The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Sampling, Chromosomal Preparations, and Genomic DNA Isolation
2.3. Genome Sequencing and Bioinformatic Analyses
2.4. Fluorescent In Situ Hybridization (FISH)
3. Results
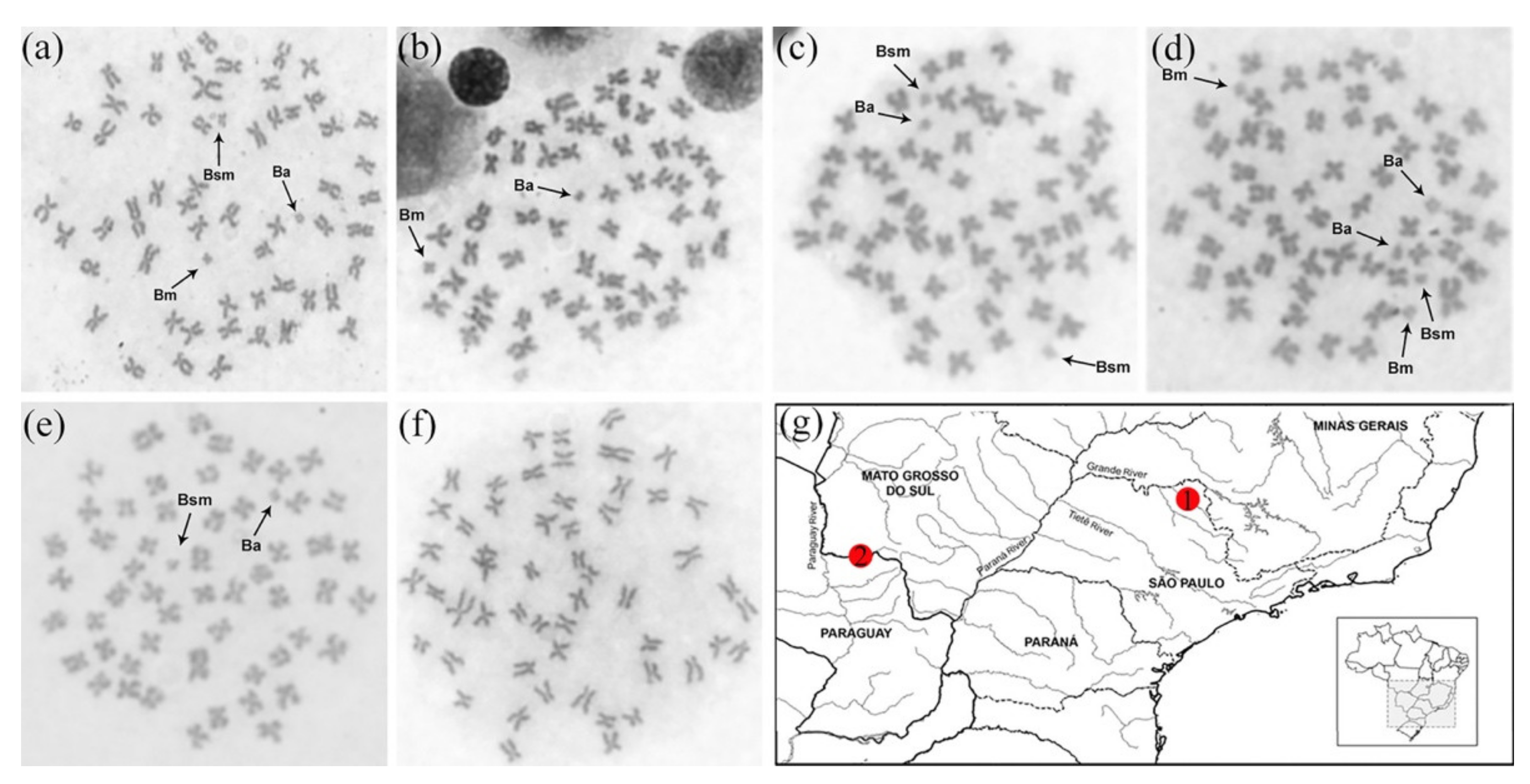
3.1. Karyotypes and the First Description of a Population without B Chromosomes
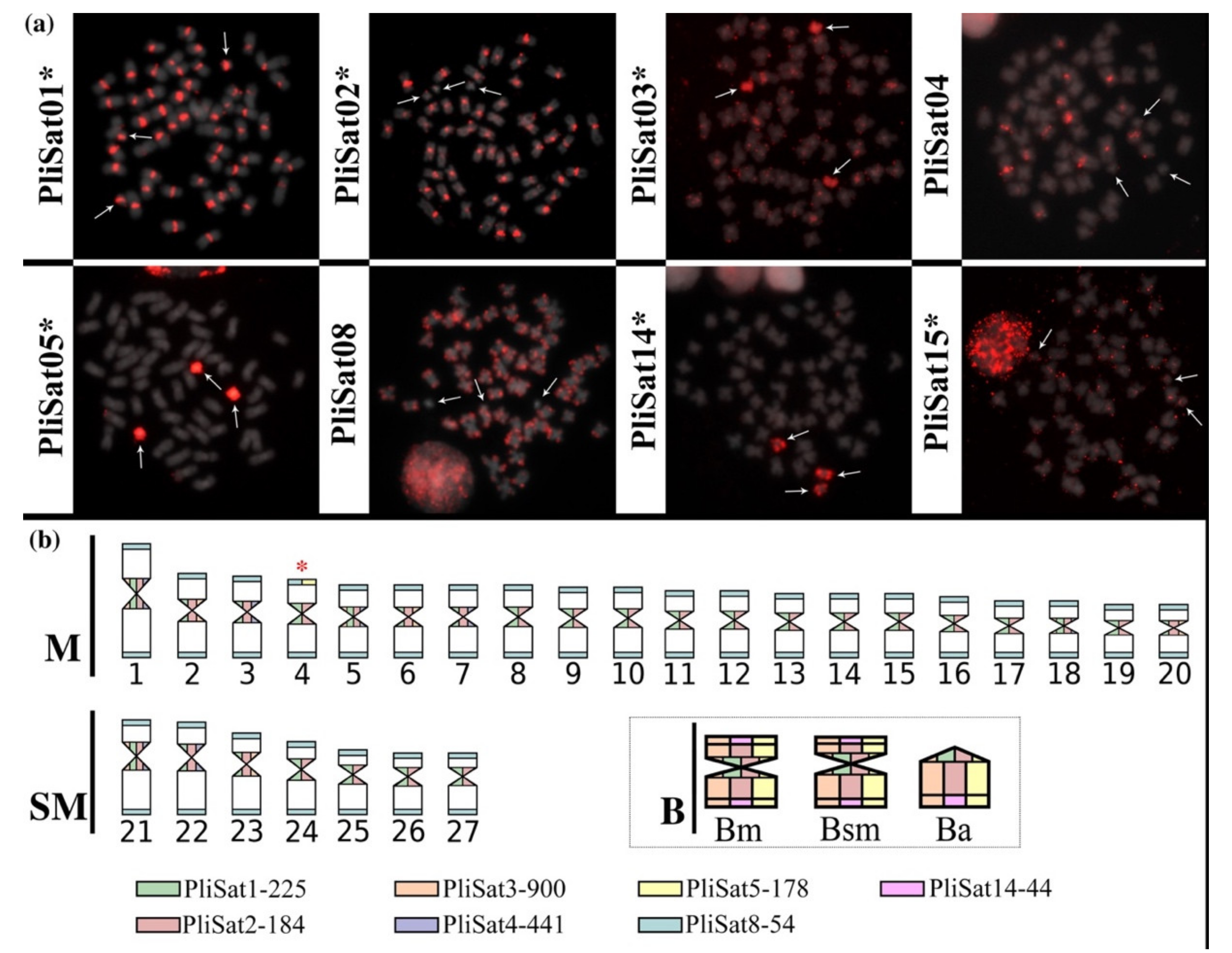
3.2. The B chromosomes of P. lineatus Are Highly-Enriched in Satellite DNAs
3.3. P. lineatus and M. macrocephalus Share Several satDNAs
4. Discussion
4.1. The Karyotypes of P. lineatus Populations
4.2. The Satellitome of Prochilodus lineatus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, P. Repetitive DNA in eukaryotic genomes. Chromosom. Res. 2015, 23, 415–420. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Green, D.M. Muller’s ratchet and the evolution of supernumerary chromosomes. Genome 1990, 33, 818–824. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Sharbel, T.F.; Beukeboom, L.W. B-chromosome evolution. Philos. Trans. R. Soc. B 2000, 355, 163–178. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B.; Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 2005, 95, 118–128. [Google Scholar] [CrossRef]
- Houben, A. B chromosomes—A matter of chromosome drive. Front. Plant Sci. 2017, 8, 1–6. [Google Scholar] [CrossRef]
- López-León, M.D.; Neves, N.; Schwarzacher, T.; Heslop-Harrison, J.S.; Hewitt, G.M.; Camacho, J.P.M. Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosom. Res. 1994, 2, 87–92. [Google Scholar] [CrossRef]
- Mestriner, C.A.; Galetti, P.M.; Valentini, S.R.; Ruiz, I.R.G.; Abel, L.D.S.; Moreira-Filho, O.; Camacho, J.P.M. Structural and functional evidence that a B chromosome in the characid fish Astyanax scabripinnis is an isochromosome. Heredity 2000, 85, 1–9. [Google Scholar] [CrossRef]
- Artoni, R.F.; Vicari, M.R.; Endler, A.L.; Cavallaro, Z.I.; Jesus, C.M.; De Almeida, M.C.; Moreira-Filho, O.; Bertollo, L.A.C. Banding pattern of A and B chromosomes of Prochilodus lineatus (Characiformes, Prochilodontidae), with comments on B chromosomes evolution. Genetica 2006, 127, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Cabrero, J.; Perfectti, F.; Camacho, J.P.M. B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma 2010, 119, 217–225. [Google Scholar] [CrossRef]
- Silva, D.M.Z.A.; Pansonato-Alves, J.C.; Utsunomia, R.; Araya-Jaime, C.; Ruiz-Ruano, F.J.; Daniel, S.N.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Porto-Foresti, F.; et al. Delimiting the origin of a B chromosome by FISH mapping, chromosome painting and DNA sequence analysis in Astyanax paranae (Teleostei, Characiformes). PLoS ONE 2014, 9, e94896. [Google Scholar]
