Neuroinflammation and Alzheimer’s Disease: A Machine Learning Approach to CSF Proteomics
Abstract
:1. Introduction
2. Patients and Methods
2.1. Patients Selection
2.2. CSF Collection and Storage
2.3. CSF ATN Profile
2.4. PEA Testing
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Patients
3.2. PEA Testing
3.3. Univariate Diagnostic Performance of PEA-Tested Proteins
3.4. Correlation Analysis of PEA-Tested Proteins
3.5. Multivariate Diagnostic Performance of PEA-Tested Proteins
3.6. Correlation Analysis between PEA-Tested Proteins and CSF AD Biomarkers
4. Discussion
5. Conclusions
- We present promising findings using PEA biomarker technology coupled with a machine-learning statistical approach for the discovery of novel markers and pathophysiological mechanisms of AD.
- Our findings suggest an association between AD pathology and the SIRT2 pathway, astrocyte and microglia activation, and BBB dysfunction, as reflected by concentration changes in the CSF of SIRT2, HGF, MMP10, and CXCL5.
- Studies confirming these results in larger cohorts also by means of different assays, such as the single-molecule array and ELISA, might be needed to validate PEA as a promising tool for biomarker discovery in the field of neurological diseases.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheltens, P.; Blennow, K.; Breteler, M.M.B.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s Disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Jellinger, K.A. Neuropathological Aspects of Alzheimer Disease, Parkinson Disease and Frontotemporal Dementia. Neuro-Degener. Dis. 2008, 5, 118–121. [Google Scholar] [CrossRef]
- Leng, F.; Edison, P. Neuroinflammation and Microglial Activation in Alzheimer Disease: Where Do We Go from Here? Nat. Rev. Neurol. 2020, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. How Neuroinflammation Contributes to Neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Ransohoff, R.M.; Brown, M.A. Innate Immunity in the Central Nervous System. J. Clin. Investig. 2012, 122, 1164–1171. [Google Scholar] [CrossRef]
- Calsolaro, V.; Edison, P. Neuroinflammation in Alzheimer’s Disease: Current Evidence and Future Directions. Alzheimer’s Dement. 2016, 12, 719–732. [Google Scholar] [CrossRef]
