The MicroRNA Landscape of Acute Beta Cell Destruction in Type 1 Diabetic Recipients of Intraportal Islet Grafts
Abstract
:1. Introduction
2. Material and Methods
2.1. Intrahepatic Transplantation of Human Beta Cells in Type 1 Diabetic Recipients
2.2. Recovery—Linearity Experiments with Human Beta Cell Lysate
2.3. GAD65 Cytometric Bead Array and Calibrated miR-375 Assay
2.4. microRNA Array Profiling
2.5. microRNA Array Data Analysis
2.6. Targeted PCR in Validation Cohorts
2.7. Statistical Analyses
3. Results
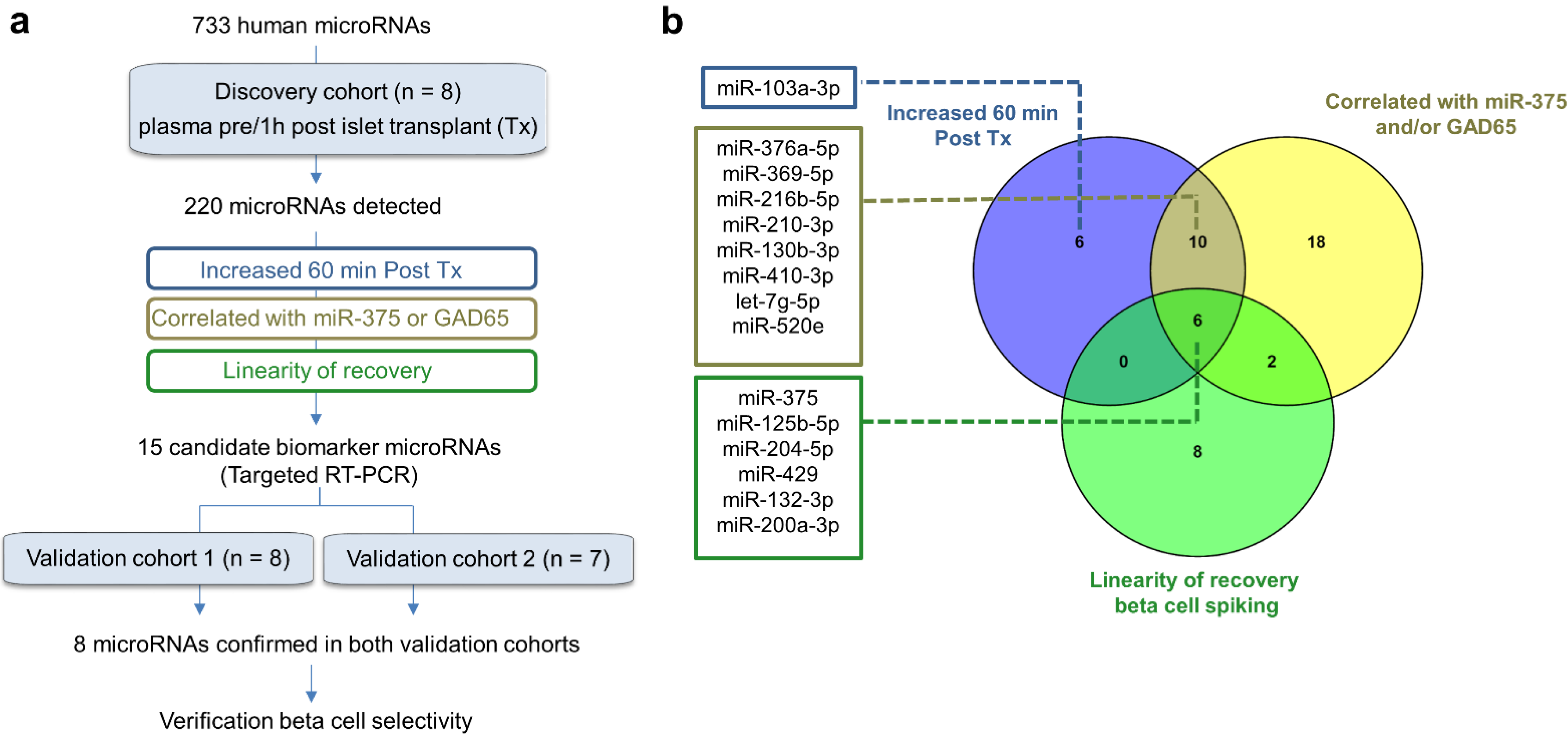
3.1. Study Design
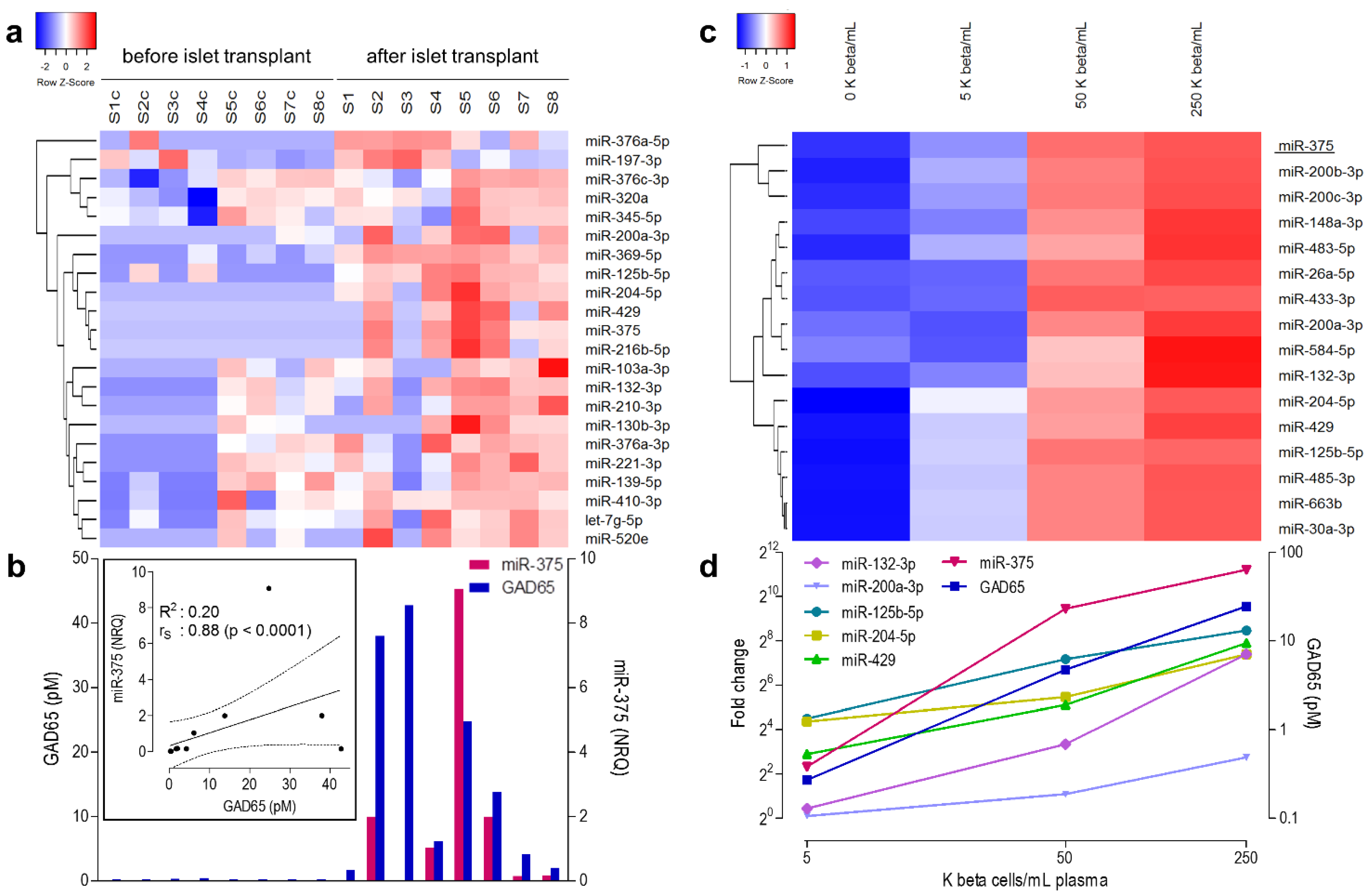
3.2. microRNA Profiling in the Discovery Cohort
3.3. microRNAs Correlating Linearly to Beta Cell Number in Recovery Experiment
3.4. Selection of Candidate Biomarker MicroRNAs for Validation Studies
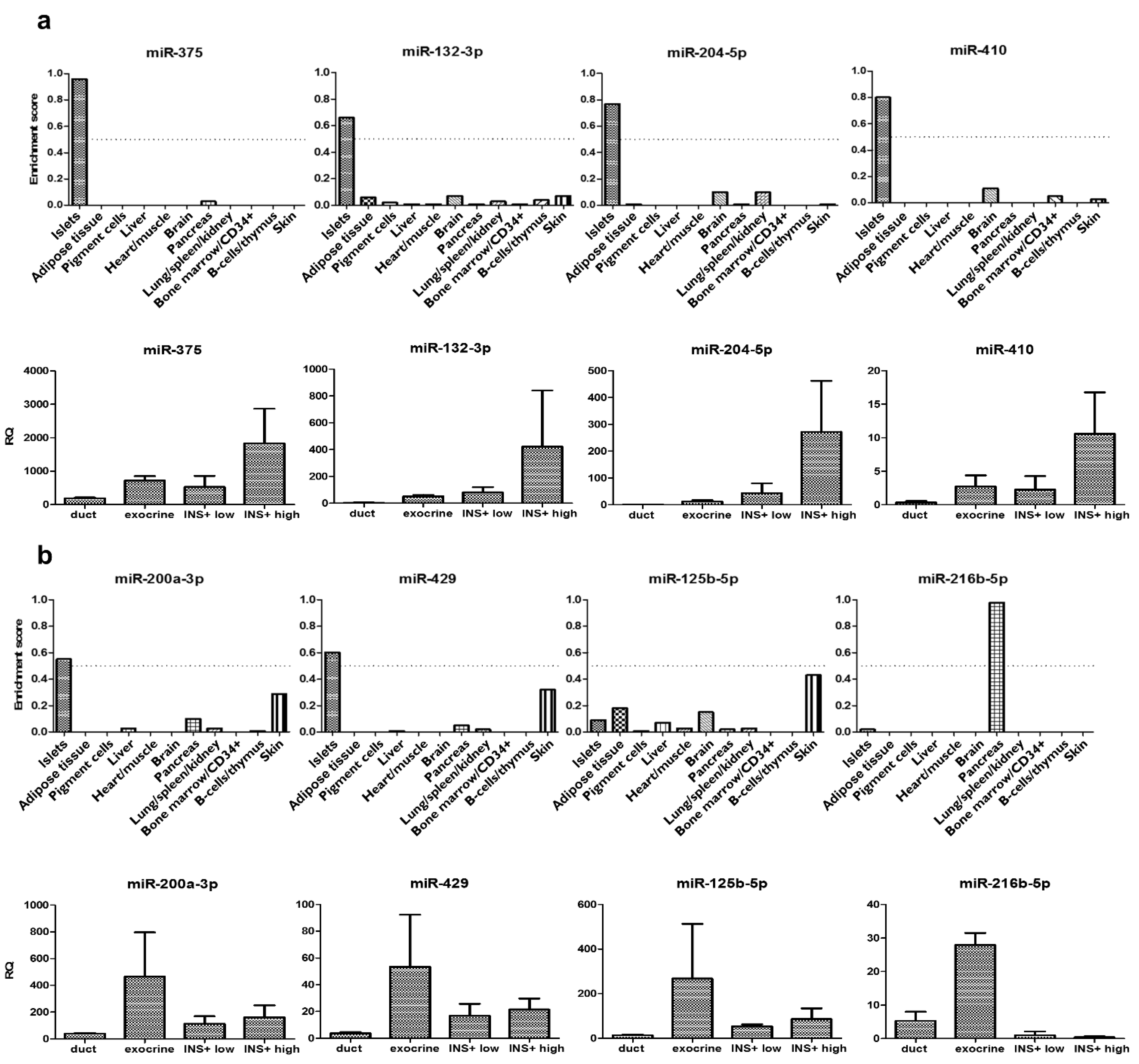
3.5. Verification of Pancreatic Cell Type and Tissue Tropism of the Eight Micrornas Consistently Increased after Islet Transplantation
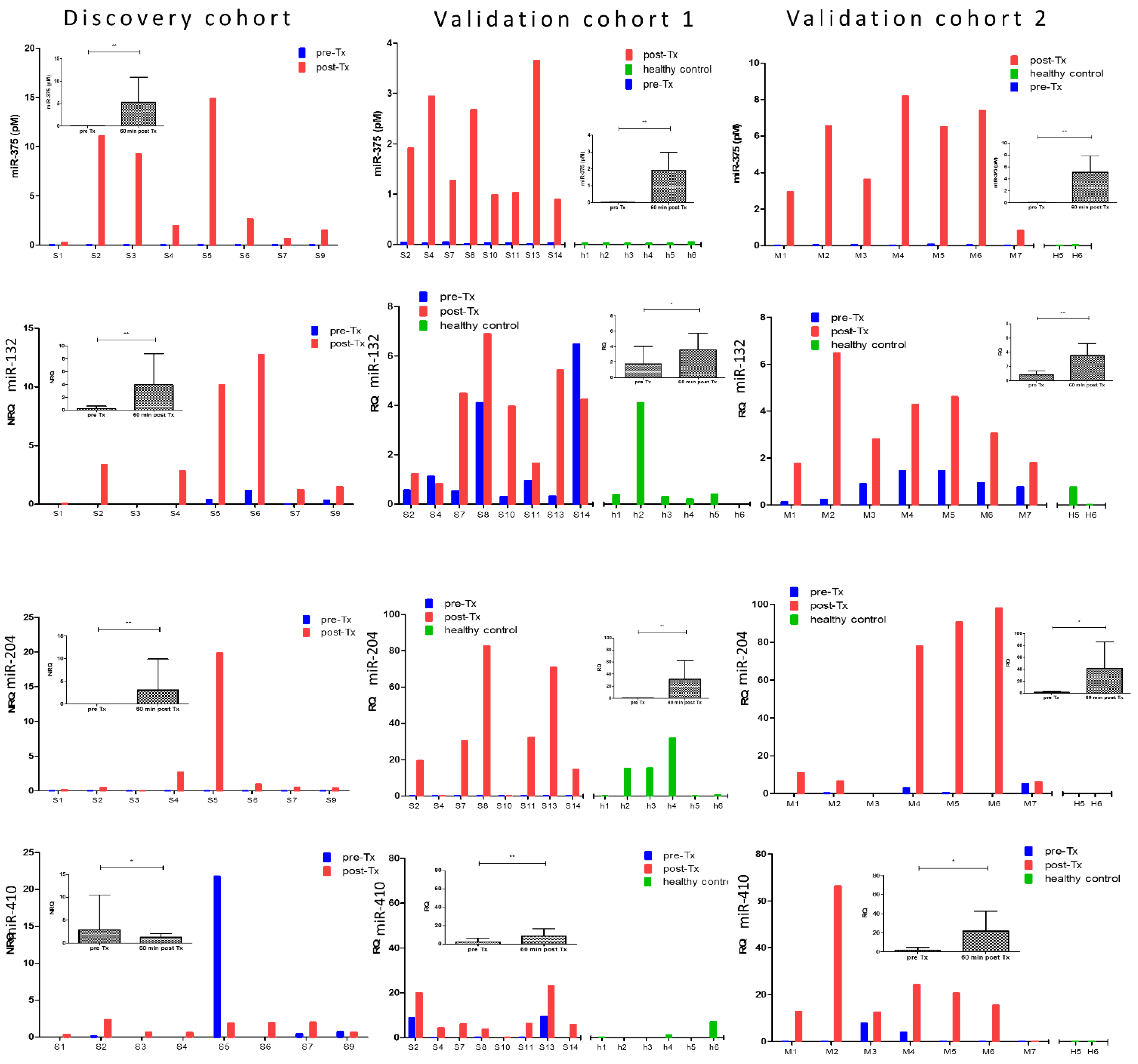
3.6. Clinical Validation: Correlation of early Post-Transplant Microrna Levels to Late-Stage Graft Function
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ling, Z.; De Pauw, P.; Jacobs-Tulleneers-Thevissen, D.; Mao, R.; Gillard, P.; Hampe, C.S.; Martens, G.A.; In’t Veld, P.; Lernmark, A.; Keymeulen, B.; et al. Plasma GAD65, a marker for early beta-Cell Loss After Intraportal Islet Cell Transplantation in Diabetic Patients. J. Clin. Endocrinol. Metab. 2015, 100, 2314–2321. [Google Scholar] [CrossRef] [Green Version]