- Utsunomia, R.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Araya-Jaime, C.; Pansonato-Alves, J.C.; Scacchetti, P.C.; Hashimoto, D.T.; Oliveira, C.; Trifonov, V.A.; Porto-Foresti, F.; et al. Uncovering the ancestry of B chromosomes in Moenkhausia sanctaefilomenae (Teleostei, Characidae). PLoS ONE 2016, 11, e0150573. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Freitas, E.A.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Utsunomia, R.; Araya-Jaime, C.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. Satellite DNA content of B chromosomes in the characid fish Characidium gomesi supports their origin from sex chromosomes. Mol. Genet. Genom. 2020, 295, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Maistro, E.L.; Oliveira, C.; Foresti, F. Cytogenetic analysis of A- and B-chromosomes of Prochilodus lineatus (Teleostei, Prochilodontidae) using different restriction enzyme banding and staining methods. Genetica 2000, 108, 119–125. [Google Scholar] [CrossRef]
- Voltolin, T.A.; Laudicina, A.; Senhorini, J.A.; Bortolozzi, J.; Oliveira, C.; Foresti, F.; Porto-Foresti, F. Origin and molecular organization of supernumerary chromosomes of Prochilodus lineatus (Characiformes, Prochilodontidae) obtained by DNA probes. Genetica 2010, 138, 1133–1139. [Google Scholar] [CrossRef]
- Penitente, M.; Voltolin, T.A.; Senhorini, J.A.; Bortolozzi, J.; Foresti, F.; Porto-Foresti, F. Transmission rate variation among three B chromosome variants in the fish Prochilodus lineatus (Characiformes, Prochilodontidae). An. Acad. Bras. Cienc. 2013, 85, 1371–1377. [Google Scholar] [CrossRef]
- Voltolin, T.A.; Pansonato-Alves, J.C.; Senhorini, J.A.; Foresti, F.; Camacho, J.P.M.; Porto-Foresti, F. Common descent of B Chromosomes in two species of the fish genus Prochilodus (Characiformes, Prochilodontidae). Cytogenet. Genome Res. 2013, 141, 206–211. [Google Scholar] [CrossRef]
- Melo, S.; Utsunomia, R.; Penitente, M.; Sobrinho-Scudeler, P.E.; Porto-Foresti, F.; Oliveira, C.; Foresti, F.; Dergam, J.A. B chromosome dynamics in Prochilodus costatus (Teleostei, Characiformes) and comparisons with supernumerary chromosome system in other Prochilodus species. Comp. Cytogenet. 2017, 11, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Jesus, C.M.; Galetti, P.M.; Valentini, S.R.; Moreira-Filho, O. Molecular characterization and chromosomal localization of two families of satellite DNA in Prochilodus lineatus (Pisces, Prochilodontidae), a species with B chromosomes. Genetica 2003, 118, 25–32. [Google Scholar] [CrossRef]