- Jaturapatporn, D.; Isaac, M.G.E.K.N.; McCleery, J.; Tabet, N. Aspirin, Steroidal and Non-Steroidal Anti-Inflammatory Drugs for the Treatment of Alzheimer’s Disease. Cochrane Database Syst. Rev. 2012, 15, CD006378. [Google Scholar] [CrossRef]
- Elahi, F.M.; Casaletto, K.B.; Joie, R.L.; Walters, S.M.; Harvey, D.; Wolf, A.; Edwards, L.; Rivera-Contreras, W.; Karydas, A.; Cobigo, Y.; et al. Plasma Biomarkers of Astrocytic and Neuronal Dysfunction in Early- and Late-Onset Alzheimer’s Disease. Alzheimer’s Dement. 2020, 16, 681–695. [Google Scholar] [CrossRef]
- Higginbotham, L.; Ping, L.; Dammer, E.B.; Duong, D.M.; Zhou, M.; Gearing, M.; Hurst, C.; Glass, J.D.; Factor, S.A.; Johnson, E.C.B.; et al. Integrated Proteomics Reveals Brain-Based Cerebrospinal Fluid Biomarkers in Asymptomatic and Symptomatic Alzheimer’s Disease. Sci. Adv. 2020, 6, eaaz9360. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-Plex PEA Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabbari, E.; Woodside, J.; Guo, T.; Magdalinou, N.K.; Chelban, V.; Athauda, D.; Lees, A.J.; Foltynie, T.; Houlden, H.; Church, A.; et al. Proximity Extension Assay Testing Reveals Novel Diagnostic Biomarkers of Atypical Parkinsonian Syndromes. J. Neurol. Neurosurg. Psychiatry 2019, 90, 768–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellegaard Nielsen, J.; Sofie Pedersen, K.; Vestergård, K.; Georgiana Maltesen, R.; Christiansen, G.; Lundbye-Christensen, S.; Moos, T.; Risom Kristensen, S.; Pedersen, S. Novel Blood-Derived Extracellular Vesicle-Based Biomarkers in Alzheimer’s Disease Identified by Proximity Extension Assay. Biomedicines 2020, 8, 199. [Google Scholar] [CrossRef]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a Biological Definition of Alzheimer’s Disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Teunissen, C.E.; Petzold, A.; Bennett, J.L.; Berven, F.S.; Brundin, L.; Comabella, M.; Franciotta, D.; Frederiksen, J.L.; Fleming, J.O.; Furlan, R.; et al. A Consensus Protocol for the Standardization of Cerebrospinal Fluid Collection and Biobanking. Neurology 2009, 73, 1914–1922. [Google Scholar] [CrossRef] [Green Version]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Machado e Costa, F.; Braga, A.C. Adjusting ROC Curve for Covariates with AROC R Package. In Computational Science and Its Applications, Proceedings of the ICCSA 2020, Cagliari, Italy, 1–4 July 2020; Springer: Cham, Switzerland, 2020; Volume 12251, pp. 185–198. [Google Scholar] [CrossRef]
