HER Tyrosine Kinase Family and Rhabdomyosarcoma: Role in Onset and Targeted Therapy
Abstract
1. Introduction
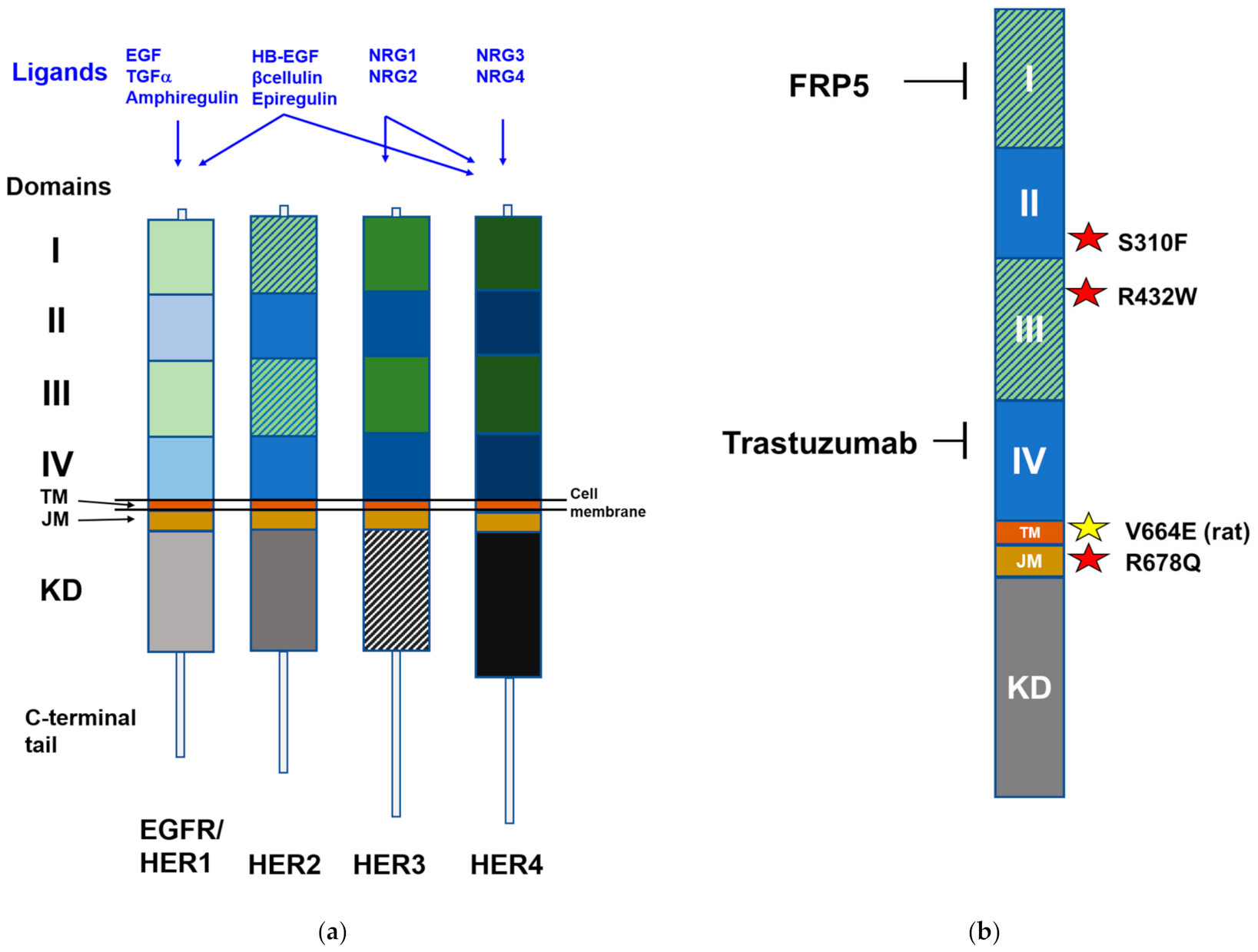
2. HER Family
3. HER Family in Myogenesis
4. Expression of HER Family Members in Human RMS Subtypes
5. The Role of HER Family in the Onset and Malignancy of RMS
5.1. Human RMS
5.2. HER2-Driven Murine RMS Model
6. HER Family Members as Therapeutic Targets
6.1. Antibodies and Inhibitors in Preclinical Models
6.2. Inhibitors in Clinical Trials
6.3. Immunotoxins
6.4. Chimeric Antigen Receptors
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1. [Google Scholar] [CrossRef]
- Leiner, J.; Le Loarer, F. The current landscape of rhabdomyosarcomas: An update. Virchows Arch. 2020, 476, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Rudzinski, E.R.; Kelsey, A.; Vokuhl, C.; Linardic, C.M.; Shipley, J.; Hettmer, S.; Koscielniak, E.; Hawkins, D.S.; Bisogno, G. Pathology of childhood rhabdomyosarcoma: A consensus opinion document from the Children’s Oncology Group, European Paediatric Soft Tissue Sarcoma Study Group, and the Cooperative Weichteilsarkom Studiengruppe. Pediatr. Blood Cancer 2021, 68, e28798. [Google Scholar] [CrossRef] [PubMed]
- Pappo, A.; Gartrell, J. Recent advances in understanding and managing pediatric rhabdomyosarcoma. F1000Research 2020, 9, F1000. [Google Scholar]
- Skapek, S.X.; Anderson, J.; Barr, F.G.; Bridge, J.A.; Gastier-Foster, J.M.; Parham, D.M.; Rudzinski, E.R.; Triche, T.; Hawkins, D.S. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: A children’s oncology group report. Pediatr. Blood Cancer 2013, 60, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Shern, J.F.; Selfe, J.; Izquierdo, E.; Patidar, R.; Chou, H.-C.; Song, Y.K.; Yohe, M.E.; Sindiri, S.; Wei, J.; Wen, X.; et al. Genomic Classification and Clinical Outcome in Rhabdomyosarcoma: A Report from an International Consortium. J. Clin. Oncol. 2021, JCO2003060. [Google Scholar] [CrossRef]
- Wachtel, M.; Runge, T.; Leuschner, I.; Stegmaier, S.; Koscielniak, E.; Treuner, J.; Odermatt, B.; Behnke, S.; Niggli, F.K.; Schäfer, B.W. Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. J. Clin. Oncol. 2006, 24, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014, 4, 216–231. [Google Scholar] [CrossRef]
- Chen, C.; Dorado Garcia, H.; Scheer, M.; Henssen, A.G. Current and Future Treatment Strategies for Rhabdomyosarcoma. Front. Oncol. 2019, 9, 1458. [Google Scholar] [CrossRef]
- Robbins, K.M.; Stabley, D.L.; Holbrook, J.; Sahraoui, R.; Sadreameli, A.; Conard, K.; Baker, L.; Gripp, K.W.; Sol-Church, K. Paternal uniparental disomy with segmental loss of heterozygosity of chromosome 11 are hallmark characteristics of syndromic and sporadic embryonal rhabdomyosarcoma. Am. J. Med. Genet. Part A 2016, 170, 3197–3206. [Google Scholar] [CrossRef]
- Chen, L.; Shern, J.F.; Wei, J.S.; Yohe, M.E.; Song, Y.K.; Hurd, L.; Liao, H.; Catchpoole, D.; Skapek, S.X.; Barr, F.G.; et al. Clonality and Evolutionary History of Rhabdomyosarcoma. PLoS Genet. 2015, 11, e1005075. [Google Scholar] [CrossRef]
