Fibroblast Growth Factor Receptor (FGFR) Signaling in GIST and Soft Tissue Sarcomas
Abstract
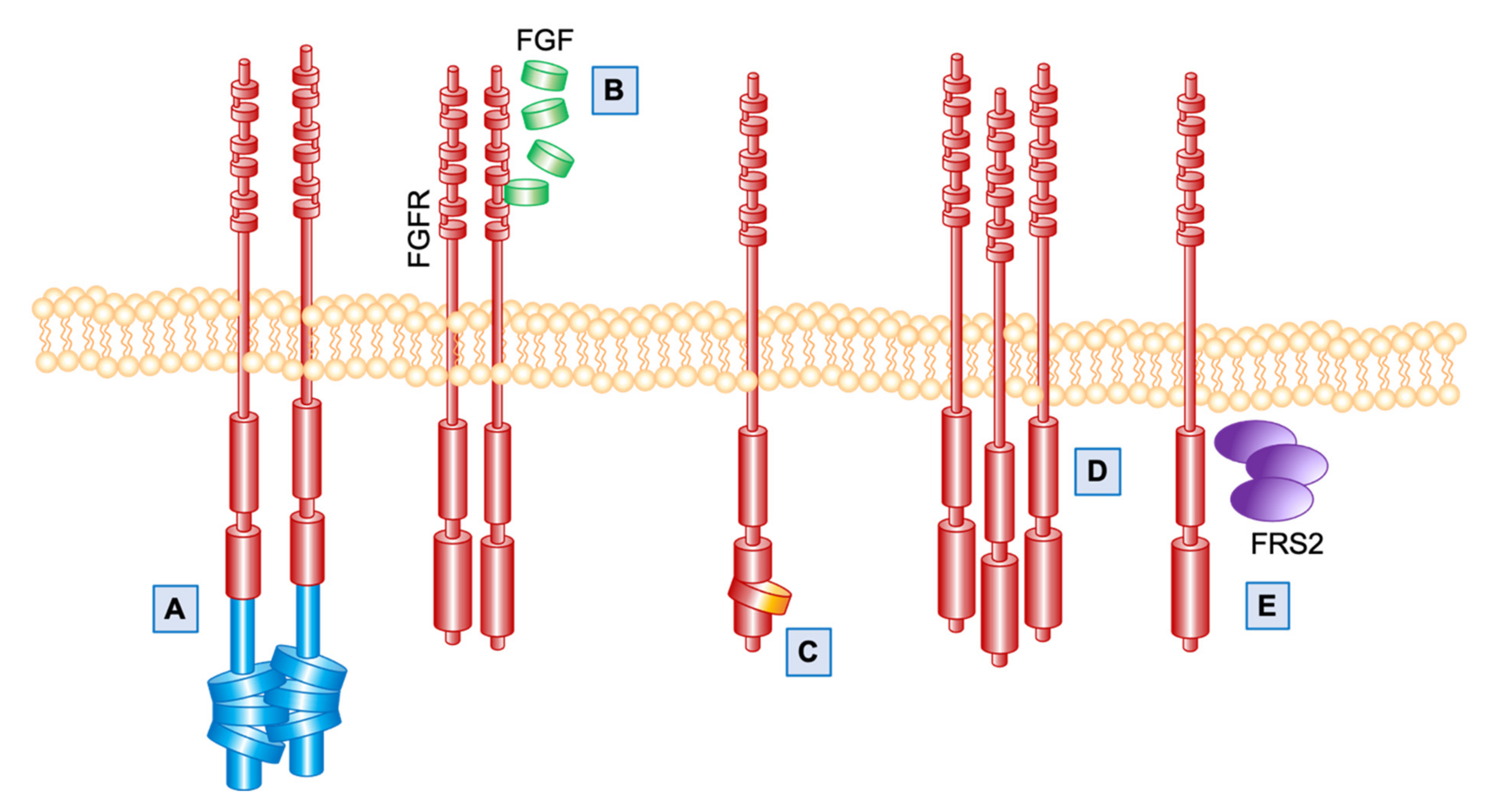
1. Introduction
2. The Roles of FGF/FGFR Pathway in GIST and STS
2.1. Gastrointestinal Stromal Tumors
2.2. Rhabdomyosarcoma
2.3. Liposarcomas
2.4. Phosphaturic Mesenchymal Tumor (PMT)
2.5. Malignant Rhabdoid Tumors
2.6. Other Sarcomas
3. FGFR Inhibition in the Therapy of Gist and STS
3.1. Preclinical Evidence
3.2. Clinical Evidence: Multi-Target TKIs
3.3. Clinical Evidence: Selective FGFR TKIs
3.4. Clinical Evidence: Non-TKI FGFR-Specific Inhibitors
4. Conclusions
Funding
Conflicts of Interest
References
- Antonescu, C.R.; WHO Classification of Tumours Editorial Board. WHO Classification of Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Gamboa, A.C.; Gronchi, A.; Cardona, K. Soft-tissue sarcoma in adults: An update on the current state of histiotype-specific management in an era of personalized medicine. CA Cancer J. Clin. 2020, 70, 200–229. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaf, W.T.A.; Orbach, D.; Judson, I.R.; Ferrari, A. Soft tissue sarcomas in adolescents and young adults: A comparison with their paediatric and adult counterparts. Lancet Oncol. 2017, 18, e166–e175. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Wesche, J.; Haglund, K.; Haugsten, E.M. Fibroblast growth factors and their receptors in cancer. Biochem. J. 2011, 437, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.K.; Xu, H.; Li, H.C.; Goldfarb, M. Broadly expressed SNT-like proteins link FGF receptor stimulation to activators of Ras. Oncogene 1996, 13, 721–729. [Google Scholar]
- de Almeida Carvalho, L.M.; de Oliveira Sapori Avelar, S.; Haslam, A.; Gill, J.; Prasad, V. Estimation of Percentage of Patients with Fibroblast Growth Factor Receptor Alterations Eligible for Off-label Use of Erdafitinib. JAMA Netw. Open 2019, 2, e1916091. [Google Scholar] [CrossRef] [PubMed]
- Shi, E.; Chmielecki, J.; Tang, C.M.; Wang, K.; Heinrich, M.C.; Kang, G.; Corless, C.L.; Hong, D.; Fero, K.E.; Murphy, J.D.; et al. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. J. Transl Med. 2016, 14, 339. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, M.A.; Urbini, M.; Indio, V.; Ravegnini, G.; Nannini, M.; De Luca, M.; Tarantino, G.; Angelini, S.; Gronchi, A.; Vincenzi, B.; et al. Genome-Wide Analysis Identifies MEN1 and MAX Mutations and a Neuroendocrine-Like Molecular Heterogeneity in Quadruple WT GIST. Mol. Cancer Res. 2017, 15, 553–562. [Google Scholar] [CrossRef]
- Urbini, M.; Indio, V.; Tarantino, G.; Ravegnini, G.; Angelini, S.; Nannini, M.; Saponara, M.; Santini, D.; Ceccarelli, C.; Fiorentino, M.; et al. Gain of FGF4 is a frequent event in KIT/PDGFRA/SDH/RAS-P WT GIST. Genes Chromosomes Cancer 2019, 58, 636–642. [Google Scholar] [CrossRef]
- Taylor, J.G.t.; Cheuk, A.T.; Tsang, P.S.; Chung, J.Y.; Song, Y.K.; Desai, K.; Yu, Y.; Chen, Q.R.; Shah, K.; Youngblood, V.; et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Investig. 2009, 119, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Shukla, N.; Ameur, N.; Yilmaz, I.; Nafa, K.; Lau, C.Y.; Marchetti, A.; Borsu, L.; Barr, F.G.; Ladanyi, M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin. Cancer Res. 2012, 18, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014, 4, 216–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Asmann, Y.W.; Erickson-Johnson, M.R.; Oliveira, J.L.; Zhang, H.; Moura, R.D.; Lazar, A.J.; Lev, D.; Bill, K.; Lloyd, R.V.; et al. High-resolution genomic mapping reveals consistent amplification of the fibroblast growth factor receptor substrate 2 gene in