Deficiency in MT5-MMP Supports Branching of Human iPSCs-Derived Neurons and Reduces Expression of GLAST/S100 in iPSCs-Derived Astrocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line
2.2. hiPSC Culture
2.3. CRISPR-Cas9 Plasmid Production
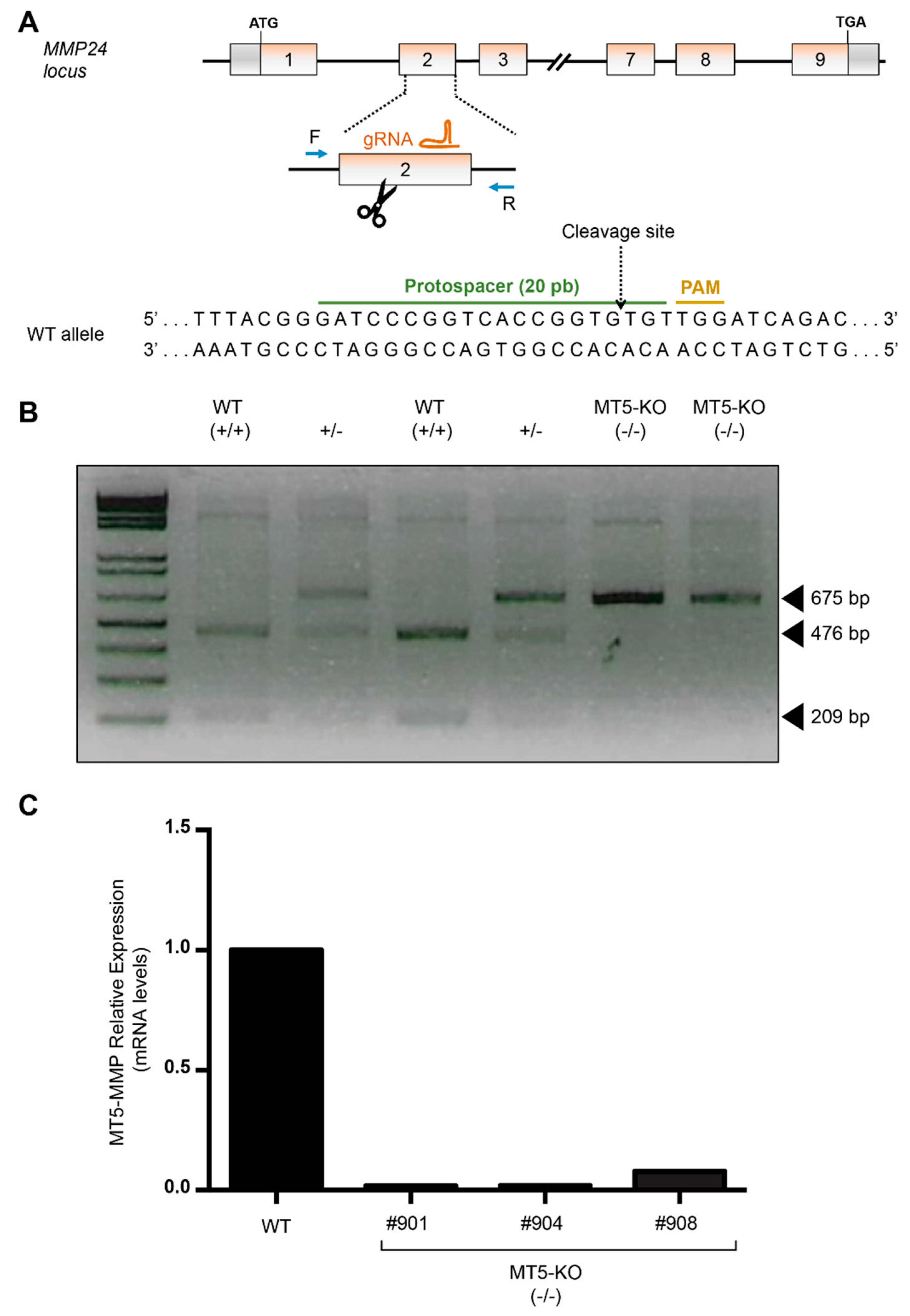
2.4. CRISPR-Cas9-Mediated Edition for the Generation of MMP24 Knockout hiPSCs
2.5. Derivation of NPCs from hiPSCs
2.6. Neuronal Differentiation
2.7. Astrocyte Differentiation
2.8. Immunocytochemistry
2.9. Confocal Microscopy and Data Collection
2.10. Image Analysis
2.11. Patch-Clamp Recordings
2.12. Statistical Analysis
3. Results
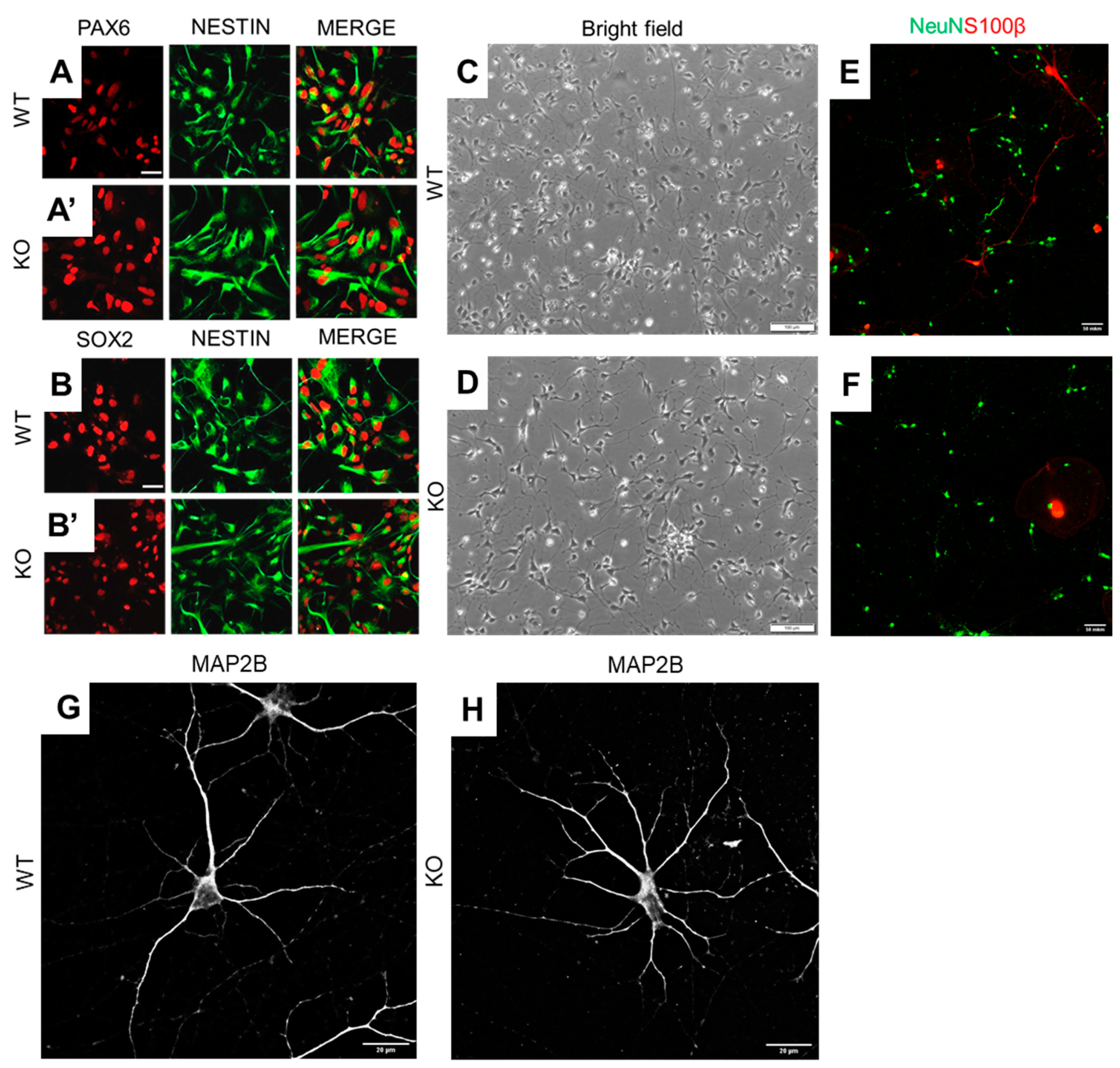
3.1. MT5-MMP Deficient hiPSCs Preserve the Ability to Differentiate into Neuronal Cells
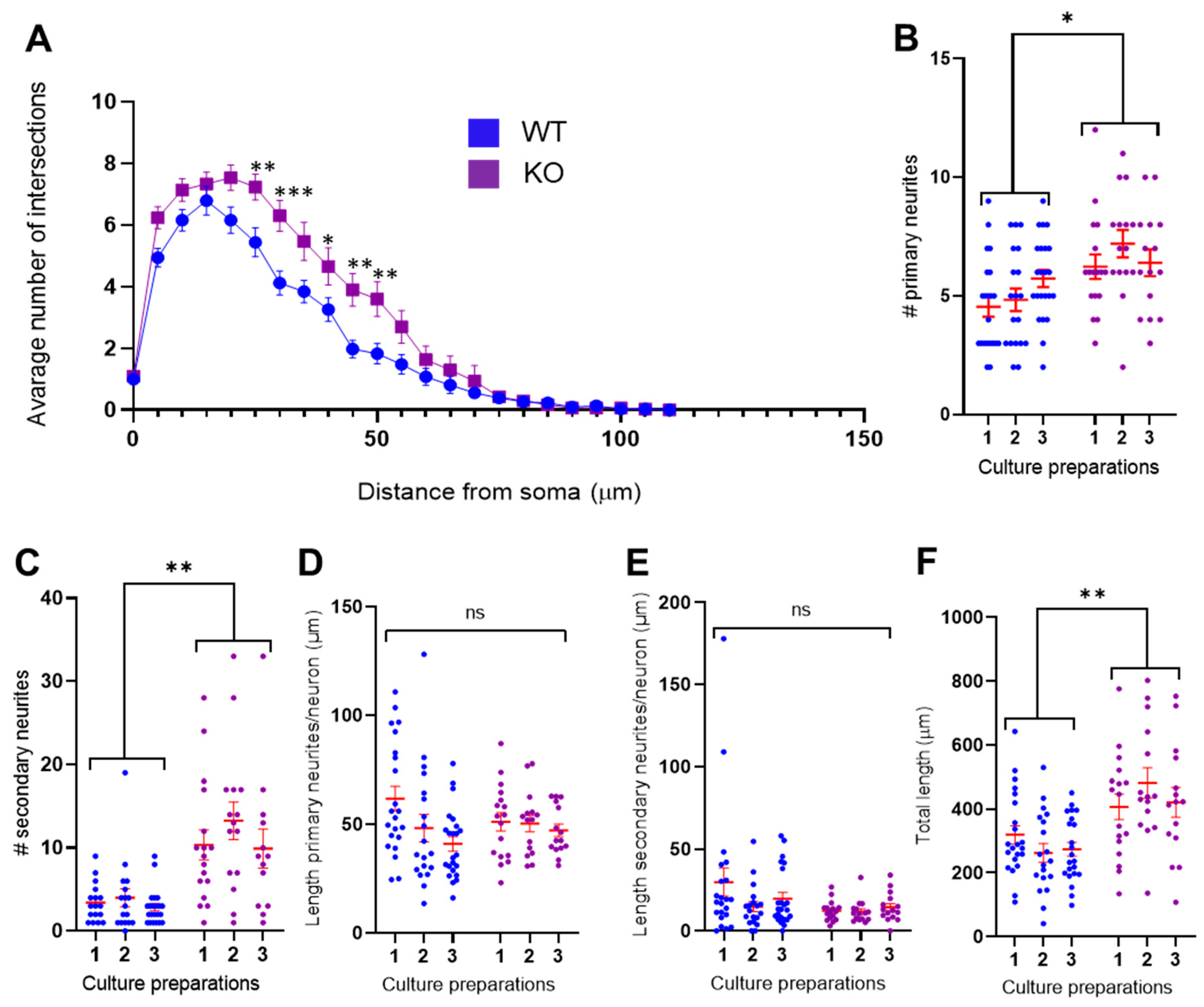
3.2. MT5-MMP Deficient Neurons Exhibited Increased Branching of Primary and Secondary Neurites
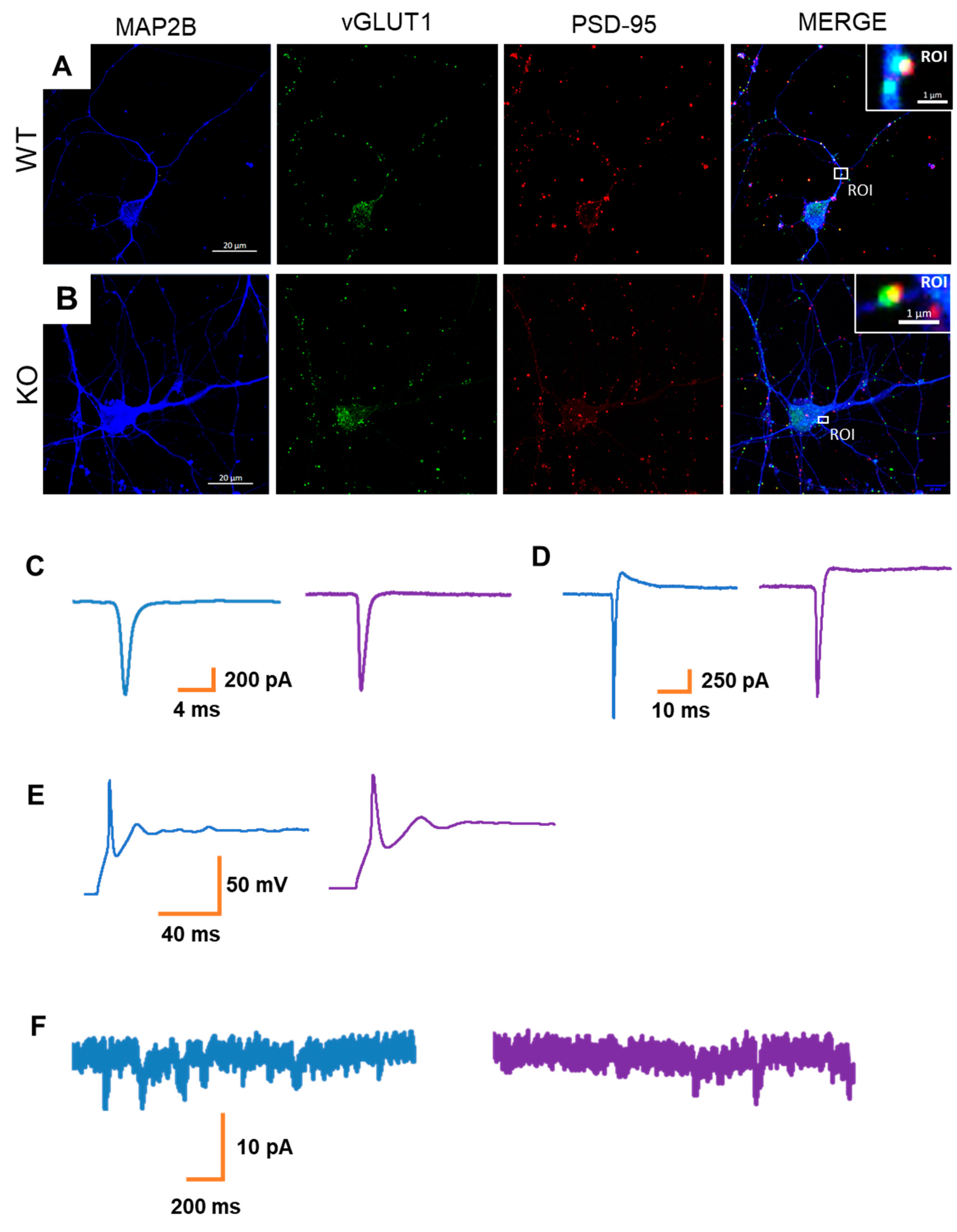
3.3. MT5-MMP Deficiency Does Not Alter Neuronal Activity in hiPSC-Derived Neurons
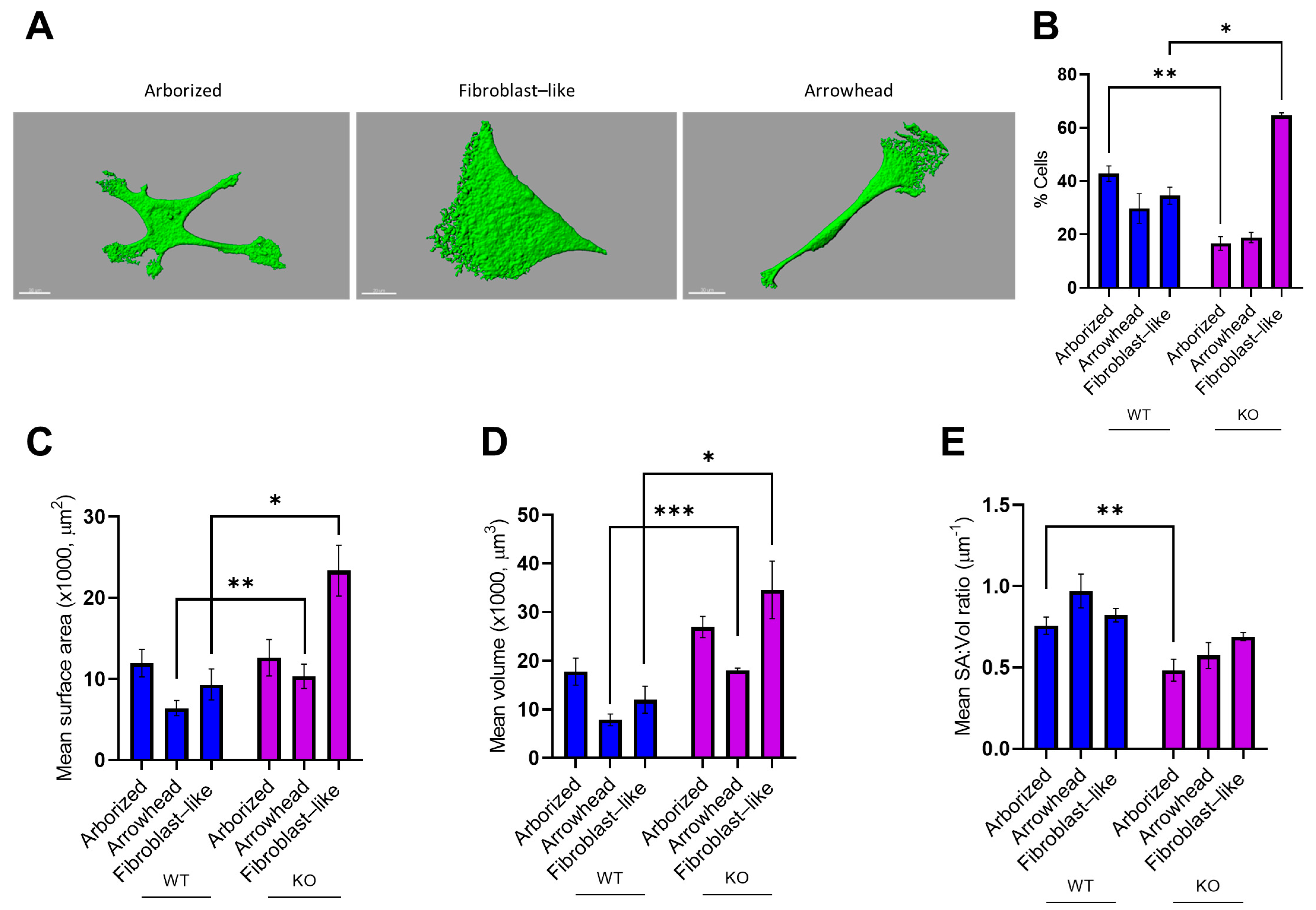
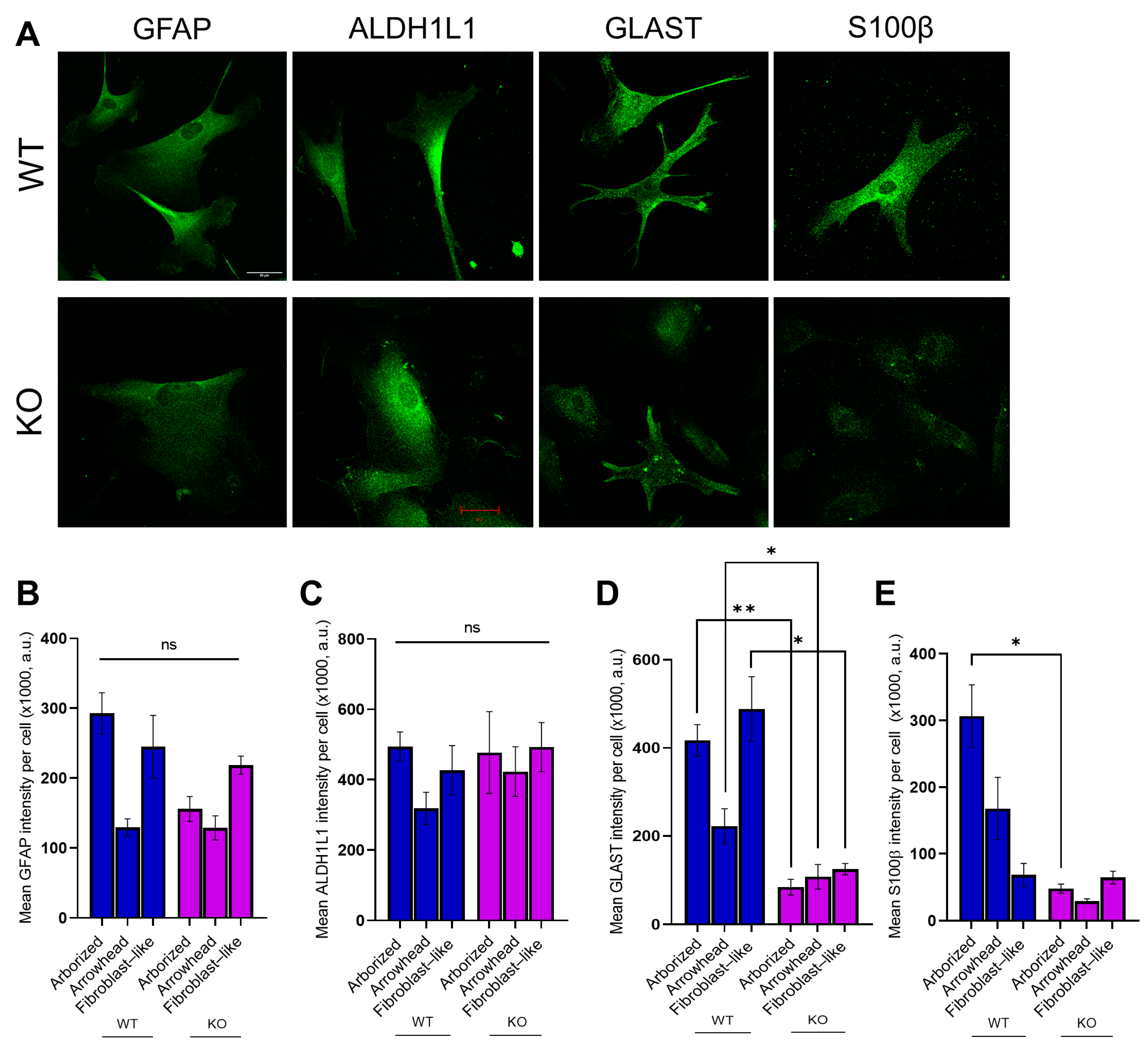
3.4. MT5-MMP Deficiency Induced Morphological Changes and Reduced the Expression of GLAST and S100β in Astrocyte-Like Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moussa-Pacha, N.M.; Abdin, S.M.; Omar, H.A.; Alniss, H.; Al-Tel, T.H. BACE1 inhibitors: Current status and future directions in treating Alzheimer’s disease. Med. Res. Rev. 2020, 40, 339–384. [Google Scholar] [CrossRef] [PubMed]
- Sanabria-Castro, A.; Alvarado-Echeverría, I.; Monge-Bonilla, C. Molecular Pathogenesis of Alzheimer’s Disease: An Update. Ann. Neurosci. 2017, 24, 46–54. [Google Scholar] [CrossRef] [PubMed]