- Van Erp, A.E.M.; Versleijen-Jonkers, Y.M.H.; Van Der Graaf, W.T.A.; Fleuren, E.D.G. Targeted therapy-based combination treatment in rhabdomyosarcoma. Mol. Cancer Ther. 2018, 17, 1365–1380. [Google Scholar] [CrossRef] [PubMed]
- Yohe, M.E.; Heske, C.M.; Stewart, E.; Adamson, P.C.; Ahmed, N.; Antonescu, C.R.; Chen, E.; Collins, N.; Ehrlich, A.; Galindo, R.L.; et al. Insights into pediatric rhabdomyosarcoma research: Challenges and goals. Pediatr. Blood Cancer 2019, 66, e27869. [Google Scholar] [CrossRef] [PubMed]
- Miwa, S.; Yamamoto, N.; Hayashi, K.; Takeuchi, A.; Igarashi, K.; Tsuchiya, H. Recent advances and challenges in the treatment of rhabdomyosarcoma. Cancers 2020, 12, 1758. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.D.; Hristova, K. The RTK Interactome: Overview and Perspective on RTK Heterointeractions. Chem. Rev. 2019, 119, 5881–5921. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019, 139, 395–411. [Google Scholar] [CrossRef]
- Kumagai, S.; Koyama, S.; Nishikawa, H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 2021, 21, 181–197. [Google Scholar] [CrossRef]
- Pahuja, K.B.; Nguyen, T.T.; Jaiswal, B.S.; Prabhash, K.; Thaker, T.M.; Senger, K.; Chaudhuri, S.; Kljavin, N.M.; Antony, A.; Phalke, S.; et al. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell 2018, 34, 792–806.e5. [Google Scholar] [CrossRef]
- Oh, D.Y.; Bang, Y.J. HER2-targeted therapies—A role beyond breast cancer. Nat. Rev. Clin. Oncol. 2020, 17, 33–48. [Google Scholar]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Connell, C.M.; Doherty, G.J. Activating HER2 mutations as emerging targets in multiple solid cancers. ESMO Open 2017, 2, e000279. [Google Scholar] [CrossRef]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Triulzi, T.; Bianchi, G.V.; Tagliabue, E. Predictive biomarkers in the treatment of HER2-positive breast cancer: An ongoing challenge. Futur. Oncol. 2016, 12, 1413–1428. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, J.; Schüßler-Lenz, M.; Bondanza, A.; Buchholz, C.J. Clinical development of CAR T cells—challenges and opportunities in translating innovative treatment concepts. EMBO Mol. Med. 2017, 9, 1183–1197. [Google Scholar] [CrossRef] [PubMed]
- Charville, G.W.; Cheung, T.H.; Yoo, B.; Santos, P.J.; Lee, G.K.; Shrager, J.B.; Rando, T.A. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Rep. 2015, 5, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.T.V.; Blau, H.M. Muscling toward therapy with ERBB3 and NGFR. Nat. Cell Biol. 2018, 20, 6–7. [Google Scholar] [CrossRef]
- Leroy, M.C.; Perroud, J.; Darbellay, B.; Bernheim, L.; Konig, S. Epidermal Growth Factor Receptor Down-Regulation Triggers Human Myoblast Differentiation. PLoS ONE 2013, 8, e71770. [Google Scholar] [CrossRef]
- Wang, Y.X.; Feige, P.; Brun, C.E.; Hekmatnejad, B.; Dumont, N.A.; Renaud, J.M.; Faulkes, S.; Guindon, D.E.; Rudnicki, M.A. EGFR-Aurka Signaling Rescues Polarity and Regeneration Defects in Dystrophin-Deficient Muscle Stem Cells by Increasing Asymmetric Divisions. Cell Stem Cell 2019, 24, 419–432.e6. [Google Scholar] [CrossRef]
- LeBrasseur, N.K.; Mizer, K.C.; Parkington, J.D.; Sawyer, D.B.; Fielding, R.A. The expression of neuregulin and erbB receptors in human skeletal muscle: Effects of progressive resistance training. Eur. J. Appl. Physiol. 2005, 94, 371–375. [Google Scholar] [CrossRef]