- Bonferroni, C.E. Teoria Statistica delle Classi e Calcolo delle Probabilità; Libreria Internazionale Seeber: Florence, Italy, 1936. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 23 July 2021).
- Rokach, L.; Maimon, O. Clustering Methods. In Data Mining and Knowledge Discovery Handbook; Maimon, O., Rokach, L., Eds.; Springer: Boston, MA, USA, 2005; pp. 321–352. ISBN 978-0-387-25465-4. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Tibshirani, R. The Lasso Method for Variable Selection in the Cox Model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Muthukrishnan, R.; Rohini, R. LASSO: A Feature Selection Technique in Predictive Modeling for Machine Learning. In Proceedings of the 2016 IEEE International Conference on Advances in Computer Applications (ICACA), Coimbatore, India, 24 October 2016; pp. 18–20. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, J.; Hong, T.; Chen, X.; Cui, L. SIRT2: Controversy and Multiple Roles in Disease and Physiology. Ageing Res. Rev. 2019, 55, 100961. [Google Scholar] [CrossRef]
- Wongchitrat, P.; Pakpian, N.; Kitidee, K.; Phopin, K.; Dharmasaroja, P.A.; Govitrapong, P. Alterations in the Expression of Amyloid Precursor Protein Cleaving Enzymes MRNA in Alzheimer Peripheral Blood. Curr. Alzheimer Res. 2019, 16, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Polito, L.; Kehoe, P.G.; Davin, A.; Benussi, L.; Ghidoni, R.; Binetti, G.; Quadri, P.; Lucca, U.; Tettamanti, M.; Clerici, F.; et al. The SIRT2 Polymorphism Rs10410544 and Risk of Alzheimer’s Disease in Two Caucasian Case-Control Cohorts. Alzheimer’s Dement. 2013, 9, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Biella, G.; Fusco, F.; Nardo, E.; Bernocchi, O.; Colombo, A.; Lichtenthaler, S.F.; Forloni, G.; Albani, D. Sirtuin 2 Inhibition Improves Cognitive Performance and Acts on Amyloid-β Protein Precursor Processing in Two Alzheimer’s Disease Mouse Models. J. Alzheimer’s Dis. 2016, 53, 1193–1207. [Google Scholar] [CrossRef]