- García-González, L.; Pilat, D.; Baranger, K.; Rivera, S. Emerging Alternative Proteinases in APP Metabolism and Alzheimer’s Disease Pathogenesis: A Focus on MT1-MMP and MT5-MMP. Front. Aging Neurosci. 2019, 11, 244. [Google Scholar] [CrossRef]
- Van Acker, Z.P.; Bretou, M.; Annaert, W. Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: Impact of genetic risk factors. Mol. Neurodegener. 2019, 14, 20. [Google Scholar] [CrossRef] [Green Version]
- Mullard, A. BACE inhibitor bust in Alzheimer trial. Nat. Rev. Drug Discov. 2017, 16, 155. [Google Scholar] [CrossRef]
- Piton, M.; Hirtz, C.; Desmetz, C.; Milhau, J.; Lajoix, A.D.; Bennys, K.; Lehmann, S.; Gabelle, A. Alzheimer’s Disease: Advances in Drug Development. J. Alzheimers Dis. 2018, 65, 3–13. [Google Scholar] [CrossRef]
- Haapasalo, A.; Hiltunen, M. A report from the 8th Kuopio Alzheimer symposium. Neurodegener. Dis. Manag. 2018, 8, 289–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullard, A. BACE failures lower AD expectations, again. Nat. Rev. Drug Discov. 2018, 17, 385. [Google Scholar] [CrossRef] [PubMed]
- Citron, M. Emerging Alzheimer’s disease therapies: Inhibition of beta-secretase. Neurobiol. Aging 2002, 23, 1017–1022. [Google Scholar] [CrossRef]
- De Strooper, B.; Vassar, R.; Golde, T. The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 99–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surguchev, A.A.; Emamzadeh, F.N.; Surguchov, A. Cell Responses to Extracellular α-Synuclein. Molecules 2019, 24, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera, S.; García-González, L.; Khrestchatisky, M.; Baranger, K. Metalloproteinases and their tissue inhibitors in Alzheimer’s disease and other neurodegenerative disorders. Cell Mol. Life Sci. 2019, 76, 3167–3191. [Google Scholar] [CrossRef]
