Melatonin Treatment Improves Renal Fibrosis via miR-4516/SIAH3/PINK1 Axis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Human Proximal Tubular Epithelial (TH1) Cells
2.2. Mitochondrial Fraction Preparation
2.3. Quantification of miRNA
2.4. Transmission Electron Microscopy
2.5. Western Blotting
2.6. Immunoprecipitation
2.7. OCR Measurements and Calculation of Bioenergetic Parameters
2.8. Fluorescence Staining for Flow Cytometry
2.9. CKD Mouse Model
2.10. Hematoxylin and Eosin (H&E), and Immunofluorescence Staining
2.11. Immunohistochemical Staining
2.12. Ethics Statement
2.13. Statistical Analysis
3. Results
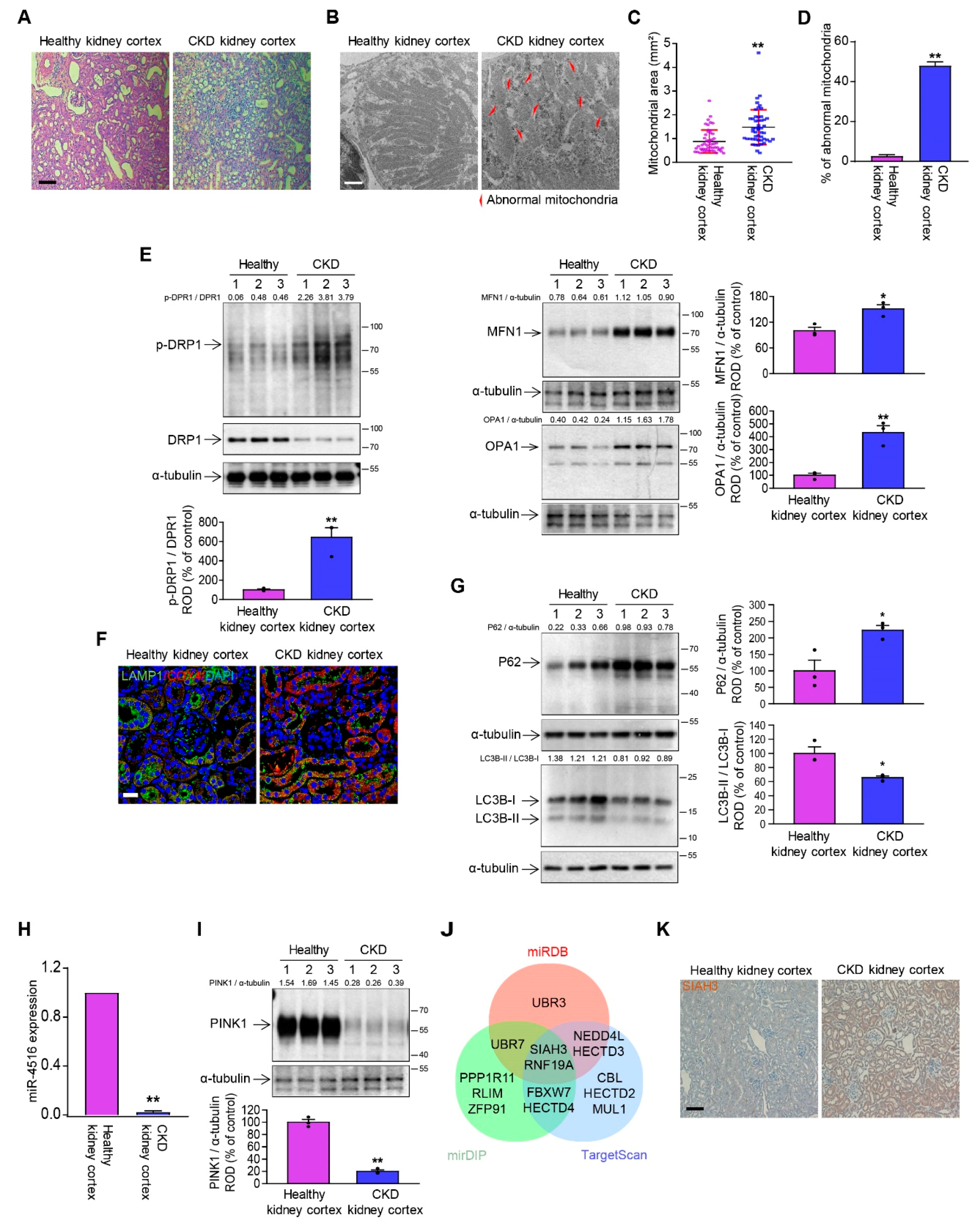
3.1. The Kidney of CKD Mice Has Dysregulated Mitochondrial Dynamics and Mitophagy
3.2. miR-4516/SIAH3 Pathway Is Involved in Dysregulated Mitophagy
3.3. Melatonin-Induced miR-4516 Targets SIAH3 to Improve Mitophagy
3.4. Melatonin-Induced miR-4516 Restore Mitochondrial Dysfunctions
3.5. Melatonin Injection Ameliorates CKD via miR-4516/SIAH3/PINK1 Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 2011, 7, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.L.; Manenti, L.; Vaglio, A. Fibrosis—A Common Pathway to Organ Injury and Failure. N. Engl. J. Med. 2015, 373, 95–96. [Google Scholar]
- Panizo, S.; Martinez-Arias, L.; Alonso-Montes, C.; Cannata, P.; Martin-Carro, B.; Fernandez-Martin, J.L.; Naves-Diaz, M.; Carrillo-Lopez, N.; Cannata-Andia, J.B. Fibrosis in Chronic Kidney Disease: Pathogenesis and Consequences. Int. J. Mol. Sci. 2021, 22, 408. [Google Scholar] [CrossRef] [PubMed]
- Eddy, A.A. Overview of the cellular and molecular basis of kidney fibrosis. Kidney. Int. Suppl. 2014, 4, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chawla, L.S.; Eggers, P.W.; Star, R.A.; Kimmel, P.L. Acute kidney injury and chronic kidney disease as interconnected syndromes. N. Engl. J. Med. 2014, 371, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Tampe, D.; Zeisberg, M. Potential approaches to reverse or repair renal fibrosis. Nat. Rev. Nephrol. 2014, 10, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Cai, J.; Yin, X.M.; Weinberg, J.M.; Venkatachalam, M.A.; Dong, Z. Mitochondrial quality control in kidney injury and repair. Nat. Rev. Nephrol. 2021, 17, 299–318. [Google Scholar] [CrossRef]
- Duann, P.; Lin, P.H. Mitochondria Damage and Kidney Disease. Adv. Exp. Med. Biol. 2017, 982, 529–551. [Google Scholar] [PubMed]
- Rangarajan, S.; Bernard, K.; Thannickal, V.J. Mitochondrial Dysfunction in Pulmonary Fibrosis. Ann. Am. Thorac. Soc. 2017, 14 (Suppl. 5), S383–S388. [Google Scholar] [CrossRef]