- Oliveira, C.; Saboya, S.M.R.; Foresti, F.; Senhorini, J.A.; Bernardino, G. Increased B chromosome frequency and absence of drive in the fish Prochilodus lineatus. Heredity 1997, 79, 473–476. [Google Scholar] [CrossRef]
- Dias, A.L.; Foresti, F.; Oliveira, C. Synapsis in supernumerary chromosomes of Prochilodus lineatus (Teleostei: Prochilodontidae). Caryologia 1998, 51, 105–113. [Google Scholar] [CrossRef][Green Version]
- Cavallaro, Z.I.; Bertollo, L.A.C.; Perfectti, F.; Camacho, J.P.M. Frequency increase and mitotic stabilization of a B chromosome in the fish Prochilodus lineatus. Chromosom. Res. 2000, 8, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Voltolin, T.A.; Senhorini, J.A.; Oliveira, C.; Foresti, F.; Bortolozzi, J.; Porto-Foresti, F. B-chromosome frequency stability in Prochilodus lineatus (Characiformes, Prochilodontidae). Genetica 2010, 138, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Penitente, M.; Daniel, S.N.; Scudeler, P.E.S.; Foresti, F.; Porto-Foresti, F. B chromosome variants in Prochilodus lineatus (Characiformes, Prochilodontidae) analyzed by microdissection and chromosome painting techniques. Caryologia 2016, 69, 181–186. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Cechova, M.; Makova, K.D. Noise-cancelling repeat finder: Uncovering tandem repeats in error-prone long-read sequencing data. Bioinformatics 2019, 35, 4809–4811. [Google Scholar] [CrossRef] [PubMed]
- Foresti, F.; Almeida-Toledo, L.F.; Toledo-Filho, S.A. Polymorphic nature of nucleolus organizer regions in fishes. Cytogenet. Cell Genet. 1981, 31, 137–144. [Google Scholar] [CrossRef]
- Fenocchio, A.S.; Bertollo, L.A.C. A simple method for fresh-weter fish lymphocyte culture. Rev. Bras. Genet 1988, 11, 847–852. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Robledillo, L.Á.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.org (accessed on 22 November 2020).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Dos Santos, R.Z.; Calegari, R.M.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Melo, S.; Oliveira, C.; Foresti, F.; Uliano-Silva, M.; Foresti, F.P.; Utsunomia, R. A long-term conserved satellite DNA that remains unexpanded in several genomes of Characiformes fish is actively transcribed. Genome Biol. Evol. 2021, 13, evab002. [Google Scholar] [CrossRef]