- Zipfel, P.; Rochais, C.; Baranger, K.; Rivera, S.; Dallemagne, P. Matrix Metalloproteinases as New Targets in Alzheimer’s Disease: Opportunities and Challenges. J. Med. Chem. 2020, 63, 10705–10725. [Google Scholar] [CrossRef] [PubMed]
- Baranger, K.; Khrestchatisky, M.; Rivera, S. MT5-MMP, just a new APP processing proteinase in Alzheimer’s disease? J. Neuroinflamm. 2016, 13, 167. [Google Scholar] [CrossRef] [Green Version]
- Sekine-Aizawa, Y.; Hama, E.; Watanabe, K.; Tsubuki, S.; Kanai-Azuma, M.; Kanai, Y.; Arai, H.; Aizawa, H.; Iwata, N.; Saido, T.C. Matrix metalloproteinase (MMP) system in brain: Identification and characterization of brain-specific MMP highly expressed in cerebellum. Eur. J. Neurosci. 2001, 13, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Pei, D. Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. J. Biol. Chem. 1999, 274, 8925–8932. [Google Scholar] [CrossRef] [Green Version]
- Jaworski, D.M. Developmental regulation of membrane type-5 matrix metalloproteinase (MT5-MMP) expression in the rat nervous system. Brain Res. 2000, 860, 174–177. [Google Scholar] [CrossRef]
- Warren, K.M.; Reeves, T.M.; Phillips, L.L. MT5-MMP, ADAM-10, and N-cadherin act in concert to facilitate synapse reorganization after traumatic brain injury. J. Neurotrauma. 2012, 29, 1922–1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monea, S.; Jordan, B.A.; Srivastava, S.; DeSouza, S.; Ziff, E.B. Membrane localization of membrane type 5 matrix metalloproteinase by AMPA receptor binding protein and cleavage of cadherins. J. Neurosci. 2006, 26, 2300–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, K.; Nonaka, T.; Okada, A.; Kinoh, H.; Hayashita-Kinoh, H.; Yoshida, N.; Yana, I.; Seiki, M. Absence of mechanical allodynia and Abeta-fiber sprouting after sciatic nerve injury in mice lacking membrane-type 5 matrix metalloproteinase. FEBS Lett. 2004, 557, 125–128. [Google Scholar] [CrossRef] [Green Version]