- Che, R.; Yuan, Y.; Huang, S.; Zhang, A. Mitochondrial dysfunction in the pathophysiology of renal diseases. Am. J. Physiol. Renal. Physiol. 2014, 306, F367–F378. [Google Scholar] [CrossRef]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Vives-Bauza, C.; Zhou, C.; Huang, Y.; Cui, M.; de Vries, R.L.; Kim, J.; May, J.; Tocilescu, M.A.; Liu, W.; Ko, H.S.; et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 378–383. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, D.; Chung, K.P.; Nakahira, K.; Patino, E.; Rice, M.C.; Torres, L.K.; Muthukumar, T.; Choi, A.M.; Akchurin, O.M.; Choi, M.E. Mitophagy-dependent macrophage reprogramming protects against kidney fibrosis. JCI Insight 2019, 4, e132826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Zhao, Z.; Feng, X.; Cheng, Z.; Xiong, Z.; Wang, T.; Lin, J.; Zhang, M.; Hu, J.; Fan, Y.; et al. Melatonin activates Parkin translocation and rescues the impaired mitophagy activity of diabetic cardiomyopathy through Mst1 inhibition. J. Cell Mol. Med. 2018, 22, 5132–5144. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ma, Z.; Jiang, S.; Fan, C.; Deng, C.; Yan, X.; Di, S.; Lv, J.; Reiter, R.J.; Yang, Y. Melatonin: The dawning of a treatment for fibrosis? J. Pineal Res. 2016, 60, 121–131. [Google Scholar] [CrossRef]
- Han, Y.S.; Lee, J.H.; Lee, S.H. Melatonin suppresses ischemia-induced fibrosis by regulating miR-149. Biochem. Biophys. Res. Commun. 2020, 525, 354–359. [Google Scholar] [CrossRef]
- Yoon, Y.M.; Go, G.; Yun, C.W.; Lim, J.H.; Lee, J.H.; Lee, S.H. Melatonin Suppresses Renal Cortical Fibrosis by Inhibiting Cytoskeleton Reorganization and Mitochondrial Dysfunction through Regulation of miR-4516. Int. J. Mol. Sci. 2020, 21, 5323. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.M.; Kim, S.; Han, Y.S.; Yun, C.W.; Lee, J.H.; Noh, H.; Lee, S.H. TUDCA-treated chronic kidney disease-derived hMSCs improve therapeutic efficacy in ischemic disease via PrP(C). Redox Biol. 2019, 22, 101144. [Google Scholar] [CrossRef]
- Jiang, M.; Bai, M.; Lei, J.; Xie, Y.; Xu, S.; Jia, Z.; Zhang, A. Mitochondrial dysfunction and the AKI-to-CKD transition. Am. J. Physiol. Renal. Physiol. 2020, 319, F1105–F1116. [Google Scholar] [CrossRef]
- Galvan, D.L.; Green, N.H.; Danesh, F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017, 92, 1051–1057. [Google Scholar] [CrossRef]
- Bhatia, D.; Choi, M.E. The Emerging Role of Mitophagy in Kidney Diseases. J. Life Sci. (Westlake Village) 2019, 1, 13–22. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Moscat, J.; Diaz-Meco, M.T. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009, 137, 1001–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasson, S.A.; Kane, L.A.; Yamano, K.; Huang, C.H.; Sliter, D.A.; Buehler, E.; Wang, C.; Heman-Ackah, S.M.; Hessa, T.; Guha, R.; et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 2013, 504, 291–295. [Google Scholar] [CrossRef]
- Bhatia, D.; Capili, A.; Choi, M.E. Mitochondrial dysfunction in kidney injury, inflammation, and disease: Potential therapeutic approaches. Kidney Res. Clin. Pract. 2020, 39, 244–258. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, C.; Cai, J.; Chen, G.; Zhang, D.; Zhang, Z.; Dong, Z. PINK1/Parkin-mediated mitophagy is activated in cisplatin nephrotoxicity to protect against kidney injury. Cell Death Dis. 2018, 9, 1113. [Google Scholar] [CrossRef]