well-differentiated and dedifferentiated liposarcoma. Genes Chromosomes Cancer 2011, 50, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chu, K.; Wu, X.; Gao, H.; Wang, J.; Yuan, Y.C.; Loera, S.; Ho, K.; Wang, Y.; Chow, W.; et al. Amplification of FRS2 and activation of FGFR/FRS2 signaling pathway in high-grade liposarcoma. Cancer Res. 2013, 73, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Lan, T.; Chen, H.; Zhang, Z.; Chen, M.; Peng, R.; He, X.; Zhang, H. Amplification of FRS2 in atypical lipomatous tumour/well-differentiated liposarcoma and de-differentiated liposarcoma: A clinicopathological and genetic study of 146 cases. Histopathology 2018, 72, 1145–1155. [Google Scholar] [CrossRef]
- Chudasama, P.; Renner, M.; Straub, M.; Mughal, S.S.; Hutter, B.; Kosaloglu, Z.; Schwessinger, R.; Scheffler, M.; Alldinger, I.; Schimmack, S.; et al. Targeting Fibroblast Growth Factor Receptor 1 for Treatment of Soft-Tissue Sarcoma. Clin. Cancer Res. 2017, 23, 962–973. [Google Scholar] [CrossRef]
- von Mehren, M.; Joensuu, H. Gastrointestinal Stromal Tumors. J. Clin. Oncol. 2018, 36, 136–143. [Google Scholar] [CrossRef]
- De Luca, A.; Esposito Abate, R.; Rachiglio, A.M.; Maiello, M.R.; Esposito, C.; Schettino, C.; Izzo, F.; Nasti, G.; Normanno, N. FGFR Fusions in Cancer: From Diagnostic Approaches to Therapeutic Intervention. Int. J. Mol. Sci. 2020, 21, 6856. [Google Scholar] [CrossRef]
- Reshmi, S.C.; Harvey, R.C.; Roberts, K.G.; Stonerock, E.; Smith, A.; Jenkins, H.; Chen, I.M.; Valentine, M.; Liu, Y.; Li, Y.; et al. Targetable kinase gene fusions in high-risk B-ALL: A study from the Children’s Oncology Group. Blood 2017, 129, 3352–3361. [Google Scholar] [CrossRef]
- Sievers, P.; Stichel, D.; Schrimpf, D.; Sahm, F.; Koelsche, C.; Reuss, D.E.; Wefers, A.K.; Reinhardt, A.; Huang, K.; Ebrahimi, A.; et al. FGFR1:TACC1 fusion is a frequent event in molecularly defined extraventricular neurocytoma. Acta Neuropathol. 2018, 136, 293–302. [Google Scholar] [CrossRef]
- Singh, D.; Chan, J.M.; Zoppoli, P.; Niola, F.; Sullivan, R.; Castano, A.; Liu, E.M.; Reichel, J.; Porrati, P.; Pellegatta, S.; et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012, 337, 1231–1235. [Google Scholar] [CrossRef]
- Devereaux, K.A.; Weiel, J.J.; Mills, A.M.; Kunder, C.A.; Longacre, T.A. Neurofibrosarcoma Revisited: An Institutional Case Series of Uterine Sarcomas Harboring Kinase-related Fusions With Report of a Novel FGFR1-TACC1 Fusion. Am. J. Surg. Pathol. 2021, 45, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Killian, J.K.; Kim, S.Y.; Miettinen, M.; Smith, C.; Merino, M.; Tsokos, M.; Quezado, M.; Smith, W.I., Jr.; Jahromi, M.S.; Xekouki, P.; et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013, 3, 648–657. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Drier, Y.; Johnstone, S.E.; Hemming, M.L.; Tarjan, D.R.; Hegazi, E.; Shareef, S.J.; Javed, N.M.; Raut, C.P.; Eschle, B.K.; et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 2019, 575, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Mazzocca, A.; Napolitano, A.; Silletta, M.; Spalato Ceruso, M.; Santini, D.; Tonini, G.; Vincenzi, B. New frontiers in the medical management of gastrointestinal stromal tumours. Ther. Adv. Med. Oncol. 2019, 11, 1758835919841946. [Google Scholar] [CrossRef]
- Javidi-Sharifi, N.; Traer, E.; Martinez, J.; Gupta, A.; Taguchi, T.; Dunlap, J.; Heinrich, M.C.; Corless, C.L.; Rubin, B.P.; Druker, B.J.; et al. Crosstalk between KIT and FGFR3 Promotes Gastrointestinal Stromal Tumor Cell Growth and Drug Resistance. Cancer Res. 2015, 75, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Huynh, H.; Li, X.; Ruddy, D.A.; Wang, Y.; Ong, R.; Chow, P.; Qiu, S.; Tam, A.; Rakiec, D.P.; et al. FGFR-Mediated Reactivation of MAPK Signaling Attenuates Antitumor Effects of Imatinib in Gastrointestinal Stromal Tumors. Cancer Discov. 2015, 5, 438–451. [Google Scholar] [CrossRef]