- Hayashita-Kinoh, H.; Kinoh, H.; Okada, A.; Komori, K.; Itoh, Y.; Chiba, T.; Kajita, M.; Yana, I.; Seiki, M. Membrane-type 5 matrix metalloproteinase is expressed in differentiated neurons and regulates axonal growth. Cell Growth Differ. 2001, 12, 573–580. [Google Scholar]
- Porlan, E.; Martí-Prado, B.; Morante-Redolat, J.M.; Consiglio, A.; Delgado, A.C.; Kypta, R.; López-Otín, C.; Kirstein, M.; Fariñas, I. MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat. Cell Biol. 2014, 16, 629–638. [Google Scholar] [CrossRef]
- Folgueras, A.R.; Valdés-Sánchez, T.; Llano, E.; Menéndez, L.; Baamonde, A.; Denlinger, B.L.; Belmonte, C.; Juárez, L.; Lastra, A.; García-Suárez, O.; et al. Metalloproteinase MT5-MMP is an essential modulator of neuro-immune interactions in thermal pain stimulation. Proc. Natl. Acad. Sci. USA 2009, 106, 16451–16456. [Google Scholar] [CrossRef] [Green Version]
- Baranger, K.; Marchalant, Y.; Bonnet, A.E.; Crouzin, N.; Carrete, A.; Paumier, J.-M.; Py, N.A.; Bernard, A.; Bauer, C.; Charrat, E.; et al. MT5-MMP is a new pro-amyloidogenic proteinase that promotes amyloid pathology and cognitive decline in a transgenic mouse model of Alzheimer’s disease. Cell Mol. Life Sci. 2016, 73, 217–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranger, K.; Bonnet, A.E.; Girard, S.D.; Paumier, J.M.; García-González, L.; Elmanaa, W.; Bernard, A.; Charrat, E.; Stephan, D.; Bauer, C.; et al. MT5-MMP Promotes Alzheimer’s Pathogenesis in the Frontal Cortex of 5xFAD Mice and APP Trafficking in vitro. Front. Mol. Neurosci. 