- Negm, S.; Greenberg, A.; Larracuente, A.M.; Sproul, J.S. RepeatProfiler: A pipeline for visualization and comparative analysis of repetitive DNA profiles. Mol. Ecol. Resour. 2021, 21, 969–981. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Ruiz-Ruano, F.J.; Silva, D.M.Z.A.; Serrano, E.A.; Rosa, I.F.; Scudeler, P.E.S.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. A glimpse into the satellite DNA library in Characidae fish (Teleostei, Characiformes). Front. Genet. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Terencio, M.L.; Schneider, C.H.; Gross, M.C.; Nogaroto, V.; Almeida, M.C.; Artoni, R.F.; Vicari, M.R.; Feldberg, E. Repetitive sequences associated with differentiation of W chromosome in Semaprochilodus taeniurus. Genetica 2012, 140, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Kolmann, M.A.; Hughes, L.C.; Hernandez, L.P.; Arcila, D.; Betancur-R, R.; Sabaj, M.H.; López-Fernández, H.; Ortí, G. Phylogenomics of Piranhas and Pacus (Serrasalmidae) Uncovers How Dietary Convergence and Parallelism Obfuscate Traditional Morphological Taxonomy. Syst. Biol. 2020, 70, 576–592. [Google Scholar] [CrossRef]
- Pauls, E.; Bertollo, L.A.C. Evidence for a system of supernumerary chromosomes in Prochilodus scrofa Steindachner, 1881 (Pisces, Prochilodontidae). Caryologia 1983, 36, 307–314. [Google Scholar] [CrossRef]
- Voltolin, T.A.; Penitente, M.; Mendonça, B.B.; Senhorini, J.A.; Foresti, F.; Porto-Foresti, F. Karyotypic conservatism in five species of Prochilodus (Characiformes, Prochilodontidae) disclosed by cytogenetic markers. Genet. Mol. Biol. 2013, 36, 347–352. [Google Scholar] [CrossRef]
- Pauls, E.; Bertollo, L.A.C. Distribution of a supernumerary chromosome system and aspects of karyotypic evolution in the genus Prochilodus (Pisces, Prochilodontidae). Genetica 1990, 81, 117–123. [Google Scholar] [CrossRef]
- Moreira-Filho, O.; Galetti, P.M.; Bertollo, L.A.C. B chromosomes in the fish Astyanax scabripinnis (Characidae, Tetragonopterinae): An overview in natural populations. Cytogenet. Genome Res. 2004, 106, 230–234. [Google Scholar] [CrossRef]
- Scacchetti, P.C.; Utsunomia, R.; Pansonato-Alves, J.C.; Vicari, M.R.; Artoni, R.F.; Oliveira, C.; Foresti, F. Chromosomal mapping of repetitive DNAs in Characidium (Teleostei, Characiformes): Genomic organization and diversification of ZW sex chromosomes. Cytogenet. Genome Res. 2015, 146, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Speranza, E.D.; Colombo, J.C. Biochemical composition of a dominant detritivorous fish Prochilodus lineatus along pollution gradients in the Paraná-Río de la Plata Basin. J. Fish Biol. 2009, 74, 1226–1244. [Google Scholar] [CrossRef]