- Boichuk, S.; Galembikova, A.; Dunaev, P.; Valeeva, E.; Shagimardanova, E.; Gusev, O.; Khaiboullina, S. A Novel Receptor Tyrosine Kinase Switch Promotes Gastrointestinal Stromal Tumor Drug Resistance. Molecules 2017, 22, 2152. [Google Scholar] [CrossRef]
- Paulson, V.; Chandler, G.; Rakheja, D.; Galindo, R.L.; Wilson, K.; Amatruda, J.F.; Cameron, S. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes Chromosomes Cancer 2011, 50, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Crose, L.E.; Etheridge, K.T.; Chen, C.; Belyea, B.; Talbot, L.J.; Bentley, R.C.; Linardic, C.M. FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clin. Cancer Res. 2012, 18, 3780–3790. [Google Scholar] [CrossRef]
- Davicioni, E.; Finckenstein, F.G.; Shahbazian, V.; Buckley, J.D.; Triche, T.J.; Anderson, M.J. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006, 66, 6936–6946. [Google Scholar] [CrossRef]
- Cao, L.; Yu, Y.; Bilke, S.; Walker, R.L.; Mayeenuddin, L.H.; Azorsa, D.O.; Yang, F.; Pineda, M.; Helman, L.J.; Meltzer, P.S. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010, 70, 6497–6508. [Google Scholar] [CrossRef] [PubMed]
- Wachtel, M.; Rakic, J.; Okoniewski, M.; Bode, P.; Niggli, F.; Schafer, B.W. FGFR4 signaling couples to Bim and not Bmf to discriminate subsets of alveolar rhabdomyosarcoma cells. Int. J. Cancer 2014, 135, 1543–1552. [Google Scholar] [CrossRef]
- Liu, J.; Guzman, M.A.; Pezanowski, D.; Patel, D.; Hauptman, J.; Keisling, M.; Hou, S.J.; Papenhausen, P.R.; Pascasio, J.M.; Punnett, H.H.; et al. FOXO1-FGFR1 fusion and amplification in a solid variant of alveolar rhabdomyosarcoma. Mod. Pathol. 2011, 24, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, M.; Setoguchi, T.; Matsunoshita, Y.; Sasaki, H.; Nagao, H.; Gao, H.; Sugimura, K.; Komiya, S. Tumour formation by single fibroblast growth factor receptor 3-positive rhabdomyosarcoma-initiating cells. Br. J. Cancer 2009, 101, 2030–2037. [Google Scholar] [CrossRef]
- Lee, A.T.J.; Thway, K.; Huang, P.H.; Jones, R.L. Clinical and Molecular Spectrum of Liposarcoma. J. Clin. Oncol 2018, 36, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shen, Y.; Ren, Y.; Liu, W.; Li, M.; Liang, W.; Liu, C.; Li, F. Oncogene mutation profiling reveals poor prognosis associated with FGFR1/3 mutation in liposarcoma. Hum. Pathol. 2016, 55, 143–150. [Google Scholar] [CrossRef]
- Dadone-Montaudie, B.; Laroche-Clary, A.; Mongis, A.; Chamorey, E.; Mauro, I.D.; Chaire, V.; Finetti, P.; Schiappa, R.; Le Loarer, F.; Birtwisle-Peyrottes, I.; et al. Novel Therapeutic Insights in Dedifferentiated Liposarcoma: A Role for FGFR and MDM2 Dual Targeting. Cancers 2020, 12, 3058. [Google Scholar] [CrossRef] [PubMed]
- Kunstlinger, H.; Fassunke, J.; Schildhaus, H.U.; Brors, B.; Heydt, C.; Ihle, M.A.; Mechtersheimer, G.; Wardelmann, E.; Buttner, R.; Merkelbach-Bruse, S. FGFR2 is overexpressed in myxoid liposarcoma and inhibition of FGFR signaling impairs tumor growth in vitro. Oncotarget 2015, 6, 20215–20230. [Google Scholar] [CrossRef][Green Version]
- Folpe, A.L. Phosphaturic mesenchymal tumors: A review and update. Semin. Diagn. Pathol. 2019, 36, 260–268. [Google Scholar] [CrossRef]
- Lee, J.C.; Jeng, Y.M.; Su, S.Y.; Wu, C.T.; Tsai, K.S.; Lee, C.H.; Lin, C.Y.; Carter, J.M.; Huang, J.W.; Chen, S.H.; et al. Identification of a novel FN1-FGFR1 genetic fusion as a frequent event in phosphaturic mesenchymal tumour. J. Pathol. 2015, 235, 539–545. [Google Scholar] [CrossRef]