2017, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Willem, M.; Tahirovic, S.; Busche, M.A.; Ovsepian, S.V.; Chafai, M.; Kootar, S.; Hornburg, D.; Evans, L.D.B.; Moore, S.; Daria, A.; et al. η-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 2015, 526, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2015, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Rontani, P.; Perche, O.; Greetham, L.; Jullien, N.; Gepner, B.; Feron, F.; Erard-Garcia, M. Impaired expression of the COSMOC/MOCOS gene unit in ASD patient stem cells. Mol. Psychiatry 2021, 26, 1606–1618. [Google Scholar] [CrossRef] [Green Version]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Bardy, C.; van den Hurk, M.; Eames, T.; Marchand, C.; Hernandez, R.V.; Kellogg, M.; Gorris, M.; Galet, B.; Palomares, V.; Brown, J.; et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. USA 2015, 112, 2725–2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tcw, J.; Wang, M.; Pimenova, A.A.; Bowles, K.R.; Hartley, B.J.; Lacin, E.; Machlovi, S.I.; Abdelaal, R.; Karch, C.M.; Phatnani, H.; et al. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 9, 600–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, V.C.; Atkinson-Dell, R.; Verkhratsky, A.; Mohamet, L. Aberrant iPSC-derived human astrocytes in Alzheimer’s disease. Cell Death Dis. 2017, 8, e2696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penney, J.; Ralvenius, W.T.; Tsai, L.-H. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry 2020, 25, 148–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef] [Green Version]
- Ye, B.; Jan, Y.N. The cadherin superfamily and dendrite development. Trends Cell Biol. 2005, 15, 64–67. [Google Scholar] [CrossRef]
- Obst-Pernberg, K.; Redies, C. Cadherins and synaptic specificity. J. Neurosci. Res. 1999, 58, 130–138. [Google Scholar] [CrossRef]