- Sivasundar, A.; Bermingham, E.; Ortí, G. Population structure and biogeography of migratory freshwater fishes (Prochilodus: Characiformes) in major South American rivers. Mol. Ecol. 2001, 10, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Garcez, R.; Calcagnotto, D.; Almeida-Toledo, L.F. Population structure of the migratory fish Prochilodus lineatus (Characiformes) from rio Grande basin (Brazil), an area fragmented by dams. Aquat. Conserv. Mar. Freshw. Ecosyst. 2011, 21, 268–275. [Google Scholar] [CrossRef]
- Ferreira, D.G.; Souza-Shibatta, L.; Shibatta, O.A.; Sofia, S.H.; Carlsson, J.; Dias, J.H.P.; Makrakis, S.; Makrakis, M.C. Genetic structure and diversity of migratory freshwater fish in a fragmented Neotropical river system. Rev. Fish Biol. Fish. 2017, 27, 209–231. [Google Scholar] [CrossRef]
- Melo, B.F.; Dorini, B.F.; Foresti, F.; Oliveira, C. Little divergence among mitochondrial lineages of Prochilodus (Teleostei, Characiformes). Front. Genet. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Hewitt, G.; Ruscoe, C. Changes in microclimate correlated with a cline for B-chromosomes in the grasshopper Myrmeleotettix maculatus (Thunb.) (Orthoptera: Acrididae). J. Anim. Ecol. 1971, 40, 753. [Google Scholar] [CrossRef]
- Néo, D.M.; Moreira-Filho, O.; Camacho, J.P.M. Altitudinal variation for B chromosome frequency in the characid fish Astyanax scabripinnis. Heredity 2000, 85, 136–141. [Google Scholar] [CrossRef][Green Version]
- Vujosevic, M.; Blagojevic, J. Does environment affect polymorphism of B chromosomes in the yellow-necked mouse Apodemus flavicollis? Z. Saugetierkd. 2000, 65, 313–317. [Google Scholar]
- Vujošević, M.; Blagojević, J. Seasonal changes of B-chromosome frequencies within the population of Apodemus flavicollis (Rodentia) on Cer mountain in Yugoslavia. Acta Theriol. 1995, 40, 131–137. [Google Scholar] [CrossRef]
- Martis, M.M.; Klemme, S.; Banaei-Moghaddam, A.M.; Blattner, F.R.; Macas, J.; Schmutzer, T.; Scholz, U.; Gundlach, H.; Wicker, T.; Sǐmková, H.; et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 2012, 109, 13343–13346. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Conte, M.A.; Fantinatti, B.E.A.; Cabral-de-Mello, D.C.; Carvalho, R.F.; Vicari, M.R.; Kocher, T.D.; Martins, C. Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol. Biol. Evol. 2014, 31, 2061–2072. [Google Scholar] [CrossRef]
- Ferreira, D.; Meles, S.; Escudeiro, A.; Mendes-da-Silva, A.; Adega, F.; Chaves, R. Satellite non-coding RNAs: The emerging players in cells, cellular pathways and cancer. Chromosom. Res. 2015, 23, 479–493. [Google Scholar] [CrossRef]