- Lee, J.C.; Su, S.Y.; Changou, C.A.; Yang, R.S.; Tsai, K.S.; Collins, M.T.; Orwoll, E.S.; Lin, C.Y.; Chen, S.H.; Shih, S.R.; et al. Characterization of FN1-FGFR1 and novel FN1-FGF1 fusion genes in a large series of phosphaturic mesenchymal tumors. Mod. Pathol. 2016, 29, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Wohrle, S.; Weiss, A.; Ito, M.; Kauffmann, A.; Murakami, M.; Jagani, Z.; Thuery, A.; Bauer-Probst, B.; Reimann, F.; Stamm, C.; et al. Fibroblast growth factor receptors as novel therapeutic targets in SNF5-deleted malignant rhabdoid tumors. PLoS ONE 2013, 8, e77652. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.P.; Todd, J.R.; Finetti, M.A.; McCarthy, F.; Broncel, M.; Vyse, S.; Luczynski, M.T.; Crosier, S.; Ryall, K.A.; Holmes, K.; et al. Dual Targeting of PDGFRalpha and FGFR1 Displays Synergistic Efficacy in Malignant Rhabdoid Tumors. Cell Rep. 2016, 17, 1265–1275. [Google Scholar] [CrossRef]
- Chauvin, C.; Leruste, A.; Tauziede-Espariat, A.; Andrianteranagna, M.; Surdez, D.; Lescure, A.; Han, Z.Y.; Anthony, E.; Richer, W.; Baulande, S.; et al. High-Throughput Drug Screening Identifies Pazopanib and Clofilium Tosylate as Promising Treatments for Malignant Rhabdoid Tumors. Cell Rep. 2017, 21, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Agelopoulos, K.; Richter, G.H.; Schmidt, E.; Dirksen, U.; von Heyking, K.; Moser, B.; Klein, H.U.; Kontny, U.; Dugas, M.; Poos, K.; et al. Deep Sequencing in Conjunction with Expression and Functional Analyses Reveals Activation of FGFR1 in Ewing Sarcoma. Clin. Cancer Res. 2015, 21, 4935–4946. [Google Scholar] [CrossRef]
- Toulmonde, M.; Lucchesi, C.; Verbeke, S.; Crombe, A.; Adam, J.; Geneste, D.; Chaire, V.; Laroche-Clary, A.; Perret, R.; Bertucci, F.; et al. High throughput profiling of undifferentiated pleomorphic sarcomas identifies two main subgroups with distinct immune profile, clinical outcome and sensitivity to targeted therapies. EBioMedicine 2020, 62, 103131. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Jo, V.Y.; Mills, A.M.; Zhu, S.X.; Lee, C.H.; Espinosa, I.; Nucci, M.R.; Varma, S.; Forgo, E.; Hastie, T.; et al. Clinically Relevant Molecular Subtypes in Leiomyosarcoma. Clin. Cancer Res. 2015, 21, 3501–3511. [Google Scholar] [CrossRef]
- Ishibe, T.; Nakayama, T.; Okamoto, T.; Aoyama, T.; Nishijo, K.; Shibata, K.R.; Shima, Y.; Nagayama, S.; Katagiri, T.; Nakamura, Y.; et al. Disruption of fibroblast growth factor signal pathway inhibits the growth of synovial sarcomas: Potential application of signal inhibitors to molecular target therapy. Clin. Cancer Res. 2005, 11, 2702–2712. [Google Scholar] [CrossRef][Green Version]
- Katoh, M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 2019, 16, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Q.; Cheuk, A.T.; Shern, J.F.; Song, Y.K.; Hurd, L.; Liao, H.; Wei, J.S.; Khan, J. Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534). PLoS ONE 2013, 8, e76551. [Google Scholar] [CrossRef]
- Hanes, R.; Grad, I.; Lorenz, S.; Stratford, E.W.; Munthe, E.; Reddy, C.C.; Meza-Zepeda, L.A.; Myklebost, O. Preclinical evaluation of potential therapeutic targets in dedifferentiated liposarcoma. Oncotarget 2016, 7, 54583–54595. [Google Scholar] [CrossRef]
- Hanes, R.; Munthe, E.; Grad, I.; Han, J.; Karlsen, I.; McCormack, E.; Meza-Zepeda, L.A.; Stratford, E.W.; Myklebost, O. Preclinical Evaluation of the Pan-FGFR Inhibitor LY2874455 in FRS2-Amplified Liposarcoma. Cells 2019, 8, 189. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.S.; Shim, J.J.; Lee, J.; Kim, A.Y.; Kim, J. Critical role of the fibroblast growth factor signalling pathway in Ewing’s sarcoma octamer-binding transcription factor 4-mediated cell proliferation and tumorigenesis. FEBS J. 2019, 286, 4443–4472. [Google Scholar] [CrossRef]