- Gal, J.; Bang, Y.; Choi, H.J. SIRT2 Interferes with Autophagy-Mediated Degradation of Protein Aggregates in Neuronal Cells under Proteasome Inhibition. Neurochem. Int. 2012, 61, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.R.; Palma, A.M.; Gomes, R.; Santos, D.; Silva, D.F.; Cardoso, S.M. Acetylation as a Major Determinant to Microtubule-Dependent Autophagy: Relevance to Alzheimer’s and Parkinson Disease Pathology. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2008–2023. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, M.; Baba, A.; Ikeda-Matsuo, Y.; Yamada, M.K.; Itamura, R.; Nishiyama, N.; Ikegaya, Y.; Matsuki, N. Hepatocyte Growth Factor as an Enhancer of Nmda Currents and Synaptic Plasticity in the Hippocampus. Neuroscience 2004, 128, 155–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, M.; Takagi, N.; Takagi, K.; Funakoshi, H.; Matsumoto, K.; Nakamura, T.; Takeo, S. Hepatocyte Growth Factor Promotes the Number of PSD-95 Clusters in Young Hippocampal Neurons. Exp. Neurol. 2007, 207, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Tyndall, S.J.; Patel, S.J.; Walikonis, R.S. Hepatocyte Growth Factor-Induced Enhancement of Dendritic Branching Is Blocked by Inhibitors of N-Methyl-D-Aspartate Receptors and Calcium/Calmodulin-Dependent Kinases. J. Neurosci. Res. 2007, 85, 2343–2351. [Google Scholar] [CrossRef]
- Yamada, T.; Tsubouchi, H.; Daikuhara, Y.; Prat, M.; Comoglio, P.M.; McGeer, P.L.; McGeer, E.G. Immunohistochemistry with Antibodies to Hepatocyte Growth Factor and Its Receptor Protein (c-MET) in Human Brain Tissues. Brain Res. 1994, 637, 308–312. [Google Scholar] [CrossRef]
- Tsuboi, Y.; Kakimoto, K.; Nakajima, M.; Akatsu, H.; Yamamoto, T.; Ogawa, K.; Ohnishi, T.; Daikuhara, Y.; Yamada, T. Increased Hepatocyte Growth Factor Level in Cerebrospinal Fluid in Alzheimer’s Disease. Acta Neurol. Scand. 2003, 107, 81–86. [Google Scholar] [CrossRef]
- Cuadrado, E.; Rosell, A.; Penalba, A.; Slevin, M.; Alvarez-Sabín, J.; Ortega-Aznar, A.; Montaner, J. Vascular MMP-9/TIMP-2 and Neuronal MMP-10 up-Regulation in Human Brain after Stroke: A Combined Laser Microdissection and Protein Array Study. J. Proteome Res. 2009, 8, 3191–3197. [Google Scholar] [CrossRef] [PubMed]