- Costa, O.R.; Stange, G.; Verhaeghen, K.; Brackeva, B.; Nonneman, E.; Hampe, C.S.; Ling, Z.; Pipeleers, D.; Gorus, F.K.; Martens, G.A. Development of an enhanced sensitivity bead-based immunoassay for real-time in vivo detection of pancreatic beta-cell death. Endocrinology 2015, 156, 4755–4760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akirav, E.M.; Lebastchi, J.; Galvan, E.M.; Henegariu, O.; Akirav, M.; Ablamunits, V.; Herold, K.C. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 19018–19023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebastchi, J.; Deng, S.; Lebastchi, A.H.; Beshar, I.; Gitelman, S.; Willi, S.; Herold, K.C. Immune therapy and beta-cell death in type 1 diabetes. Diabetes 2013, 62, 1676–1680. [Google Scholar] [CrossRef] [Green Version]
- Speake, C.; Ylescupidez, A.; Neiman, D.; Shemer, R.; Glaser, B.; A Tersey, S.; Usmani-Brown, S.; Clark, P.; Wilhelm, J.J.; Bellin, M.D.; et al. Circulating unmethylated insulin DNA as a biomarker of human beta cell death: A multi-laboratory assay comparison. J. Clin. Endocrinol. Metab. 2020, 105, 781–791. [Google Scholar] [CrossRef]
- Neiman, D.; Gillis, D.; Piyanzin, S.; Cohen, D.; Fridlich, O.; Moss, J.; Dor, Y. Multiplexing DNA methylation markers to detect circulating cell-free DNA derived from human pancreatic beta cells. JCI Insight 2020, 5, e136579. [Google Scholar] [CrossRef]