- Phanhthilath, N.; Hakim, S.; Su, C.; Liu, A.; Subramonian, D.; Lesperance, J.; Zage, P.E. Mechanisms of Efficacy of the FGFR1-3 Inhibitor AZD4547 in Pediatric Solid Tumor Models. Investig. New Drugs 2020, 38, 1677–1686. [Google Scholar] [CrossRef]
- Boichuk, S.; Galembikova, A.; Dunaev, P.; Micheeva, E.; Valeeva, E.; Novikova, M.; Khromova, N.; Kopnin, P. Targeting of FGF-Signaling Re-Sensitizes Gastrointestinal Stromal Tumors (GIST) to Imatinib In Vitro and In Vivo. Molecules 2018, 23, 2643. [Google Scholar] [CrossRef]
- Boichuk, S.; Dunaev, P.; Galembikova, A.; Mustafin, I.; Valeeva, E. Inhibition of fibroblast growth factor receptor-signaling sensitizes imatinib-resistant gastrointestinal stromal tumors to low doses of topoisomerase II inhibitors. Anticancer Drugs 2018, 29, 549–559. [Google Scholar] [CrossRef]
- Sergei, B.; Pavel, D.; Aigul, G.; Firyuza, B.; Ilmira, N.; Ilshat, M.; Aida, A.; Refat, K.; Natalia, A.; Elena, S.; et al. Inhibition of FGFR2-Signaling Attenuates a Homology-Mediated DNA Repair in GIST and Sensitizes Them to DNA-Topoisomerase II Inhibitors. Int. J. Mol. Sci. 2020, 21, 352. [Google Scholar] [CrossRef]
- Darvishi, E.; Slemmons, K.; Wan, Z.; Mitra, S.; Hou, X.; Hugues Parmentier, J.; Eddie Loh, Y.H.; Helman, L.J. Molecular mechanisms of Guadecitabine induced FGFR4 down regulation in alveolar rhabdomyosarcomas. Neoplasia 2020, 22, 274–282. [Google Scholar] [CrossRef]
- McKinnon, T.; Venier, R.; Yohe, M.; Sindiri, S.; Gryder, B.E.; Shern, J.F.; Kabaroff, L.; Dickson, B.; Schleicher, K.; Chouinard-Pelletier, G.; et al. Functional screening of FGFR4-driven tumorigenesis identifies PI3K/mTOR inhibition as a therapeutic strategy in rhabdomyosarcoma. Oncogene 2018, 37, 2630–2644. [Google Scholar] [CrossRef]
- Presta, M.; Foglio, E.; Churruca Schuind, A.; Ronca, R. Long Pentraxin-3 Modulates the Angiogenic Activity of Fibroblast Growth Factor-2. Front. Immunol. 2018, 9, 2327. [Google Scholar] [CrossRef] [PubMed]
- Presta, M.; Chiodelli, P.; Giacomini, A.; Rusnati, M.; Ronca, R. Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacol. Ther. 2017, 179, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.F.; Matarazzo, S.; Maccarinelli, F.; Foglio, E.; Giacomini, A.; Silva Nunes, J.P.; Presta, M.; Dias, A.A.M.; Ronca, R. Long Pentraxin 3-Mediated Fibroblast Growth Factor Trapping Impairs Fibrosarcoma Growth. Front. Oncol. 2018, 8, 472. [Google Scholar] [CrossRef] [PubMed]
- Alijaj, N.; Moutel, S.; Gouveia, Z.L.; Gray, M.; Roveri, M.; Dzhumashev, D.; Weber, F.; Meier, G.; Luciani, P.; Rossler, J.K.; et al. Novel FGFR4-Targeting Single-Domain Antibodies for Multiple Targeted Therapies against Rhabdomyosarcoma. Cancers 2020, 12, 3313. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.T.J.; Jones, R.L.; Huang, P.H. Pazopanib in advanced soft tissue sarcomas. Signal. Transduct. Target. Ther 2019, 4, 16. [Google Scholar] [CrossRef]
- Schutz, F.A.; Choueiri, T.K.; Sternberg, C.N. Pazopanib: Clinical development of a potent anti-angiogenic drug. Crit. Rev. Oncol. Hematol. 2011, 77, 163–171. [Google Scholar] [CrossRef] [PubMed]
- van der Graaf, W.T.; Blay, J.Y.; Chawla, S.P.; Kim, D.W.; Bui-Nguyen, B.; Casali, P.G.; Schoffski, P.; Aglietta, M.; Staddon, A.P.; Beppu, Y.; et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2012, 379, 1879–1886. [Google Scholar] [CrossRef]