- Golding, J.P.; Calderbank, E.; Partridge, T.A.; Beauchamp, J.R. Skeletal muscle stem cells express anti-apoptotic ErbB receptors during activation from quiescence. Exp. Cell Res. 2007, 313, 341–356. [Google Scholar] [CrossRef]
- Morano, M.; Ronchi, G.; Nicolò, V.; Fornasari, B.E.; Crosio, A.; Perroteau, I.; Geuna, S.; Gambarotta, G.; Raimondo, S. Modulation of the Neuregulin 1/ErbB system after skeletal muscle denervation and reinnervation. Sci. Rep. 2018, 8, 5047. [Google Scholar] [CrossRef]
- Lee, K.F.; Simon, H.; Chen, H.; Bates, B.; Hung, M.C.; Hauser, C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 1995, 378, 394–398. [Google Scholar] [CrossRef]
- Cote, G.M.; Sawyer, D.B.; Chabner, B.A. ERBB2 Inhibition and Heart Failure. N. Engl. J. Med. 2012, 367, 2150–2153. [Google Scholar] [CrossRef] [PubMed]
- D’Uva, G.; Aharonov, A.; Lauriola, M.; Kain, D.; Yahalom-Ronen, Y.; Carvalho, S.; Weisinger, K.; Bassat, E.; Rajchman, D.; Yifa, O.; et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015, 17, 627–638. [Google Scholar] [CrossRef]
- Yutzey, K.E. Regenerative biology: Neuregulin 1 makes heart muscle. Nature 2015, 520, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, D.; Seitz, G.; Warmann, S.W.; Bonin, M.; Fuchs, J.; Armeanu-Ebinger, S. Cetuximab promotes immunotoxicity against rhabdomyosarcoma in vitro. J. Immunother. 2010, 33, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Grass, B.; Wachtel, M.; Behnke, S.; Leuschner, I.; Niggli, F.K.; Schäfer, B.W. Immunohistochemical detection of EGFR, fibrillin-2, P-cadherin and AP2β as biomarkers for rhabdomyosarcoma diagnostics. Histopathology 2009, 54, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Ganti, R.; Skapek, S.X.; Zhang, J.; Fuller, C.E.; Wu, J.; Billups, C.A.; Breitfeld, P.P.; Dalton, J.D.; Meyer, W.H.; Khoury, J.D. Expression and genomic status of EGFR and ErbB-2 in alveolar and embryonal rhabdomyosarcoma. Mod. Pathol. 2006, 19, 1213–1220. [Google Scholar] [CrossRef]
- Cen, L.; Arnoczky, K.J.; Hsieh, F.C.; Lin, H.J.; Qualman, S.J.; Yu, S.; Xiang, H.; Lin, J. Phosphorylation profiles of protein kinases in alveolar and embryonal rhabdomyosarcoma. Mod. Pathol. 2007, 20, 936–946. [Google Scholar] [CrossRef]
- Nordberg, J.; Mpindi, J.P.; Iljin, K.; Pulliainen, A.T.; Kallajoki, M.; Kallioniemi, O.; Elenius, K.; Elenius, V. Systemic Analysis of Gene Expression Profiles Identifies ErbB3 as a Potential Drug Target in Pediatric Alveolar Rhabdomyosarcoma. PLoS ONE 2012, 7, e50819. [Google Scholar] [CrossRef]
- Armistead, P.M.; Salganick, J.; Roh, J.S.; Steinert, D.M.; Patel, S.; Munsell, M.; El-Naggar, A.K.; Benjamin, R.S.; Zhang, W.; Trent, J.C. Expression of receptor tyrosine kinases and apoptotic molecules in rhabdomyosarcoma: Correlation with overall survival in 105 patients. Cancer 2007, 110, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- De Andrade, C.R.; Takahama Junior, A.; Nishimoto, I.N.; Kowalski, L.P.; Lopes, M.A. Rhabdomyosarcoma of the head and neck: A clinicopathological and immunohistochemical analysis of 29 cases. Braz. Dent. J. 2010, 21, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Mark, H.F.L.; Brown, S.; Sun, C.L.; Samy, M.; Afify, A. Fluorescent in situ hybridization detection of HER-2/neu gene amplification in rhabdomyosarcoma. Pathobiology 1998, 66, 59–63. [Google Scholar] [CrossRef]