- Sklenarova, J.; Petruzelkova, L.; Kolouskova, S.; Lebl, J.; Sumnik, Z.; Cinek, O. Glucokinase gene may be a more suitable target than the insulin gene for detection of beta cell death. Endocrinology 2017, 158, 2058–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; MacDonald, P.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Roels, S.; Costa, O.R.; Tersey, S.A.; Stangé, G.; De Smet, D.; Balti, E.V.; Gillard, P.; Keymeulen, B.; Ling, Z.; Pipeleers, D.G.; et al. Combined analysis of GAD65, miR-375, and unmethylated insulin dna following islet transplantation in patients with T1D. J. Clin. Endocrinol. Metab. 2019, 104, 451–460. [Google Scholar] [CrossRef] [PubMed]
- van de Bunt, M.; Gaulton, K.; Parts, L.; Moran, I.; Johnson, P.R.; Lindgren, C.; Ferrer, J.; Gloyn, A.L.; McCarthy, M.I. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS ONE 2013, 8, e55272. [Google Scholar] [CrossRef] [PubMed]
- Erener, S.; Mojibian, M.; Fox, J.K.; Denroche, H.C.; Kieffer, T.J. Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology 2013, 154, 603–608. [Google Scholar] [CrossRef]
- Keymeulen, B.; Gillard, P.; Mathieu, C.; Movahedi, B.; Maleux, G.; Delvaux, G.; Ysebaert, D.; Roep, B.; Vandemeulebroucke, E.; Marichal, M.; et al. Correlation between beta cell mass and glycemic control in type 1 diabetic recipients of islet cell graft. Proc. Natl. Acad. Sci. USA 2006, 103, 17444–17449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilbrands, R.; Huurman, V.A.L.; Gillard, P.; Velthuis, J.H.L.; De