- Navarro-Domínguez, B.; Ruiz-Ruano, F.J.; Cabrero, J.; Corral, J.M.; López-León, M.D.; Sharbel, T.F.; Camacho, J.P.M. Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.P.; Hattori, R.S.; Yoshinaga, T.T.; Silva, D.M.Z.A.; Foresti, F.; Santos, M.H.; Almeida, M.C.; Artoni, R.F. Differential expression of dmrt1 in Astyanax scabripinnis (Teleostei, Characidade) is correlated with B chromosome occurrence. Zebrafish 2019, 16, 182–188. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the role of satellite DNA in genome architecture and plasticity—An evolutionary and clinical affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Robles, F.; De La Herrán, R.; Ludwig, A.; Ruiz Rejón, C.; Ruiz Rejón, M.; Garrido-Ramos, M.A. Evolution of ancient satellite DNAs in sturgeon genomes. Gene 2004, 338, 133–142. [Google Scholar] [CrossRef]
- Plohl, M.; Petrović, V.; Luchetti, A.; Ricci, A.; Šatović, E.; Passamonti, M.; Mantovani, B. Long-term conservation vs high sequence divergence: The case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 2010, 104, 543–551. [Google Scholar] [CrossRef]
- Chaves, R.; Ferreira, D.; Mendes-Da-Silva, A.; Meles, S.; Adega, F. FA-SAT is an old satellite DNA frozen in several bilateria genomes. Genome Biol. Evol. 2017, 9, 3073–3087. [Google Scholar] [CrossRef]
- Lorite, P.; Muñoz-López, M.; Carrillo, J.A.; Sanllorente, O.; Vela, J.; Mora, P.; Tinaut, A.; Torres, M.I.; Palomeque, T. Concerted evolution, a slow process for ant satellite DNA: Study of the satellite DNA in the Aphaenogaster genus (Hymenoptera, Formicidae). Org. Divers. Evol. 2017, 17, 595–606. [Google Scholar] [CrossRef]
- Dover, G. Molecular drive: A cohesive mode of species evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA in plants: More than just Rubbish. Cytogenet. Genome Res. 2015, 146, 153–170. [Google Scholar] [CrossRef] [PubMed]

| SF | satDNA Family | RUL | A+T (%) | Abundance (%) | Divergence (%) |
|---|---|---|---|---|---|
| 1 | PliSat01-225 | 225 | 64.5 | 1.332 × 10−2 | 7.25 |
| PliSat02-184 | 184 | 54.9 | 9.882 × 10−3 | 10.89 | |
| PliSat03-900 | 900 | 56.5 | 4.829 × 10−3 | 1.84 | |
| PliSat04-441 | 441 | 57.9 | 3.191 × 10−3 | 6.43 | |
| PliSat05-178 | 178 | 64.6 | 3.097 × 10−3 | 8.99 | |
| PliSat06-44 | 44 | 47.8 | 2.021 × 10−3 | 13.85 | |
| PliSat07-3008 | 3008 | 63.4 | 1.920 × 10−3 | 3.31 | |
| PliSat08-54 | 54 | 62.9 | 1.847 × 10−3 | 4.04 | |
| 1 | PliSat09-495 | 495 | 58.0 | 1.809 × 10−3 | 5.78 |
| PliSat10-25 | 25 | 40.0 | 1.215 × 10−3 | 11.61 | |
| PliSat11-709 | 709 | 50.4 | 1.120 × 10−3 | 9.90 | |
| PliSat12-42 | 42 | 52.4 | 9.841 × 10−4 | 13.58 | |
| PliSat13-1928 | 1928 | 62.4 | 8.420 × 10−4 | 7.86 | |
| PliSat14-44 | 44 | 50.0 | 5.822 × 10−4 | 7.52 | |
| PliSat15-75 | 75 | 61.3 | 4.322 × 10−4 | 3.33 | |