- Walther, C.; Mayrhofer, M.; Nilsson, J.; Hofvander, J.; Jonson, T.; Mandahl, N.; Øra, I.; Gisselsson, D.; Mertens, F. Genetic heterogeneity in rhabdomyosarcoma revealed by SNP array analysis. Genes Chromosom. Cancer 2016, 55, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, E.; Turina, C.B.; Kikuchi, K.; Langenau, D.M.; Keller, C. Proof-of-concept rare cancers in drug development: The case for rhabdomyosarcoma. Oncogene 2014, 33, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Ricci, C.; Landuzzi, L.; Rossi, I.; De Giovanni, C.; Nicoletti, G.; Astolfi, A.; Pupa, S.; Menard, S.; Scotlandi, K.; Nanni, P.; et al. Expression of HER/erbB family of receptor tyrosine kinases and induction of differentiation by glial growth factor 2 in human rhabdomyosarcoma cells. Int. J. Cancer 2000, 87, 29–36. [Google Scholar] [CrossRef]
- Astolfi, A.; De Giovanni, C.; Landuzzi, L.; Nicoletti, G.; Ricci, C.; Croci, S.; Scopece, L.; Nanni, P.; Lollini, P.L. Identification of new genes related to the myogenic differentiation arrest of human rhabdomyosarcoma cells. Gene 2001, 274, 139–149. [Google Scholar] [CrossRef]
- Kendall, G.C.; Watson, S.; Xu, L.; Lavigne, C.A.; Murchison, W.; Rakheja, D.; Skapek, S.X.; Tirode, F.; Delattre, O.; Amatruda, J.F. PAX3-FOXO1 transgenic zebrafish models identify HES3 as a mediator of rhabdomyosarcoma tumorigenesis. eLife 2018, 7, e33800. [Google Scholar] [CrossRef]
- Bersani, F.; Lingua, M.F.; Morena, D.; Foglizzo, V.; Miretti, S.; Lanzetti, L.; Carrà, G.; Morotti, A.; Ala, U.; Provero, P.; et al. Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res. 2016, 76, 6095–6106. [Google Scholar] [CrossRef]
- Nanni, P.; Nicoletti, G.; De Giovanni, C.; Croci, S.; Astolfi, A.; Landuzzi, L.; Di Carlo, E.; Iezzi, M.; Musiani, P.; Lollini, P.L. Development of rhabdomyosarcoma in HER-2/neu transgenic p53 mutant mice. Cancer Res. 2003, 63, 2728–2732. [Google Scholar]
- Kashi, V.P.; Hatley, M.E.; Galindo, R.L. Probing for a deeper understanding of rhabdomyosarcoma: Insights from complementary model systems. Nat. Rev. Cancer 2015, 15, 426–439. [Google Scholar] [CrossRef]
- Croci, S.; Nicoletti, G.; Landuzzi, L.; De Giovanni, C.; Astolfi, A.; Marini, C.; Di Carlo, E.; Musiani, P.; Forni, G.; Nanni, P.; et al. Immunological prevention of a multigene cancer syndrome. Cancer Res. 2004, 64, 8428–8434. [Google Scholar] [CrossRef]
- De Giovanni, C.; Landuzzi, L.; Palladini, A.; Ianzano, M.L.; Nicoletti, G.; Ruzzi, F.; Amici, A.; Croci, S.; Nanni, P.; Lollini, P.L. Cancer vaccines co-targeting HER2/NEU and IGF1R. Cancers 2019, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Ianzano, M.L.; Croci, S.; Nicoletti, G.; Palladini, A.; Landuzzi, L.; Grosso, V.; Ranieri, D.; Dall’Ora, M.; Santeramo, I.; Urbini, M.; et al. Tumor suppressor genes promote rhabdomyosarcoma progression in p53 heterozygous, HER-2/neu transgenic mice. Oncotarget 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- De Giovanni, C.; Nanni, P.; Landuzzi, L.; Ianzano, M.L.; Nicoletti, G.; Croci, S.; Palladini, A.; Lollini, P.L. Immune targeting of autocrine IGF2 hampers rhabdomyosarcoma growth and metastasis. BMC Cancer 2019, 19, 126. [Google Scholar] [CrossRef] [PubMed]