- Könnecke, H.; Bechmann, I. The Role of Microglia and Matrix Metalloproteinases Involvement in Neuroinflammation and Gliomas. Clin. Dev. Immunol. 2013, 2013, 914104. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Guillamon, M.; Delgado, P.; Ortega, L.; Pares, M.; Rosell, A.; García-Bonilla, L.; Fernández-Cadenas, I.; Borrell-Pagès, M.; Boada, M.; Montaner, J. Neuronal TIMP-1 Release Accompanies Astrocytic MMP-9 Secretion and Enhances Astrocyte Proliferation Induced by Beta-Amyloid 25-35 Fragment. J. Neurosci. Res. 2009, 87, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Backstrom, J.R.; Lim, G.P.; Cullen, M.J.; Tökés, Z.A. Matrix Metalloproteinase-9 (MMP-9) Is Synthesized in Neurons of the Human Hippocampus and Is Capable of Degrading the Amyloid-Beta Peptide (1-40). J. Neurosci. 1996, 16, 7910–7919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-M.; Yin, K.-J.; Hsin, I.; Chen, S.; Fryer, J.D.; Holtzman, D.M.; Hsu, C.Y.; Xu, J. Matrix Metalloproteinase-9 and Spontaneous Hemorrhage in an Animal Model of Cerebral Amyloid Angiopathy. Ann. Neurol. 2003, 54, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Duits, F.H.; Hernandez-Guillamon, M.; Montaner, J.; Goos, J.D.C.; Montañola, A.; Wattjes, M.P.; Barkhof, F.; Scheltens, P.; Teunissen, C.E.; van der Flier, W.M. Matrix Metalloproteinases in Alzheimer’s Disease and Concurrent Cerebral Microbleeds. J. Alzheimer’s Dis. 2015, 48, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Whelan, C.D.; Mattsson, N.; Nagle, M.W.; Vijayaraghavan, S.; Hyde, C.; Janelidze, S.; Stomrud, E.; Lee, J.; Fitz, L.; Samad, T.A.; et al. Multiplex Proteomics Identifies Novel CSF and Plasma Biomarkers of Early Alzheimer’s Disease. Acta Neuropathol. Commun. 2019, 7, 169. [Google Scholar] [CrossRef]
- Nuttall, R.K.; Silva, C.; Hader, W.; Bar-Or, A.; Patel, K.D.; Edwards, D.R.; Yong, V.W. Metalloproteinases Are Enriched in Microglia Compared with Leukocytes and They Regulate Cytokine Levels in Activated Microglia. Glia 2007, 55, 516–526. [Google Scholar] [CrossRef]
- Chandrasekar, B.; Melby, P.C.; Sarau, H.M.; Raveendran, M.; Perla, R.P.; Marelli-Berg, F.M.; Dulin, N.O.; Singh, I.S. Chemokine-Cytokine Cross-Talk. The ELR+ CXC Chemokine LIX (CXCL5) Amplifies a Proinflammatory Cytokine Response via a Phosphatidylinositol 3-Kinase-NF-Kappa B Pathway. J. Biol. Chem. 2003, 278, 4675–4686. [Google Scholar] [CrossRef] [Green Version]