- Lin, Q.; Li, S.; Jiang, N.; Shao, X.; Zhang, M.; Jin, H.; Zhang, Z.; Shen, J.; Zhou, Y.; Zhou, W.; et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019, 26, 101254. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.S.; Chen, X.M.; Hua, W.; He, J.L.; Liu, T.; Li, X.J.; Wan, J.M.; Gan, H.; Du, X.G. PINK1/Parkin mediated mitophagy ameliorates palmitic acid-induced apoptosis through reducing mitochondrial ROS production in podocytes. Biochem. Biophys. Res. Commun. 2020, 525, 954–961. [Google Scholar] [CrossRef]
- Li, W.; Du, M.; Wang, Q.; Ma, X.; Wu, L.; Guo, F.; Ji, H.; Huang, F.; Qin, G. FoxO1 Promotes Mitophagy in the Podocytes of Diabetic Male Mice via the PINK1/Parkin Pathway. Endocrinology 2017, 158, 2155–2167. [Google Scholar] [CrossRef] [Green Version]
- Klinkhammer, B.M.; Goldschmeding, R.; Floege, J.; Boor, P. Treatment of Renal Fibrosis-Turning Challenges into Opportunities. Adv. Chronic Kidney Dis. 2017, 24, 117–129. [Google Scholar] [CrossRef]
- Liu, T.; Yang, Q.; Zhang, X.; Qin, R.; Shan, W.; Zhang, H.; Chen, X. Quercetin alleviates kidney fibrosis by reducing renal tubular epithelial cell senescence through the SIRT1/PINK1/mitophagy axis. Life Sci. 2020, 257, 118116. [Google Scholar] [CrossRef]
- Qiao, H.; Ren, H.; Du, H.; Zhang, M.; Xiong, X.; Lv, R. Liraglutide repairs the infarcted heart: The role of the SIRT1/Parkin/mitophagy pathway. Mol. Med. Rep. 2018, 17, 3722–3734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsujimoto, T.; Mori, T.; Houri, K.; Onodera, Y.; Takehara, T.; Shigi, K.; Nakao, S.; Teramura, T.; Fukuda, K. miR-155 inhibits mitophagy through suppression of BAG5, a partner protein of PINK1. Biochem. Biophys. Res. Commun. 2020, 523, 707–712. [Google Scholar] [CrossRef]
- Cheng, M.; Liu, L.; Lao, Y.; Liao, W.; Liao, M.; Luo, X.; Wu, J.; Xie, W.; Zhang, Y.; Xu, N. MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 2016, 7, 42274–42287. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Fiesel, F.C.; Belmonte, K.C.; Hudec, R.; Wang, W.X.; Kim, C.; Nelson, P.T.; Springer, W.; Kim, J. miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener 2016, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, Y.; Xu, J.; Tian, F.; Hu, S.; Chen, Y.; Fu, Z. Melatonin attenuates myocardial ischemia-reperfusion injury via improving mitochondrial fusion/mitophagy and activating the AMPK-OPA1 signaling pathways. J. Pineal Res. 2019, 66, e12542. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, A.; Javad-Moosavi, S.A.; Reiter, R.J.; Yarahmadi, R.; Ghaznavi, H.; Mehrzadi, S. Oxidative/nitrosative stress, autophagy and apoptosis as therapeutic targets of melatonin in idiopathic pulmonary fibrosis. Expert Opin. Ther. Targets 2018, 22, 1049–1061. [Google Scholar] [CrossRef]