- Croci, S.; Landuzzi, L.; Nicoletti, G.; Palladini, A.; Antognoli, A.; De Giovanni, C.; Nanni, P.; Lollini, P.L. Expression of connective tissue growth factor (CTGF/CCN2) in a mouse model of rhabdomyosarcomagenesis. Pathol. Oncol. Res. 2007, 13, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Hinson, A.R.P.; Jones, R.; Lisa, L.E.; Belyea, B.C.; Barr, F.G.; Linardic, C.M. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: Utility and pitfalls. Front. Oncol. 2013, 115, 4218–4226. [Google Scholar] [CrossRef] [PubMed]
- De Giovanni, C.; Landuzzi, L.; Nicoletti, G.; Lollini, P.L.; Nanni, P. Molecular and cellular biology of rhabdomyosarcoma. Futur. Oncol. 2009, 5, 1449–1475. [Google Scholar] [CrossRef]
- Martinelli, S.; McDowell, H.P.; Delle Vigne, S.; Kokai, G.; Uccini, S.; Tartaglia, M.; Dominici, C. RAS signaling dysregulation in human embryonal rhabdomyosarcoma. Genes Chromosom. Cancer 2009, 48, 975–982. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Fukuda, K.; Fuchimoto, Y.; Matsuzaki, Y.; Saikawa, Y.; Kitagawa, Y.; Morikawa, Y.; Kuroda, T. Cetuximab promotes anticancer drug toxicity in rhabdomyosarcomas with EGFR amplification in vitro. Oncol. Rep. 2013, 30, 1081–1086. [Google Scholar] [CrossRef]
- Niesen, J.; Hehmann-Titt, G.; Woitok, M.; Fendel, R.; Barth, S.; Fischer, R.; Stein, C. A novel fully-human cytolytic fusion protein based on granzyme B shows in vitro cytotoxicity and ex vivo binding to solid tumors overexpressing the epidermal growth factor receptor. Cancer Lett. 2016, 374, 229–240. [Google Scholar] [CrossRef]
- Lollini, P.-L. (University of Bologna, Bologna, BO, Italy). Unpublished work. 2021. [Google Scholar]
- Hou, J.; Dong, J.; Sun, L.; Geng, L.; Wang, J.; Zheng, J.; Li, Y.; Bridge, J.; Hinrichs, S.H.; Ding, S.J. Inhibition of phosphorylated c-Met in rhabdomyosarcoma cell lines by a small molecule inhibitor SU11274. J. Transl. Med. 2011, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Granados, V.A.; Avirneni-Vadlamudi, U.; Dalal, P.; Scarborough, S.R.; Galindo, K.A.; Mahajan, P.; Galindo, R.L. Selective targeting of myoblast fusogenic signaling and differentiation-arrest antagonizes rhabdomyosarcoma cells. Cancer Res. 2019, 79, 4585–4591. [Google Scholar] [CrossRef] [PubMed]
- De Giovanni, C.; Melani, C.; Nanni, P.; Landuzzi, L.; Nicoletti, G.; Frabetti, F.; Griffoni, C.; Colombo, M.P. Redundancy of autocrine loops in human rhabdomyosarcoma cells: Induction of differentiation by suramin. Br. J. Cancer 1995, 72, 1224–1229. [Google Scholar] [CrossRef][Green Version]