- Tanriover, G.; Kayisli, U.A.; Demir, R.; Pestereli, E.; Karaveli, S.; Demir, N. Distribution of N-cadherin in human cerebral cortex during prenatal development. Histochem. Cell Biol. 2004, 122, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Gonzalez, P.; Mato, S.; Chara, J.C.; Verkhratsky, A.; Matute, C.; Cavaliere, F. Astrocytic atrophy as a pathological feature of Parkinson’s disease with LRRK2 mutation. NPJ Parkinsons Dis. 2021, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zuo, Y.X.; Jiang, R.T. Astrocyte morphology: Diversity, plasticity, and role in neurological diseases. CNS Neurosci. Ther. 2019, 25, 665–673. [Google Scholar] [CrossRef]
- Chung, W.-S.; Allen, N.J.; Eroglu, C. Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb. Perspect. Biol. 2015, 7, a020370. [Google Scholar] [CrossRef] [Green Version]
- Clarke, L.E.; Barres, B.A. Emerging roles of astrocytes in neural circuit development. Nat. Rev. Neurosci. 2013, 14, 311–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.A.; Weick, J.P.; Pearce, R.A.; Zhang, S.-C. Functional neural development from human embryonic stem cells: Accelerated synaptic activity via astrocyte coculture. J. Neurosci. 2007, 27, 3069–3077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escartin, C.; Galea, E.; Lakatos, A.; O’Callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Verkhratsky, A. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Escartin, C.; Guillemaud, O.; Carrillo-de Sauvage, M.A. Questions and (some) answers on reactive astrocytes. Glia 2019, 67, 2221–2247. [Google Scholar] [CrossRef]
- Michetti, F.; D’Ambrosi, N.; Toesca, A.; Puglisi, M.A.; Serrano, A.; Marchese, E.; Geloso, M.C. The S100B story: From biomarker to active factor in neural injury. J. Neurochem. 2019, 148, 168–187. [Google Scholar] [CrossRef] [Green Version]