- Martin-Broto, J.; Stacchiotti, S.; Lopez-Pousa, A.; Redondo, A.; Bernabeu, D.; de Alava, E.; Casali, P.G.; Italiano, A.; Gutierrez, A.; Moura, D.S.; et al. Pazopanib for treatment of advanced malignant and dedifferentiated solitary fibrous tumour: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2019, 20, 134–144. [Google Scholar] [CrossRef]
- Martin-Broto, J.; Cruz, J.; Penel, N.; Le Cesne, A.; Hindi, N.; Luna, P.; Moura, D.S.; Bernabeu, D.; de Alava, E.; Lopez-Guerrero, J.A.; et al. Pazopanib for treatment of typical solitary fibrous tumours: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2020, 21, 456–466. [Google Scholar] [CrossRef]
- Stacchiotti, S.; Ferrari, S.; Redondo, A.; Hindi, N.; Palmerini, E.; Vaz Salgado, M.A.; Frezza, A.M.; Casali, P.G.; Gutierrez, A.; Lopez-Pousa, A.; et al. Pazopanib for treatment of advanced extraskeletal myxoid chondrosarcoma: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2019, 20, 1252–1262. [Google Scholar] [CrossRef]
- Wilding, C.P.; Elms, M.L.; Judson, I.; Tan, A.C.; Jones, R.L.; Huang, P.H. The landscape of tyrosine kinase inhibitors in sarcomas: Looking beyond pazopanib. Expert. Rev. Anticancer Ther. 2019, 19, 971–991. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.L.; Ratain, M.J.; O’Dwyer, P.J.; Siu, L.L.; Jassem, J.; Medioni, J.; DeJonge, M.; Rudin, C.; Sawyer, M.; Khayat, D.; et al. Phase II randomised discontinuation trial of brivanib in patients with advanced solid tumours. Eur. J. Cancer 2019, 120, 132–139. [Google Scholar] [CrossRef]
- Rodon, J.; Postel-Vinay, S.; Hollebecque, A.; Nuciforo, P.; Azaro, A.; Cattan, V.; Marfai, L.; Sudey, I.; Brendel, K.; Delmas, A.; et al. First-in-human phase I study of oral S49076, a unique MET/AXL/FGFR inhibitor, in advanced solid tumours. Eur. J. Cancer 2017, 81, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Hilberg, F.; Tontsch-Grunt, U.; Baum, A.; Le, A.T.; Doebele, R.C.; Lieb, S.; Gianni, D.; Voss, T.; Garin-Chesa, P.; Haslinger, C.; et al. Triple Angiokinase Inhibitor Nintedanib Directly Inhibits Tumor Cell Growth and Induces Tumor Shrinkage via Blocking Oncogenic Receptor Tyrosine Kinases. J. Pharmacol. Exp. Ther. 2018, 364, 494–503. [Google Scholar] [CrossRef]
- Schoffski, P.; Toulmonde, M.; Estival, A.; Marquina, G.; Dudzisz-Sledz, M.; Brahmi, M.; Steeghs, N.; Karavasilis, V.; de Haan, J.; Wozniak, A.; et al. Randomised phase 2 study comparing the efficacy and safety of the oral tyrosine kinase inhibitor nintedanib with single agent ifosfamide in patients with advanced, inoperable, metastatic soft tissue sarcoma after failure of first-line chemotherapy: EORTC-1506-STBSG “ANITA”. Eur. J. Cancer 2021, 152, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Song, X.; Yang, D.; Bai, D.; Yao, Y.; Lu, N. Anlotinib inhibits angiogenesis via suppressing the activation of VEGFR2, PDGFRbeta and FGFR1. Gene 2018, 654, 77–86. [Google Scholar] [CrossRef]
- Tohyama, O.; Matsui, J.; Kodama, K.; Hata-Sugi, N.; Kimura, T.; Okamoto, K.; Minoshima, Y.; Iwata, M.; Funahashi, Y. Antitumor activity of lenvatinib (e7080): An angiogenesis inhibitor that targets multiple receptor tyrosine kinases in preclinical human thyroid cancer models. J. Thyroid. Res. 2014, 2014, 638747. [Google Scholar] [CrossRef]
- Porta, C.; Giglione, P.; Liguigli, W.; Paglino, C. Dovitinib (CHIR258, TKI258): Structure, development and preclinical and clinical activity. Future Oncol. 2015, 11, 39–50. [Google Scholar] [CrossRef]
- Joensuu, H.; Blay, J.Y.; Comandone, A.; Martin-Broto, J.; Fumagalli, E.; Grignani, G.; Del Muro, X.G.; Adenis, A.; Valverde, C.; Pousa, A.L.; et al. Dovitinib in patients with gastrointestinal stromal tumour refractory and/or intolerant to imatinib. Br. J. Cancer 2017, 117, 1278–1285. [Google Scholar] [CrossRef]