Waele, M.; Mathieu, C.; Kaufman, L.; Pipeleers-Marichal, M.; Ling, Z.; Movahedi, B.; et al. Differences in baseline lymphocyte counts and autoreactivity are associated with differences in outcome of islet cell transplantation in type 1 diabetic patients. Diabetes 2009, 58, 2267–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citro, A.; Cantarelli, E.; Maffi, P.; Nano, R.; Melzi, R.; Mercalli, A.; Piemonti, L. CXCR1/2 inhibition enhances pancreatic islet survival after transplantation. J. Clin. Investig. 2012, 122, 3647–3651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, O.R.; Verhaeghen, K.; Roels, S.; Stangé, G.; Ling, Z.; Pipeleers, D.; Gorus, F.K.; Martens, G.A. An analytical comparison of three immunoassay platforms for subpicomolar detection of protein biomarker GAD65. PLoS ONE 2018, 13, e0193670. [Google Scholar] [CrossRef] [PubMed]
- Lefever, S.; Hellemans, J.; Pattyn, F.; Przybylski, D.R.; Taylor, C.; Geurts, R.; Untergasser, A.; Vandesompele, J. RDML: Structured language and reporting guidelines for real-time quantitative PCR data. Nucleic Acids Res. 2009, 37, 2065–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnes, G.; Bolker, B.; Lumley, T.; Warnes, G.R.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Maechler, M.; Magnusson, A.; Moeller, S. gplots: Various R Programming Tools for Plotting Data. Available online: https://cran.r-project.org/package=gplots2015 (accessed on 4 May 2021).