- Ricci, C.; Polito, L.; Nanni, P.; Landuzzi, L.; Astolfi, A.; Nicoletti, G.; Rossi, I.; De Giovanni, C.; Bolognesi, A.; Lollini, P.L. HER/erbB receptors as therapeutic targets of immunotoxins in human rhabdomyosarcoma cells. J. Immunother. 2002, 25, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Favours, E.; Phelps, D.A.; Del Pozo, V.; Ghilu, S.; Kurmashev, D.; Michalek, J.; Trevino, A.; Guttridge, D.; London, C.; et al. Evaluation of patritumab with or without erlotinib in combination with standard cytotoxic agents against pediatric sarcoma xenograft models. Pediatr. Blood Cancer 2018, 65, e26870. [Google Scholar] [CrossRef]
- Merker, M.; Pfirrmann, V.; Oelsner, S.; Fulda, S.; Klingebiel, T.; Wels, W.S.; Bader, P.; Rettinger, E. Generation and characterization of ErbB2-CAR-engineered cytokine-induced killer cells for the treatment of high-risk soft tissue sarcoma in children. Oncotarget 2017, 8, 66137–66153. [Google Scholar] [CrossRef]
- Pilbeam, K.; Wang, H.; Taras, E.; Bergerson, R.J.; Ettestad, B.; DeFor, T.; Borgatti, A.; Vallera, D.A.; Verneris, M.R. Targeting pediatric sarcoma with a bispecific ligand immunotoxin targeting urokinase and epidermal growth factor receptors. Oncotarget 2018, 9, 11938–11947. [Google Scholar] [CrossRef]
- Frascella, E.; Lenzini, E.; Schafer, B.W.; Brecevic, L.; Dorigo, E.; Toffolatti, L.; Nanni, P.; De Giovanni, C.; Rosolen, A. Concomitant amplification and expression of PAX7-FKHR and MYCN in a human Rhabdomyosarcoma cell line carrying a cryptic t(1;13)(p36;q14). Cancer Genet. Cytogenet. 2000, 121, 139–145. [Google Scholar] [CrossRef]
- De Giovanni, C.; Landuzzi, L.; Frabetti, F.; Nicoletti, G.; Griffoni, C.; Rossi, I.; Mazzotti, M.; Scotto, L.; Nanni, P.; Lollini, P.L. Antisense epidermal growth factor receptor transfection impairs the proliferative ability of human rhabdomyosarcoma cells. Cancer Res. 1996, 56, 3898–3901. [Google Scholar]
- Huang, F.; Greer, A.; Hurlburt, W.; Han, X.; Hafezi, R.; Wittenberg, G.M.; Reeves, K.; Chen, J.; Robinson, D.; Li, A.; et al. The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Res. 2009, 69, 161–170. [Google Scholar] [CrossRef]
- Abraham, J.; Prajapati, S.I.; Nishijo, K.; Schaffer, B.S.; Taniguchi, E.; Kilcoyne, A.; McCleish, A.T.; Nelon, L.D.; Giles, F.G.; Efstratiadis, A.; et al. Evasion mechanisms to Igf1r inhibition in rhabdomyosarcoma. Mol. Cancer Ther. 2011, 10, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Jakacki, R.I.; Hamilton, M.; Gilbertson, R.J.; Blaney, S.M.; Tersak, J.; Krailo, M.D.; Ingle, A.M.; Voss, S.D.; Dancey, J.E.; Adamson, P.C. Pediatric phase I and pharmacokinetic study of erlotinib followed by the combination of erlotinib and temozolomide: A children’s oncology group phase I consortium study. J. Clin. Oncol. 2008, 26, 4921–4927. [Google Scholar] [CrossRef] [PubMed]
- Robichaux, J.P.; Elamin, Y.Y.; Vijayan, R.S.K.; Nilsson, M.B.; Hu, L.; He, J.; Zhang, F.; Pisegna, M.; Poteete, A.; Sun, H.; et al. Pan-Cancer Landscape and Analysis of ERBB2 Mutations Identifies Poziotinib as a Clinically Active Inhibitor and Enhancer of T-DM1 Activity. Cancer Cell 2019, 36, 444–457.e7. [Google Scholar] [CrossRef]
- Sadelain, M.; Rivière, I.; Riddell, S. Therapeutic T cell engineering. Nature 2017, 545, 423–431. [Google Scholar] [CrossRef]
- June, C.H.; Sadelain, M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018, 379, 64–73. [Google Scholar] [CrossRef]