- Pajarillo, E.; Rizor, A.; Lee, J.; Aschner, M.; Lee, E. The role of astrocytic glutamate transporters GLT-1 and GLAST in neurological disorders: Potential targets for neurotherapeutics. Neuropharmacology 2019, 161, 107559. [Google Scholar] [CrossRef]
- Schousboe, A.; Scafidi, S.; Bak, L.K.; Waagepetersen, H.S.; McKenna, M.C. Glutamate Metabolism in the Brain Focusing on Astrocytes. In Glutamate and ATP at the Interface of Metabolism and Signaling in the Brain; Parpura, V., Schousboe, A., Verkhratsky, A., Eds.; Springer: Cham, Switzerland, 2014; Volume 11. [Google Scholar] [CrossRef] [Green Version]
- Llano, E.; Pendás, A.M.; Freije, J.P.; Nakano, A.; Knäuper, V.; Murphy, G.; López-Otin, C. Identification and characterization of human MT5-MMP, a new membrane-bound activator of progelatinase an overexpressed in brain tumors. Cancer Res. 1999, 59, 2570–2576. [Google Scholar]
- García-González, L.; Paumier, J.M.; Louis, L.; Pilat, D.; Bernard, A.; Stephan, D.; Rivera, S. MT5-MMP controls APP and β-CTF/C99 metabolism through proteolytic-dependent and -independent mechanisms relevant for Alzheimer’s disease. FASEB J. 2021, 35, e21727. [Google Scholar] [CrossRef]
- Ogier, C.; Bernard, A.; Chollet, A.M.; Le Diguardher, T.; Hanessian, S.; Charton, G.; Rivera, S. Matrix metalloproteinase-2 (MMP-2) regulates astrocyte motility in connection with the actin cytoskeleton and integrins. Glia 2006, 54, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Hannocks, M.J.; Zhang, X.; Gerwien, H.; Chashchina, A.; Burmeister, M.; Korpos, E.; Sorokin, L. The gelatinases, MMP-2 and MMP-9, as fine tuners of neuroinflammatory processes. Matrix Biol. 2017, 75–76, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Sorci, G.; Bianchi, R.; Riuzzi, F.; Tubaro, C.; Arcuri, C.; Giambanco, I.; Donato, R. S100B Protein, A Damage-Associated Molecular Pattern Protein in the Brain and Heart, and Beyond. Cardiovasc. Psychiatry Neurol. 2010, 2010, 656481. [Google Scholar] [CrossRef] [PubMed]