- Heinrich, M.C.; Mehren, M.v.; Demetri, G.D.; Fletcher, J.A.; Sun, J.G.; Kerstein, D.; Conlan, M.G.; George, S. Ponatinib efficacy and safety in patients (pts) with advanced gastrointestinal stromal tumors (GIST) after tyrosine kinase inhibitor (TKI) failure: Results from a phase 2 study. J. Clin. Oncol. 2015, 33, 10535. [Google Scholar] [CrossRef]
- Weaver, A.; Bossaer, J.B. Fibroblast growth factor receptor (FGFR) inhibitors: A review of a novel therapeutic class. J. Oncol. Pharm. Pract. 2021, 27, 702–710. [Google Scholar] [CrossRef]
- European Medicines Agency. Available online: https://www.ema.europa.eu/en/documents/smop-initial/chmp-summary-positive-opinion-pemazyre_en.pdf (accessed on 1 May 2021).
- Dai, S.; Zhou, Z.; Chen, Z.; Xu, G.; Chen, Y. Fibroblast Growth Factor Receptors (FGFRs): Structures and Small Molecule Inhibitors. Cells 2019, 8, 614. [Google Scholar] [CrossRef] [PubMed]
- Bahleda, R.; Italiano, A.; Hierro, C.; Mita, A.; Cervantes, A.; Chan, N.; Awad, M.; Calvo, E.; Moreno, V.; Govindan, R.; et al. Multicenter Phase I Study of Erdafitinib (JNJ-42756493), Oral Pan-Fibroblast Growth Factor Receptor Inhibitor, in Patients with Advanced or Refractory Solid Tumors. Clin. Cancer Res. 2019, 25, 4888–4897. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.; Cho, B.C.; Sayehli, C.M.; Navarro, A.; Soo, R.A.; Richly, H.; Cassier, P.A.; Tai, D.; Penel, N.; Nogova, L.; et al. Rogaratinib in patients with advanced cancers selected by FGFR mRNA expression: A phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019, 20, 1454–1466. [Google Scholar] [CrossRef]
- Voss, M.H.; Hierro, C.; Heist, R.S.; Cleary, J.M.; Meric-Bernstam, F.; Tabernero, J.; Janku, F.; Gandhi, L.; Iafrate, A.J.; Borger, D.R.; et al. A Phase I, Open-Label, Multicenter, Dose-escalation Study of the Oral Selective FGFR Inhibitor Debio 1347 in Patients with Advanced Solid Tumors Harboring FGFR Gene Alterations. Clin. Cancer Res. 2019, 25, 2699–2707. [Google Scholar] [CrossRef] [PubMed]
- Bahleda, R.; Meric-Bernstam, F.; Goyal, L.; Tran, B.; He, Y.; Yamamiya, I.; Benhadji, K.A.; Matos, I.; Arkenau, H.T. Phase I, first-in-human study of futibatinib, a highly selective, irreversible FGFR1-4 inhibitor in patients with advanced solid tumors. Ann. Oncol. 2020, 31, 1405–1412. [Google Scholar] [CrossRef]
- Yamamoto, N.; Ryoo, B.Y.; Keam, B.; Kudo, M.; Lin, C.C.; Kunieda, F.; Ball, H.A.; Moran, D.; Komatsu, K.; Takeda, K.; et al. A phase 1 study of oral ASP5878, a selective small-molecule inhibitor of fibroblast growth factor receptors 1-4, as a single dose and multiple doses in patients with solid malignancies. Investig. New Drugs 2020, 38, 445–456. [Google Scholar] [CrossRef]
- Kelly, C.M.; Shoushtari, A.N.; Qin, L.X.; D’Angelo, S.P.; Dickson, M.A.; Gounder, M.M.; Keohan, M.L.; McFadyen, C.; Sjoberg, A.; Singer, S.; et al. A phase Ib study of BGJ398, a pan-FGFR kinase inhibitor in combination with imatinib in patients with advanced gastrointestinal stromal tumor. Investig. New Drugs 2019, 37, 282–290. [Google Scholar] [CrossRef]