- Martinez, M.; Moon, E.K. CAR T cells for solid tumors: New strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 2019, 10, 128. [Google Scholar] [CrossRef]
- Morgan, R.A.; Yang, J.C.; Kitano, M.; Dudley, M.E.; Laurencot, C.M.; Rosenberg, S.A. Case report of a serious adverse event following the administration of t cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 2010, 18, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, E.; Barok, M.; Szöllosi, J.; Vereb, G. ErbB-directed immunotherapy: Antibodies in current practice and promising new agents. Immunol. Lett. 2008, 116, 126–140. [Google Scholar] [CrossRef]
- DeRenzo, C.; Krenciute, G.; Gottschalk, S. The Landscape of CAR T Cells Beyond Acute Lymphoblastic Leukemia for Pediatric Solid Tumors. Am. Soc. Clin. Oncol. Educ. B 2018, 38, 830–837. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Patrizio, M.P.; Magagnoli, F.; Luppi, S.; Serra, M. An update on emerging drugs in osteosarcoma: Towards tailored therapies? Expert Opin. Emerg. Drugs 2019, 24, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Thanindratarn, P.; Dean, D.C.; Nelson, S.D.; Hornicek, F.J.; Duan, Z. Chimeric antigen receptor T (CAR-T) cell immunotherapy for sarcomas: From mechanisms to potential clinical applications. Cancer Treat. Rev. 2020, 82, 101934. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Salsman, V.S.; Yvon, E.; Louis, C.U.; Perlaky, L.; Wels, W.S.; Dishop, M.K.; Kleinerman, E.E.; Pule, M.; Rooney, C.M.; et al. Immunotherapy for osteosarcoma: Genetic modification of T cells overcomes low levels of tumor antigen expression. Mol. Ther. 2009, 17, 1779–1787. [Google Scholar] [CrossRef]
- Ahmed, N.; Brawley, V.S.; Hegde, M.; Robertson, C.; Ghazi, A.; Gerken, C.; Liu, E.; Dakhova, O.; Ashoori, A.; Corder, A.; et al. Human epidermal growth factor receptor 2 (HER2)—Specific chimeric antigen receptor—Modified T cells for the immunotherapy of HER2-positive sarcoma. J. Clin. Oncol. 2015, 33, 1688–1696. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.; Joseph, S.K.; Pashankar, F.; DeRenzo, C.; Sanber, K.; Navai, S.; Byrd, T.T.; Hicks, J.; Xu, M.L.; Gerken, C.; et al. Tumor response and endogenous immune reactivity after administration of HER2 CAR T cells in a child with metastatic rhabdomyosarcoma. Nat. Commun. 2020, 11, 3549. [Google Scholar] [CrossRef]
- Merker, M.; Wagner, J.; Kreyenberg, H.; Heim, C.; Moser, L.M.; Wels, W.S.; Bonig, H.; Ivics, Z.; Ullrich, E.; Klingebiel, T.; et al. ERBB2-CAR-Engineered Cytokine-Induced Killer Cells Exhibit Both CAR-Mediated and Innate Immunity Against High-Risk Rhabdomyosarcoma. Front. Immunol. 2020, 11, 581468. [Google Scholar] [CrossRef]
- Siegler, E.L.; Zhu, Y.; Wang, P.; Yang, L. Off-the-Shelf CAR-NK Cells for Cancer Immunotherapy. Cell Stem Cell 2018, 23, 160–161. [Google Scholar] [CrossRef] [PubMed]