- Kang, J.W.; Hong, J.M.; Lee, S.M. Melatonin enhances mitophagy and mitochondrial biogenesis in rats with carbon tetrachloride-induced liver fibrosis. J. Pineal Res. 2016, 60, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Dominguez, N.; Ordonez, R.; Fernandez, A.; Mendez-Blanco, C.; Baulies, A.; Garcia-Ruiz, C.; Fernandez-Checa, J.C.; Mauriz, J.L.; Gonzalez-Gallego, J. Melatonin-induced increase in sensitivity of human hepatocellular carcinoma cells to sorafenib is associated with reactive oxygen species production and mitophagy. J. Pineal Res. 2016, 61, 396–407. [Google Scholar] [CrossRef]
- Pourhanifeh, M.H.; Mehrzadi, S.; Hosseinzadeh, A. Melatonin and regulation of miRNAs: Novel targeted therapy for cancerous and noncancerous disease. Epigenomics 2021, 13, 65–81. [Google Scholar] [CrossRef]
- Yoon, Y.M.; Lee, J.H.; Song, K.H.; Noh, H.; Lee, S.H. Melatonin-stimulated exosomes enhance the regenerative potential of chronic kidney disease-derived mesenchymal stem/stromal cells via cellular prion proteins. J. Pineal Res. 2020, 68, e12632. [Google Scholar] [CrossRef]
- Rukov, J.L.; Wilentzik, R.; Jaffe, I.; Vinther, J.; Shomron, N. Pharmaco-miR: Linking microRNAs and drug effects. Brief. Bioinform. 2014, 15, 648–659. [Google Scholar] [CrossRef]
- Costa, P.M.; Cardoso, A.L.; Nobrega, C.; Pereira de Almeida, L.F.; Bruce, J.N.; Canoll, P.; Pedroso de Lima, M.C. MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum. Mol. Genet. 2013, 22, 904–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.M.; Chou, W.C.; Hu, L.Y.; Hsiung, C.N.; Chu, H.W.; Huang, Y.L.; Hsu, H.M.; Yu, J.C.; Shen, C.Y. The Effect of MicroRNA-124 Overexpression on Anti-Tumor Drug Sensitivity. PLoS ONE 2015, 10, e0128472. [Google Scholar] [CrossRef] [PubMed]
- Che, H.; Wang, Y.; Li, H.; Li, Y.; Sahil, A.; Lv, J.; Liu, Y.; Yang, Z.; Dong, R.; Xue, H.; et al. Melatonin alleviates cardiac fibrosis via inhibiting lncRNA MALAT1/miR-141-mediated NLRP3 inflammasome and TGF-beta1/Smads signaling in diabetic cardiomyopathy. FASEB J. 2020, 34, 5282–5298. [Google Scholar] [CrossRef]
- Shin, N.R.; Park, J.W.; Lee, I.C.; Ko, J.W.; Park, S.H.; Kim, J.S.; Kim, J.C.; Ahn, K.S.; Shin, I.S. Melatonin suppresses fibrotic responses induced by cigarette smoke via downregulation of TGF-beta1. Oncotarget 2017, 8, 95692–95703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.S.; Yoon, Y.M.; Go, G.; Lee, J.H.; Lee, S.H. Melatonin Protects Human Renal Proximal Tubule Epithelial Cells Against High Glucose-Mediated Fibrosis via the Cellular Prion Protein-TGF-beta-Smad Signaling Axis. Int. J. Med. Sci. 2020, 17, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.Y.; Liu, X.S.; Huang, X.R.; Yu, X.Q.; Lan, H.Y. Diverse Role of TGF-beta in Kidney Disease. Front Cell Dev. Biol. 2020, 8, 123. [Google Scholar] [CrossRef]
- Chung, A.C.; Dong, Y.; Yang, W.; Zhong, X.; Li, R.; Lan, H.Y. Smad7 suppresses renal fibrosis via altering expression of TGF-beta/Smad3-regulated microRNAs. Molecular therapy. J. Am. Soc. Gene Ther. 2013, 21, 388–398. [Google Scholar] [CrossRef] [Green Version]