- Hartley, I.R.; Miller, C.B.; Papadakis, G.Z.; Bergwitz, C.; Del Rivero, J.; Blau, J.E.; Florenzano, P.; Berglund, J.A.; Tassone, J.; Roszko, K.L.; et al. Targeted FGFR Blockade for the Treatment of Tumor-Induced Osteomalacia. N. Engl. J. Med. 2020, 383, 1387–1389. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Papadopoulos, K.P.; Patnaik, A.; Wilson, K.; Thayer, S.; Zanghi, J.; Gemo, A.T.; Kavanaugh, W.M.; Keer, H.N.; LoRusso, P.M. A phase I, first in human study of FP-1039 (GSK3052230), a novel FGF ligand trap, in patients with advanced solid tumors. Ann. Oncol. 2016, 27, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Meric-Bernstam, F.; Kalyan, A.; Babich, A.; Liu, R.; Tanigawa, T.; Sommer, A.; Osada, M.; Reetz, F.; Laurent, D.; et al. First-in-Human Phase I Study of Aprutumab Ixadotin, a Fibroblast Growth Factor Receptor 2 Antibody-Drug Conjugate (BAY 1187982) in Patients with Advanced Cancer. Target. Oncol. 2019, 14, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, H.; Roberts, P.J.; Sarlomo-Rikala, M.; Andersson, L.C.; Tervahartiala, P.; Tuveson, D.; Silberman, S.; Capdeville, R.; Dimitrijevic, S.; Druker, B.; et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N. Engl. J. Med. 2001, 344, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Horak, P.; Frohling, S.; Glimm, H. Integrating next-generation sequencing into clinical oncology: Strategies, promises and pitfalls. ESMO Open 2016, 1, e000094. [Google Scholar] [CrossRef]
- Babina, I.S.; Turner, N.C. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 2017, 17, 318–332. [Google Scholar] [CrossRef]
- Krook, M.A.; Reeser, J.W.; Ernst, G.; Barker, H.; Wilberding, M.; Li, G.; Chen, H.Z.; Roychowdhury, S. Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br. J. Cancer 2021, 124, 880–892. [Google Scholar] [CrossRef]
| Disease | Alterations | Estimated Frequency | Ref. |
|---|---|---|---|
| GIST | Translocation, duplication, activating mutations | ~1–2% | [9,10,11] |
| Rhabdomyosarcoma | Activating mutations | ~7–8% | [12,13,14] |
| Dedifferentiated liposarcoma | Amplification, activating mutations | >90% | [15,16,17,18] |
| Undifferentiated pleomorphic sarcoma | Amplification | 8–16% | [18] |
| Leiomyosarcoma | Amplification | ~30% | [18] |
| Malignant peripheral nerve sheath tumors | Amplification | 11–20% | [18] |
| Drug | FGFR Selectivity | Phase | Sarcoma Subtypes Enrolled | Best Response of Sarcoma pts. | National Clinical Trial (NCT) Number | Ref. |
|---|---|---|---|---|---|---|
| Erdafinitib | 1–4 | I | Any subtype | N/A | NCT01703481 | [86] |
| Debio1347 | 1–3 | I | Any subtype harboring FGFR gene alterations | 1 PD | NCT01948297 | [88] |
| Futibatinib | 1–4 | I | Any subtype harboring FGFR gene alterations | 1 SD, 1 PD | NCT02052778 | [89] |
| ASP5878 | 1–4 | I | Any subtype | N/A | NCT02038673 | [90] |
| Infigratinib + imatinib | 1–3 | Ib | Imatinib refractory advanced GIST | 3/12 SD | NCT02257541 | [91] |
| Infigratinib | 1–3 | II | Any subtype associated to TIO | N/A | NCT03510455 | |
| Rogaratinib | 1–4 | II | Any subtype with FGFR1-4 gene alterations + advanced SDH-deficient GIST | Currently enrolling | NCT04595747 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Napolitano, A.; Ostler, A.E.; Jones, R.L.; Huang, P.H. Fibroblast Growth Factor Receptor (FGFR) Signaling in GIST and Soft Tissue Sarcomas. Cells 2021, 10, 1533. https://doi.org/10.3390/cells10061533
Napolitano A, Ostler AE, Jones RL, Huang PH. Fibroblast Growth Factor Receptor (FGFR) Signaling in GIST and Soft Tissue Sarcomas. Cells. 2021; 10(6):1533. https://doi.org/10.3390/cells10061533
Chicago/Turabian StyleNapolitano, Andrea, Alexandra E. Ostler, Robin L. Jones, and Paul H. Huang. 2021. "Fibroblast Growth Factor Receptor (FGFR) Signaling in GIST and Soft Tissue Sarcomas" Cells 10, no. 6: 1533. https://doi.org/10.3390/cells10061533
APA StyleNapolitano, A., Ostler, A. E., Jones, R. L., & Huang, P. H. (2021). Fibroblast Growth Factor Receptor (FGFR) Signaling in GIST and Soft Tissue Sarcomas. Cells, 10(6), 1533. https://doi.org/10.3390/cells10061533