- Megraw, M.; Sethupathy, P.; Corda, B.; Hatzigeorgiou, A.G. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007, 35, D149–D155. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Joglekar, M.V.; Joglekar, V.M.; Hardikar, A.A. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns 2009, 9, 109–113. [Google Scholar] [CrossRef]
- Martens, G.A.; Jiang, L.; Hellemans, K.H.; Stangé, G.; Heimberg, H.; Nielsen, F.C.; Sand, O.; Van Helden, J.; Gorus, F.K.; Pipeleers, D.G. Clusters of conserved beta cell marker genes for assessment of beta cell phenotype. PLoS ONE 2011, 6, e24134. [Google Scholar] [CrossRef] [Green Version]
- Filios, S.R.; Shalev, A. beta-Cell MicroRNAs: Small but powerful. Diabetes 2015, 64, 3631–3644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Stoffel, M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Ouaamari, A.; Baroukh, N.; Martens, G.A.; Lebrun, P.; Pipeleers, D.; van Obberghen, E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 2008, 57, 2708–2717. [Google Scholar] [CrossRef] [Green Version]
- Dusaulcy, R.; Handgraaf, S.; Visentin, F.; Vesin, C.; Philippe, J.; Gosmain, Y. miR-132-3p is a positive regulator of alpha-cell mass and is downregulated in obese hyperglycemic mice. Mol. Metab. 2019, 22, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Nesca, V.; Guay, C.; Jacovetti, C.; Menoud, V.; Peyot, M.-L.; Laybutt, D.R.; Prentki, M.; Regazzi, R. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 2013, 56, 2203–2212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, D.; Misawa, R.; Bravo-Egana, V.; Vargas, N.; Rosero, S.; Piroso, J.; Ichii, H.; Umland, O.; Zhijie, J.; Tsinoremas, N.; et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS ONE 2013, 8, e55064. [Google Scholar] [CrossRef] [Green Version]
- Hennessy, E.; Clynes, M.; Jeppesen, P.B.; O’Driscoll, L. Identification of microRNAs with a role in glucose stimulated insulin secretion by expression profiling of MIN6 cells. Biochem. Biophys. Res. Commun. 2010, 396, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chen, J.; Jing, G.; Shalev, A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat. Med. 2013, 19, 1141–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Chen, J.; Jing, G.; Grayson, T.B.; Shalev, A. miR-204 targets PERK and regulates UPR signaling and beta-cell apoptosis. Mol. Endocrinol. 2016, 30, 917–924. [Google Scholar] [CrossRef]
