, D.J.; Ganss, R. Regulator of G-protein signaling 5 controls blood pressure homeostasis and vessel wall remodeling. Circ. Res. 2013, 112, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Juarez, C.F.; Dedhia, P.H.; Jin, S.; Ruiz-Vega, R.; Ma, D.; Liu, Y.; Yamaga, K.; Shestova, O.; Gay, D.L.; Yang, Z.; et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 2019, 10, 650. [Google Scholar] [CrossRef]
- Li, H.; He, C.; Feng, J.; Zhang, Y.; Tang, Q.; Bian, Z.; Bai, X.; Zhou, H.; Jiang, H.; Heximer, S.P.; et al. Regulator of G protein signaling 5 protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Proc. Natl. Acad. Sci. USA 2010, 107, 13818–13823. [Google Scholar] [CrossRef] [Green Version]
- Bahrami, A.J.; Gunaje, J.J.; Hayes, B.J.; Riehle, K.J.; Kenerson, H.L.; Yeung, R.S.; Stempien-Otero, A.S.; Campbell, J.S.; Mahoney, W.M., Jr. Regulator of G-protein signaling-5 is a marker of hepatic stellate cells and expression mediates response to liver injury. PLoS ONE 2014, 9, e108505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhu, C.; Li, Y.; Wu, Q.; Gao, R. Mest attenuates CCl4-induced liver fibrosis in rats by inhibiting the Wnt/beta-catenin signaling pathway. Gut Liver 2014, 8, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; Van Der Weyden, L.; Fikrig, E.; et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.J. Insulin-like growth factor-I regulation of immune function: A potential therapeutic target in autoimmune diseases? Pharmacol. Rev. 2010, 62, 199–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Xi, D.; Ding, W.; Wang, F.; Zhou, H.; Ning, Q. Soluble FGL2, a novel effector molecule of activated hepatic stellate cells, regulates T-cell function in cirrhotic patients with hepatocellular carcinoma. Hepatol. Int. 2014, 8, 567–575. [Google Scholar] [CrossRef] [Green Version]
- Alnajar, A.; Nordhoff, C.; Schied, T.; Chiquet-Ehrismann, R.; Loser, K.; Vogl, T.; Ludwig, S.; Wixler, V. The LIM-only protein FHL2 attenuates lung inflammation during bleomycin-induced fibrosis. PLoS ONE 2013, 8, e81356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huss, S.; Stellmacher, C.; Goltz, D.; Khlistunova, I.; Adam, A.C.; Trebicka, J.; Kirfel, J.; Büttner, R.; Weiskirchen, R. Deficiency in four and one half LIM domain protein 2 (FHL2) aggravates liver fibrosis in mice. BMC Gastroenterol. 2013, 13, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ek, E.T.; Dass, C.R.; Choong, P.F. PEDF: A potential molecular therapeutic target with multiple anti-cancer activities. Trends Mol. Med. 2006, 12, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Gordon, F.E.; Nutt, C.L.; Cheunsuchon, P.; Nakayama, Y.; Provencher, K.A.; Rice, K.A.; Zhou, Y.; Zhang, X.; Klibanski, A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology 2010, 151, 2443–2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boeckmans, J.; Buyl, K.; Natale, A.; Vandenbempt, V.; Branson, S.; De Boe, V.; Rogiers, V.; De Kock, J.; Rodrigues, R.M.; Vanhaecke, T. Transcriptomics data of a human in vitro model of non-alcoholic steatohepatitis exposed to elafibranor. Data Brief 2019, 25, 104093. [Google Scholar] [CrossRef] [PubMed]
- Nagatsuma, K.; Hayashi, Y.; Hano, H.; Sagara, H.; Murakami, K.; Saito, M.; Masaki, T.; Lu, T.; Tanaka, M.; Enzan, H.; et al. Lecithin: Retinol acyltransferase protein is distributed in both hepatic stellate cells and endothelial cells of normal rodent and human liver. Liver Int. 2009, 29, 47–54. [Google Scholar] [CrossRef]
- Yang, L.; Jung, Y.; Omenetti, A.; Witek, R.P.; Choi, S.; Vandongen, H.M.; Huang, J.; Alpini, G.D.; Diehl, A.M. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells 2008, 26, 2104–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Conway, S.J.; Liu, Y.; Snider, P.; Chen, H.; Gao, H.; Liu, Y.; Isidan, K.; Lopez, K.J.; Campana, G.; et al. Heterogeneity of Hepatic Stellate Cells in Fibrogenesis of the Liver: Insights from Single-Cell Transcriptomic Analysis in Liver Injury. Cells 2021, 10, 2129. https://doi.org/10.3390/cells10082129
Zhang W, Conway SJ, Liu Y, Snider P, Chen H, Gao H, Liu Y, Isidan K, Lopez KJ, Campana G, et al. Heterogeneity of Hepatic Stellate Cells in Fibrogenesis of the Liver: Insights from Single-Cell Transcriptomic Analysis in Liver Injury. Cells. 2021; 10(8):2129. https://doi.org/10.3390/cells10082129
Chicago/Turabian StyleZhang, Wenjun, Simon J. Conway, Ying Liu, Paige Snider, Hanying Chen, Hongyu Gao, Yunlong Liu, Kadir Isidan, Kevin J. Lopez, Gonzalo Campana, and et al. 2021. "Heterogeneity of Hepatic Stellate Cells in Fibrogenesis of the Liver: Insights from Single-Cell Transcriptomic Analysis in Liver Injury" Cells 10, no. 8: 2129. https://doi.org/10.3390/cells10082129
APA StyleZhang, W., Conway, S. J., Liu, Y., Snider, P., Chen, H., Gao, H., Liu, Y., Isidan, K., Lopez, K. J., Campana, G., Li, P., Ekser, B., Francis, H., Shou, W., & Kubal, C. (2021). Heterogeneity of Hepatic Stellate Cells in Fibrogenesis of the Liver: Insights from Single-Cell Transcriptomic Analysis in Liver Injury. Cells, 10(8), 2129. https://doi.org/10.3390/cells10082129