- Raponi, E.; Agenes, F.; Delphin, C.; Assard, N.; Baudier, J.; Legraverend, C.; Deloulme, J.-C. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia 2007, 55, 165–177. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnst, N.; Belio-Mairal, P.; García-González, L.; Arnaud, L.; Greetham, L.; Nivet, E.; Rivera, S.; Dityatev, A. Deficiency in MT5-MMP Supports Branching of Human iPSCs-Derived Neurons and Reduces Expression of GLAST/S100 in iPSCs-Derived Astrocytes. Cells 2021, 10, 1705. https://doi.org/10.3390/cells10071705
Arnst N, Belio-Mairal P, García-González L, Arnaud L, Greetham L, Nivet E, Rivera S, Dityatev A. Deficiency in MT5-MMP Supports Branching of Human iPSCs-Derived Neurons and Reduces Expression of GLAST/S100 in iPSCs-Derived Astrocytes. Cells. 2021; 10(7):1705. https://doi.org/10.3390/cells10071705
Chicago/Turabian StyleArnst, Nikita, Pedro Belio-Mairal, Laura García-González, Laurie Arnaud, Louise Greetham, Emmanuel Nivet, Santiago Rivera, and Alexander Dityatev. 2021. "Deficiency in MT5-MMP Supports Branching of Human iPSCs-Derived Neurons and Reduces Expression of GLAST/S100 in iPSCs-Derived Astrocytes" Cells 10, no. 7: 1705. https://doi.org/10.3390/cells10071705
APA StyleArnst, N., Belio-Mairal, P., García-González, L., Arnaud, L., Greetham, L., Nivet, E., Rivera, S., & Dityatev, A. (2021). Deficiency in MT5-MMP Supports Branching of Human iPSCs-Derived Neurons and Reduces Expression of GLAST/S100 in iPSCs-Derived Astrocytes. Cells, 10(7), 1705. https://doi.org/10.3390/cells10071705