- Kang, H. Role of MicroRNAs in TGF-beta Signaling Pathway-Mediated Pulmonary Fibrosis. Int. J. Mol. Sci. 2017, 18, 2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.I. MicroRNA Control of TGF-β Signaling. Int. J. Mol. Sci. 2018, 19, 1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi, N.; Ishihara, N.; Jofuku, A.; Oka, T.; Mihara, K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007, 282, 11521–11529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cereghetti, G.M.; Stangherlin, A.; de Brito, O.M.; Chang, C.R.; Blackstone, C.; Bernardi, P.; Scorrano, L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. USA 2008, 105, 15803–15808. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lu, M.; Xiong, L.; Fan, J.; Zhou, Y.; Li, H.; Peng, X.; Zhong, Z.; Wang, Y.; Huang, F.; et al. Drp1-mediated mitochondrial fission promotes renal fibroblast activation and fibrogenesis. Cell Death Dis. 2020, 11, 29. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.; Brooks, C.; Liu, F.; Sun, L.; Dong, Z. Mitochondrial dynamics: Regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 2013, 83, 568–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, A.C.; Lan, H.Y. MicroRNAs in renal fibrosis. Front. Physiol. 2015, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Gholaminejad, A.; Abdul Tehrani, H.; Gholami Fesharaki, M. Identification of candidate microRNA biomarkers in renal fibrosis: A meta-analysis of profiling studies. Biomarkers 2018, 23, 713–724. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, H.; Huang, Z.; Zheng, H.; Zhou, J. Emerging role of miRNAs in renal fibrosis. RNA Biol. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Gourlay, C.W.; Carpp, L.N.; Timpson, P.; Winder, S.J.; Ayscough, K.R. A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 2004, 164, 803–809. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, Q.; Xie, C.; Zhou, H.; Wang, Y.; Zhang, N.; Shao, H.; Chan, S.C.; Peng, X.; Lin, S.C.; et al. Beta-actin is required for mitochondria clustering and ROS generation in TNF-induced, caspase-independent cell death. J. Cell Sci. 2004, 117 Pt 20, 4673–4680. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, Y.M.; Go, G.; Yoon, S.; Lim, J.H.; Lee, G.; Lee, J.H.; Lee, S.H. Melatonin Treatment Improves Renal Fibrosis via miR-4516/SIAH3/PINK1 Axis. Cells 2021, 10, 1682. https://doi.org/10.3390/cells10071682
Yoon YM, Go G, Yoon S, Lim JH, Lee G, Lee JH, Lee SH. Melatonin Treatment Improves Renal Fibrosis via miR-4516/SIAH3/PINK1 Axis. Cells. 2021; 10(7):1682. https://doi.org/10.3390/cells10071682
Chicago/Turabian StyleYoon, Yeo Min, Gyeongyun Go, Sungtae Yoon, Ji Ho Lim, Gaeun Lee, Jun Hee Lee, and Sang Hun Lee. 2021. "Melatonin Treatment Improves Renal Fibrosis via miR-4516/SIAH3/PINK1 Axis" Cells 10, no. 7: 1682. https://doi.org/10.3390/cells10071682
APA StyleYoon, Y. M., Go, G., Yoon, S., Lim, J. H., Lee, G., Lee, J. H., & Lee, S. H. (2021). Melatonin Treatment Improves Renal Fibrosis via miR-4516/SIAH3/PINK1 Axis. Cells, 10(7), 1682. https://doi.org/10.3390/cells10071682