- Grieco, F.A.; Schiavo, A.A.; Brozzi, F.; Juan-Mateu, J.; Bugliani, M.; Marchetti, P.; Eizirik, D.L. The miRNAs miR-211-5p and miR-204-5p modulate, E.R. stress in human beta cells. J. Mol. Endocrinol. 2019, 63, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Thielen, L.A.; Chen, J.; Grayson, T.B.; Grimes, T.; Bridges, S.L.; Tse, H.M.; Smith, B.; Patel, R.; Li, P.; et al. Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E723–E730. [Google Scholar] [CrossRef] [PubMed]
- Erdos, Z.; E Barnum, J.; Wang, E.; DeMaula, C.; Dey, P.M.; Forest, T.; Bailey, W.J.; E Glaab, W. Evaluation of the relative performance of pancreas-specific microRNAs in rat plasma as biomarkers of pancreas injury. Toxicol. Sci. 2020, 173, 5–18. [Google Scholar] [CrossRef]
- Sewer, A.; Iovino, N.; Aravin, A.; Tuschl, T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Belgardt, B.-F.; Ahmed, K.; Spranger, M.; Latreille, M.; Denzler, R.; Kondratiuk, N.; Von Meyenn, F.; Villena, F.N.; Herrmanns, K.; Bosco, D.; et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015, 21, 619–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coenen-Stass, A.M.L.; Pauwels, M.J.; Hanson, B.; Perez, C.M.; Conceição, M.; Wood, M.J.A.; Mäger, I.; Roberts, T.C. Extracellular microRNAs exhibit sequence-dependent stability and cellular release kinetics. RNA Biol. 2019, 16, 696–706. [Google Scholar] [CrossRef] [Green Version]

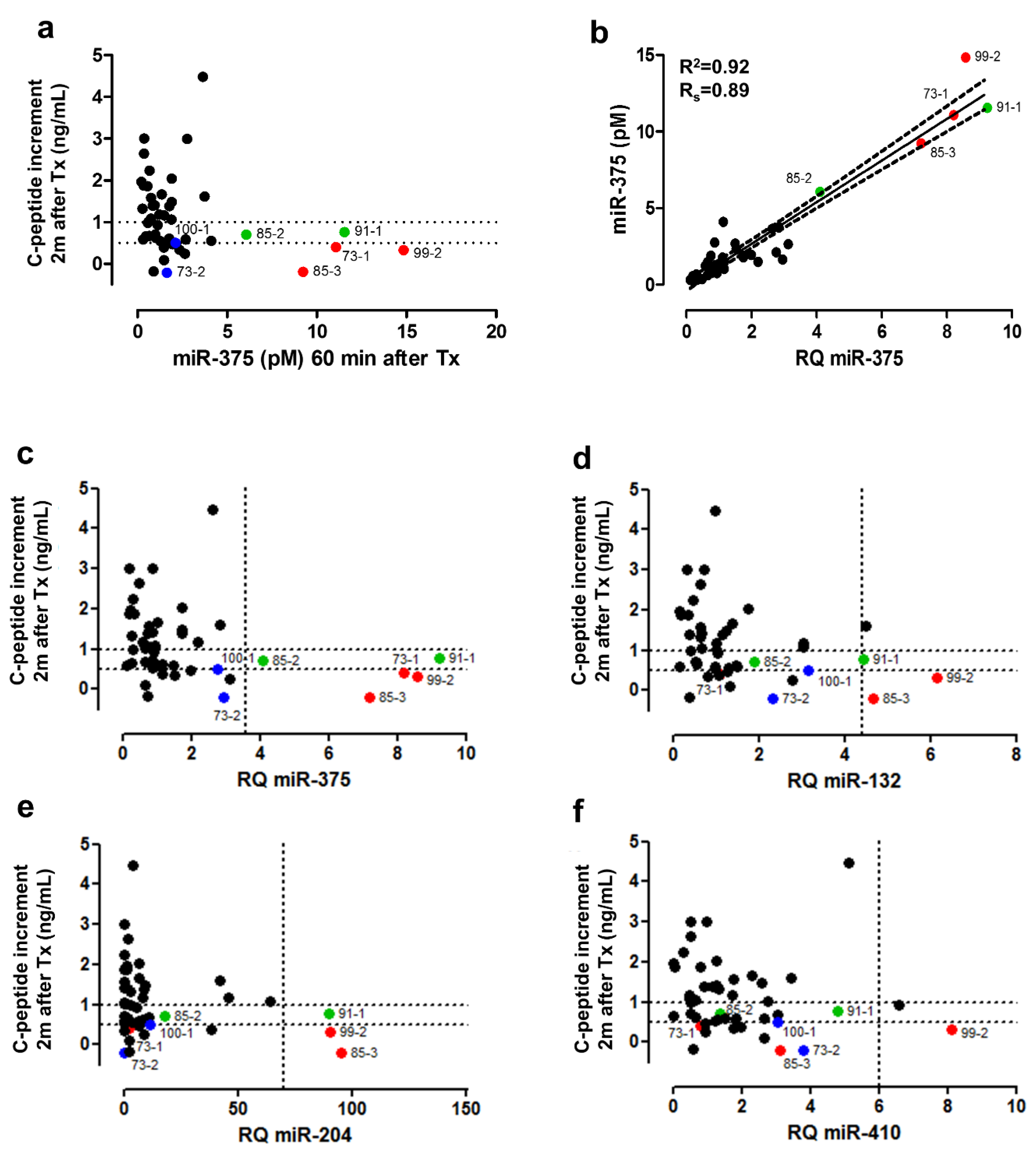
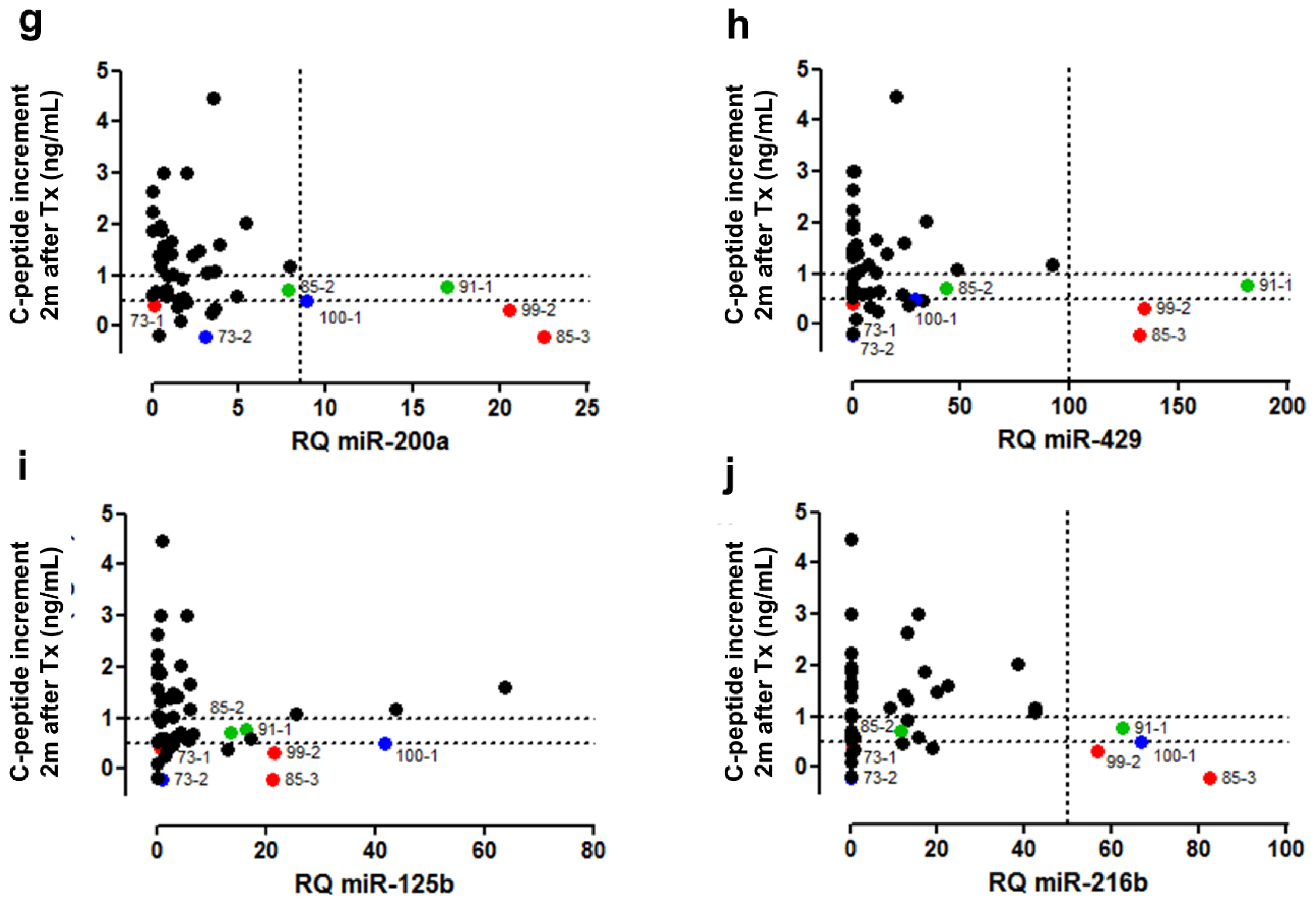
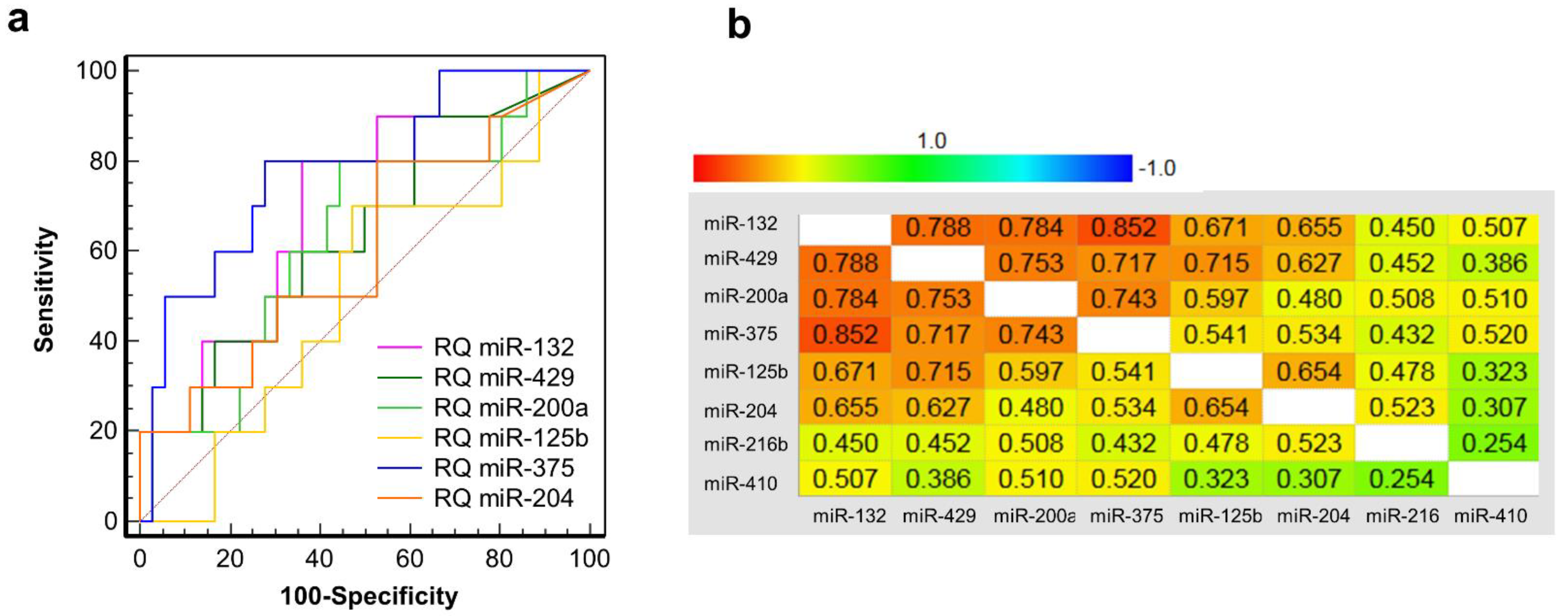
| Discovery Cohort (n = 8) | Validation Cohort 1 (n = 8) | Validation Cohort 2 (n = 7) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR Level Post/Pre-Tx | Correlation GAD65 | Correlation miR-375 | miR Level Post/Pre-Tx | miR Level Post/Pre-Tx | |||||||