- Gossel, L.D.H.; Heim, C.; Pfeffermann, L.M.; Moser, L.M.; Bönig, H.B.; Klingebiel, T.E.; Bader, P.; Wels, W.S.; Merker, M.; Rettinger, E. Retargeting of nk-92 cells against high-risk rhabdomyosarcomas by means of an erbb2 (Her2/neu)-specific chimeric antigen receptor. Cancers 2021, 13, 1443. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.; Chen, A.; Li, S.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Mitchell, E.P.; John Iafrate, A.; Sklar, J.; et al. The molecular analysis for therapy choice (NCI-MATCH) trial: Lessons for genomic trial design. J. Natl. Cancer Inst. 2020, 112, 1021–1029. [Google Scholar] [CrossRef]
| HER Family Member | ARMS | ERMS | Other Subtypes | Intensity at Immuno- Histochemistry | Amplification/Mutation | Reference |
|---|---|---|---|---|---|---|
| EGFR | 16% | 76% | Moderate to strong | No amplification a at 7p11.2 | [38] | |
| 13% | 84% | 42% b | Strong in ERMS | [7] | ||
| 32% | 55% | 73% c | [41] | |||
| 29% | 93% | [37] | ||||
| HER2 | 41% | 26% | No amplification a | [38] | ||
| 6% | 6% | 27% c | [41] | |||
| 70% d | [42] |
| RMS Cell Line | RMS Subtype | RAS Mutation/Translocation | Method | EGFR | HER2 | HER3 | HER4 |
|---|---|---|---|---|---|---|---|
| RD | ERMS | NRAS Q61H [59] | FC | +++ [36,60,61] | + [62] | ++ [62] | − [62] |
| WB | ++ a [63,64] | + a [63] | + a [40] | ||||
| RD/18 b | ERMS | NRAS Q61H [59] | FC | +++ [46,65,66] | + [46,66] | ++ [46,66] | − [46] |
| WB | +/− a [49] | ||||||
| RD/12 c | ERMS | FC | +++ [62] | + [62] | − [62] | ||
| RMS-YM | ERMS | FC | + [60] | ||||
| KYM-1 | ERMS d | FC | − [60] | ||||
| CCA | ERMS | KRAS Q61L [59] | FC | ++ [46,65] | ++ [46] | ++ [46] | − [46] |
| RH36 | ERMS | HRAS Q61K [10] | WB | ++ a [64,67] | + [67] | ||
| RH4/RH41 e | ARMS | PAX3/FOXO1 [45] | FC | − [46] | +/− 46,68] | + [46] | − [46] |
| WB | ++ [67] | ++ [67] | |||||
| RH5 | ARMS | PAX3/FOXO1 [45] | WB | − [67] | +++ [67] | ||
| RH10 | ARMS | PAX3/FOXO1 [45,57] | WB | +/− [67] | +++ [67] | ||
| RH18 f | ARMSf | PAX3/FOXO1 f [57] | WB | +++ a [67] | − [67] | ||
| RH28 | ARMS | PAX3/FOXO1 [45,57] | WB | +/− [67] | +/− [67] | ||
| RH30 | ARMS | PAX3/FOXO1 [45,57] | FC | ++ [36,46,60,69] | + [46,68] | + [46] | − [46] |
| WB | ++ a [63,67] | + a [63] | + [63,67] | ||||
| RC2 or RMZ-RC2 | ARMS | PAX7/FOXO1 [70] | FC | − [46,65] | + [46,65] | ++ [46] | − [46] |
| CW9019 | ARMS | PAX7/FOXO1 [45,57] | WB | + a [63] | + a [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Giovanni, C.; Landuzzi, L.; Palladini, A.; Nicoletti, G.; Nanni, P.; Lollini, P.-L. HER Tyrosine Kinase Family and Rhabdomyosarcoma: Role in Onset and Targeted Therapy. Cells 2021, 10, 1808. https://doi.org/10.3390/cells10071808
De Giovanni C, Landuzzi L, Palladini A, Nicoletti G, Nanni P, Lollini P-L. HER Tyrosine Kinase Family and Rhabdomyosarcoma: Role in Onset and Targeted Therapy. Cells. 2021; 10(7):1808. https://doi.org/10.3390/cells10071808
Chicago/Turabian StyleDe Giovanni, Carla, Lorena Landuzzi, Arianna Palladini, Giordano Nicoletti, Patrizia Nanni, and Pier-Luigi Lollini. 2021. "HER Tyrosine Kinase Family and Rhabdomyosarcoma: Role in Onset and Targeted Therapy" Cells 10, no. 7: 1808. https://doi.org/10.3390/cells10071808
APA StyleDe Giovanni, C., Landuzzi, L., Palladini, A., Nicoletti, G., Nanni, P., & Lollini, P.-L. (2021). HER Tyrosine Kinase Family and Rhabdomyosarcoma: Role in Onset and Targeted Therapy. Cells, 10(7), 1808. https://doi.org/10.3390/cells10071808