- Haarmann, A.; Schuhmann, M.K.; Silwedel, C.; Monoranu, C.-M.; Stoll, G.; Buttmann, M. Human Brain Endothelial CXCR2 Is Inflammation-Inducible and Mediates CXCL5- and CXCL8-Triggered Paraendothelial Barrier Breakdown. Int. J. Mol. Sci. 2019, 20, 602. [Google Scholar] [CrossRef] [Green Version]

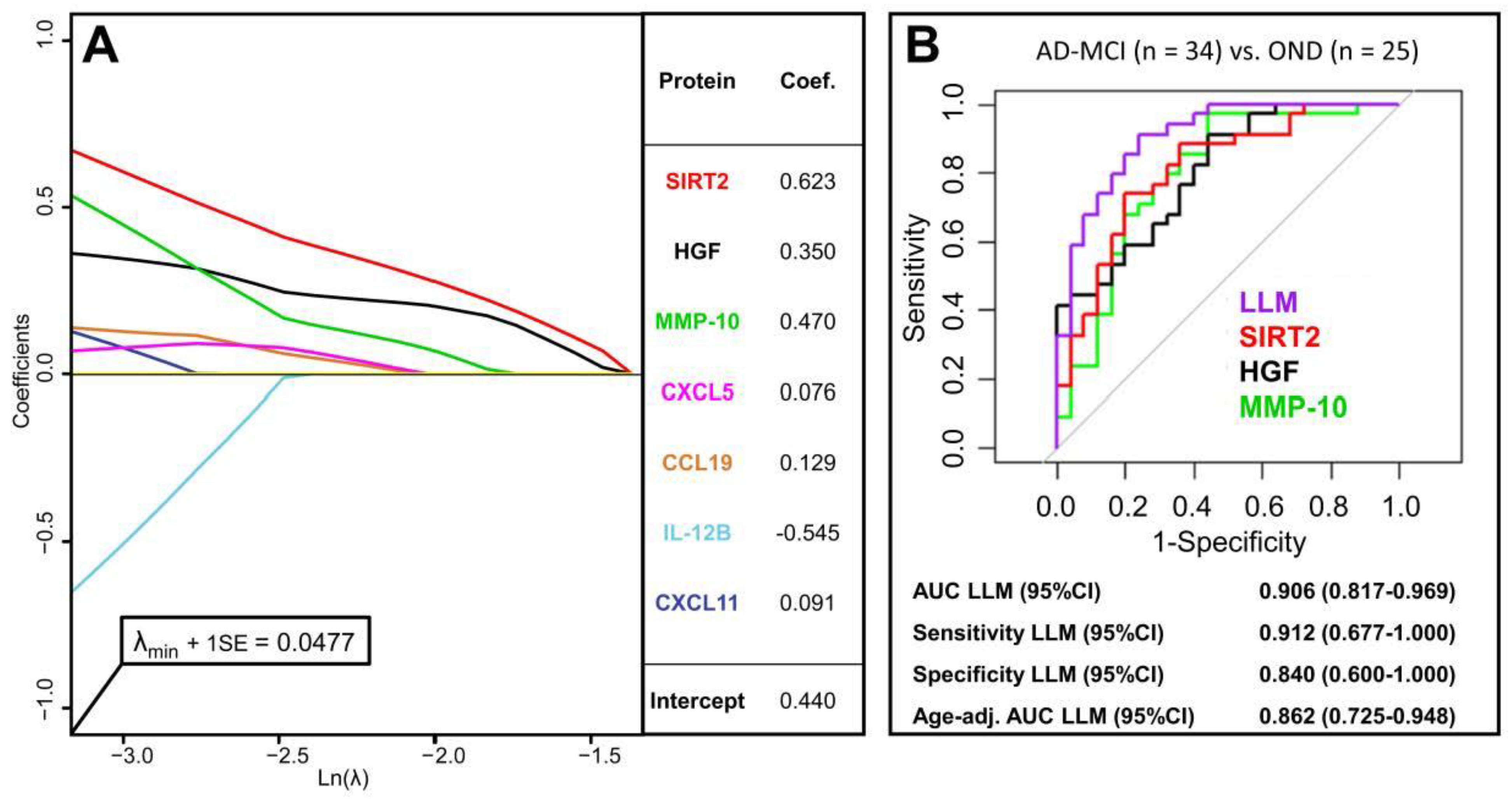
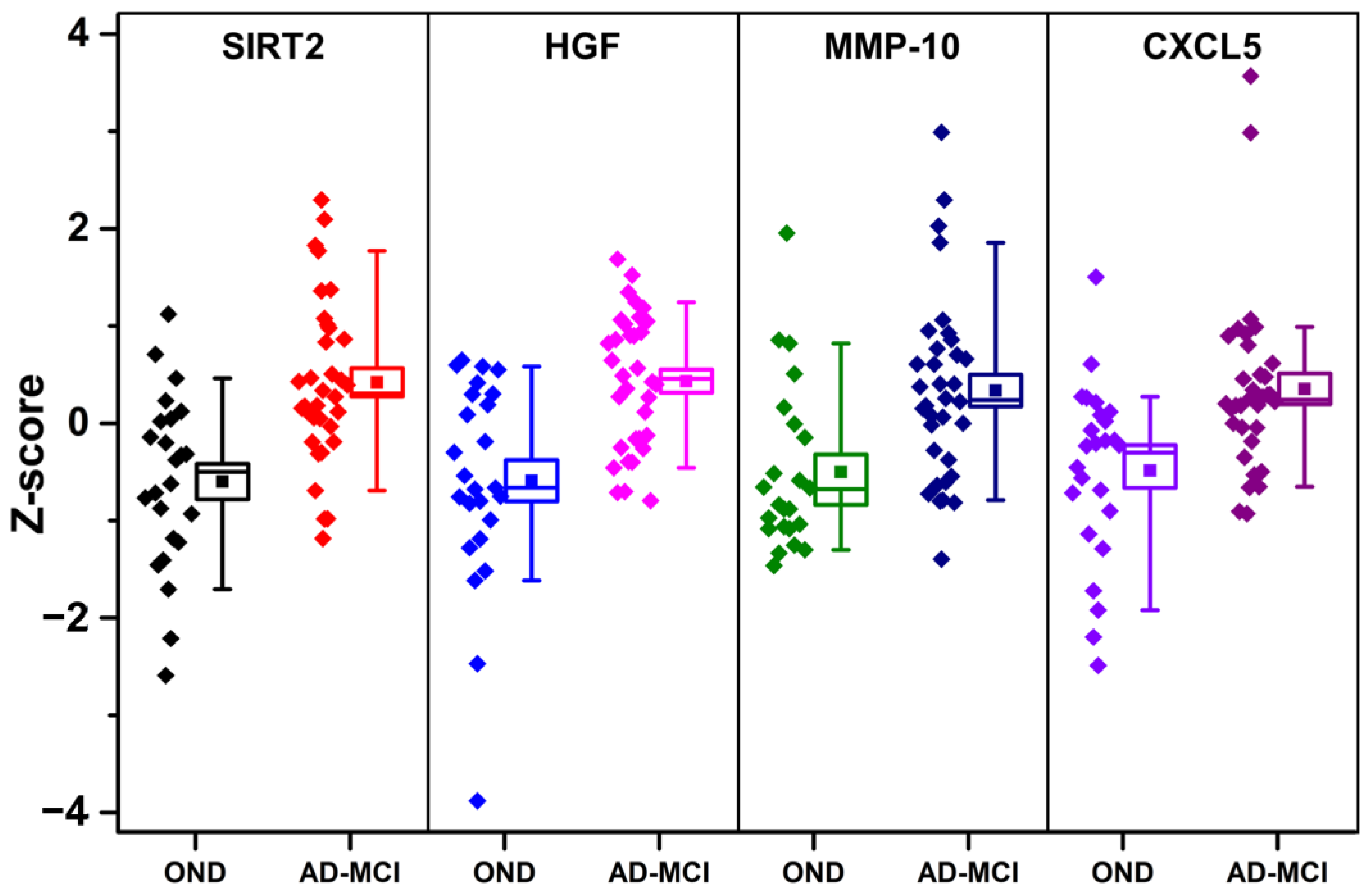
| Protein Name | AUC 1 | p-Value 2 |
|---|---|---|
| SIRT2 | 0.809 | 5.2 × 10−4 |
| HGF | 0.802 | 6.4 × 10−4 |
| MMP-10 | 0.793 | 3.2 × 10−3 |
| pIL-10RB | 0.786 | 1.3 × 10−3 |
| uPA | 0.771 | 4.0 × 10−3 |
| CXCL5 | 0.761 | 2.3 × 10−3 |
| LIF-R | 0.760 | 3.5 × 10−3 |
| CX3CL1 | 0.757 | 6.2 × 10−3 |
| SCF | 0.752 | 3.4 × 10−3 |
| Flt3L | 0.752 | 4.0 × 10−3 |
| TWEAK | 0.747 | 7.7 × 10−3 |
| Protein Name | Aβ42 | p-tau | t-tau |
|---|---|---|---|
| SIRT2 | 0.07 | 0.48 ** | 0.33 |
| HGF | −0.17 | 0.37 * | 0.35 |
| MMP-10 | −0.37 * | 0.20 | 0.09 |
| IL-10RB | −0.11 | 0.29 | 0.17 |
| uPA | 0.00 | 0.37 * | 0.29 |
| CXCL5 | −0.17 | −0.02 | 0.07 |
| LIF-R | −0.03 | 0.51 ** | 0.35 * |
| CX3CL1 | 0.01 | 0.42 * | 0.30 |
| SCF | −0.03 | 0.25 | 0.17 |
| Flt3L | −0.14 | 0.25 | 0.17 |
| TWEAK | −0.09 | 0.43 ** | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaetani, L.; Bellomo, G.; Parnetti, L.; Blennow, K.; Zetterberg, H.; Di Filippo, M. Neuroinflammation and Alzheimer’s Disease: A Machine Learning Approach to CSF Proteomics. Cells 2021, 10, 1930. https://doi.org/10.3390/cells10081930
Gaetani L, Bellomo G, Parnetti L, Blennow K, Zetterberg H, Di Filippo M. Neuroinflammation and Alzheimer’s Disease: A Machine Learning Approach to CSF Proteomics. Cells. 2021; 10(8):1930. https://doi.org/10.3390/cells10081930
Chicago/Turabian StyleGaetani, Lorenzo, Giovanni Bellomo, Lucilla Parnetti, Kaj Blennow, Henrik Zetterberg, and Massimiliano Di Filippo. 2021. "Neuroinflammation and Alzheimer’s Disease: A Machine Learning Approach to CSF Proteomics" Cells 10, no. 8: 1930. https://doi.org/10.3390/cells10081930
APA StyleGaetani, L., Bellomo, G., Parnetti, L., Blennow, K., Zetterberg, H., & Di Filippo, M. (2021). Neuroinflammation and Alzheimer’s Disease: A Machine Learning Approach to CSF Proteomics. Cells, 10(8), 1930. https://doi.org/10.3390/cells10081930