| MicroRNA | Assay ID | NRQ | P | rs | P | rs | P | RQ | P | RQ | P |
| miR-375 | hsa-miR-375-000564 | 118.1 | 0.007 | 0.88 | <0.0001 | - | - | 249.5 | 0.000 | 178.0 | 0.002 |
| miR-132-3p | hsa-miR-132-000457 | 210.9 | 0.005 | 0.50 | 0.051 | 0.87 | <0.0001 | 2.0 | 0.047 | 4.2 | 0.004 |
| miR-204-5p | hsa-miR-204-000508 | 172.0 | 0.002 | 0.71 | 0.002 | 0.93 | <0.0001 | 315.4 | 0.003 | 33.0 | 0.016 |
| miR-410-3p | hsa-miR-410-001274 | 49.1 | 0.034 | 0.46 | 0.072 | 0.57 | 0.022 | 3.7 | 0.002 | 12.6 | 0.014 |
| miR-200a-3p | hsa-miR-200a-000502 | 6249.4 | 0.044 | 0.49 | 0.054 | 0.82 | <0.0001 | 397.0 | 0.000 | 70.4 | 0.000 |
| miR-429 | hsa-miR-429-001024 | 36.5 | 0.016 | 0.64 | 0.008 | 0.92 | <0.0001 | 287.0 | 0.007 | 53.5 | 0.002 |
| miR-125b-5p | hsa-miR-125b-000449 | 52.1 | 0.001 | 0.83 | 0.000 | 0.79 | 0.000 | 6.8 | 0.039 | 6.5 | 0.282 |
| miR-216b-5p | hsa-miR-216b-002326 | 85.9 | 0.019 | 0.66 | 0.005 | 0.94 | <0.0001 | 21.6 | 0.275 | 191.4 | 0.084 |
| let-7g-5p | hsa-let-7g-002282 | 90.5 | 0.036 | 0.29 | 0.274 | 0.69 | 0.003 | 0.6 | 0.109 | 1.3 | 0.650 |
| miR-103a-3p | hsa-miR-103-000439 | 130.3 | 0.020 | 0.18 | 0.494 | 0.49 | 0.054 | 1.1 | 0.497 | 1.2 | 0.274 |
| miR-130b-3p | hsa-miR-130b-000456 | 97.1 | 0.021 | 0.26 | 0.336 | 0.62 | 0.010 | 0.8 | 0.337 | 1.5 | 0.086 |
| miR-210-3p | hsa-miR-210-000512 | 156.7 | 0.010 | 0.35 | 0.188 | 0.76 | 0.001 | 0.0 | 0.766 | 0.2 | 0.317 |
| miR-376a-5p | hsa-miR-376a-5p-002127 | 9707.1 | 0.014 | 0.52 | 0.040 | 0.26 | 0.322 | 0.0 | 0.351 | 2.7 | 0.115 |
| miR-369-5p | hsa-miR-369-5p-001021 | 1705.0 | 0.000 | 0.93 | <0.0001 | 0.78 | 0.000 | ND | ND | ||
| miR-520e | hsa-miR-520e-001119 | 167.6 | 0.027 | 0.45 | 0.079 | 0.75 | 0.001 | ND | ND | ||
| Correlation with Graft Characteristics | Functional Outcome | |||||||
|---|---|---|---|---|---|---|---|---|
| Beta Cells | C-Peptide Content | Alpha Cells | Exocrine Cells | Dead Cells | C-Peptide Increment after 2 Months (ng/mL) | |||
| Spearman (P) | Spearman (P) | Linear Regression (P) | Spearman (P) | Spearman (P) | Spearman (P) | Spearman (P) | Linear Regression (P) | |
| miR-375 | 0.0541 | p < 0.0001 | 0.0001 | 0.0441 | 0.0023 | 0.0424 | 0.0059 | 0.0535 |
| miR-132-3p | 0.1418 | 0.0198 | 0.0037 | 0.0782 | 0.0271 | 0.0262 | 0.0102 | 0.0432 |
| miR-204-5p | 0.2528 | 0.0230 | 0.0074 | 0.0384 | 0.5109 | 0.4291 | 0.1371 | / |
| miR-410 | 0.3239 | 0.1175 | / | 0.9689 | 0.0491 | 0.6401 | 0.0432 | / |
| miR-200a-3p | 0.2320 | 0.0245 | 0.0040 | 0.0859 | 0.0221 | 0.0839 | 0.0828 | / |
| miR-429 | 0.6479 | 0.0158 | 0.0006 | 0.0778 | 0.0603 | 0.6225 | 0.1249 | / |
| miR-125b-5p | 0.5322 | 0.0328 | / | 0.0043 | 0.0900 | 0.8033 | 0.1644 | / |
| miR-216b-5p | 0.7800 | 0.1152 | / | 0.6951 | 0.3092 | 0.4530 | 0.9714 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martens, G.A.; Stangé, G.; Piemonti, L.; Anckaert, J.; Ling, Z.; Pipeleers, D.G.; Gorus, F.K.; Mestdagh, P.; De Smet, D.; Vandesompele, J.; et al. The MicroRNA Landscape of Acute Beta Cell Destruction in Type 1 Diabetic Recipients of Intraportal Islet Grafts. Cells 2021, 10, 1693. https://doi.org/10.3390/cells10071693
Martens GA, Stangé G, Piemonti L, Anckaert J, Ling Z, Pipeleers DG, Gorus FK, Mestdagh P, De Smet D, Vandesompele J, et al. The MicroRNA Landscape of Acute Beta Cell Destruction in Type 1 Diabetic Recipients of Intraportal Islet Grafts. Cells. 2021; 10(7):1693. https://doi.org/10.3390/cells10071693
Chicago/Turabian StyleMartens, Geert A., Geert Stangé, Lorenzo Piemonti, Jasper Anckaert, Zhidong Ling, Daniel G. Pipeleers, Frans K. Gorus, Pieter Mestdagh, Dieter De Smet, Jo Vandesompele, and et al. 2021. "The MicroRNA Landscape of Acute Beta Cell Destruction in Type 1 Diabetic Recipients of Intraportal Islet Grafts" Cells 10, no. 7: 1693. https://doi.org/10.3390/cells10071693
APA StyleMartens, G. A., Stangé, G., Piemonti, L., Anckaert, J., Ling, Z., Pipeleers, D. G., Gorus, F. K., Mestdagh, P., De Smet, D., Vandesompele, J., Keymeulen, B., & Roels, S. (2021). The MicroRNA Landscape of Acute Beta Cell Destruction in Type 1 Diabetic Recipients of Intraportal Islet Grafts. Cells, 10(7), 1693. https://doi.org/10.3390/cells10071693