| PliSat16-16 | 67 | 58.2 | 3.913 × 10−4 | 3.79 | |
| PliSat17-21 | 21 | 76.2 | 3.087 × 10−4 | 15.05 | |
| PliSat18-40 | 40 | 57.5 | 2.978 × 10−4 | 10.47 | |
| PliSat19-30 | 30 | 56.7 | 2.642 × 10−4 | 12.96 | |
| PliSat20-54 | 54 | 57.4 | 2.558 × 10−4 | 9.63 | |
| PliSat21-59 | 59 | 49.2 | 2.382 × 10−4 | 4.69 | |
| PliSat22-37 | 37 | 45.9 | 2.120 × 10−4 | 4.59 | |
| PliSat23-39 | 39 | 53.8 | 1.860 × 10−4 | 10.87 | |
| PliSat24-31 | 31 | 51.6 | 1.846 × 10−4 | 17.13 | |
| PliSat25-34 | 34 | 47.1 | 1.611 × 10−4 | 5.42 | |
| PliSat26-52 | 52 | 59.6 | 1.569 × 10−4 | 8.83 | |
| PliSat27-1683 | 1683 | 52.0 | 1.368 × 10−4 | 2.25 | |
| PliSat28-32 | 32 | 56.8 | 1.361 × 10−4 | 14.45 | |
| PliSat29-60 | 60 | 45.0 | 1.251 × 10−4 | 3.31 | |
| PliSat30-31 | 31 | 54.8 | 1.225 × 10−4 | 16.69 | |
| PliSat31-707 | 707 | 53.6 | 1.188 × 10−4 | 4.27 | |
| PliSat32-30 | 30 | 66.3 | 1.185 × 10−4 | 12.19 | |
| 2 | PliSat33-6 | 6 | 57.1 | 1.040 × 10−4 | 25.51 |
| PliSat34-39 | 39 | 53.8 | 1.013 × 10−4 | 7.97 | |
| PliSat35-1128 | 1128 | 61.0 | 9.474 × 10−5 | 10.43 | |
| PliSat36-68 | 68 | 63.2 | 9.039 × 10−5 | 5.55 | |
| PliSat37-554 | 554 | 50.0 | 8.823 × 10−5 | 5.47 | |
| PliSat38-32 | 32 | 53.1 | 8.486 × 10−5 | 6.29 | |
| PliSat39-915 | 915 | 54.3 | 7.846 × 10−5 | 1.35 | |
| PliSat40-27 | 27 | 48.8 | 7.708 × 10−5 | 14.00 | |
| 2 | PliSat41-162 | 162 | 45.8 | 7.486 × 10−5 | 1.75 |
| PliSat42-24 | 24 | 55.6 | 6.696 × 10−5 | 7.75 | |
| PliSat43-32 | 32 | 53.1 | 6.279 × 10−5 | 9.61 | |
| PliSat44-1134 | 1134 | 51.6 | 6.220 × 10−5 | 3.43 | |
| PliSat45-340 | 340 | 50.6 | 6.194 × 10−5 | 4.83 | |
| PliSat46-29 | 29 | 65.5 | 6.009 × 10−5 | 13.97 | |
| PliSat47-574 | 574 | 53.8 | 5.280 × 10−5 | 6.89 | |
| PliSat48-387 | 387 | 48.8 | 4.802 × 10−5 | 5.00 | |
| PliSat49-67 | 67 | 49.3 | 4.517 × 10−5 | 4.30 | |
| PliSat50-48 | 48 | 58.3 | 4.280 × 10−5 | 9.07 | |
| PliSat51-911 | 911 | 55.0 | 3.897 × 10−5 | 5.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stornioli, J.H.F.; Goes, C.A.G.; Calegari, R.M.; dos Santos, R.Z.; Giglio, L.M.; Foresti, F.; Oliveira, C.; Penitente, M.; Porto-Foresti, F.; Utsunomia, R. The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs. Cells 2021, 10, 1527. https://doi.org/10.3390/cells10061527
Stornioli JHF, Goes CAG, Calegari RM, dos Santos RZ, Giglio LM, Foresti F, Oliveira C, Penitente M, Porto-Foresti F, Utsunomia R. The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs. Cells. 2021; 10(6):1527. https://doi.org/10.3390/cells10061527
Chicago/Turabian StyleStornioli, José Henrique Forte, Caio Augusto Gomes Goes, Rodrigo Milan Calegari, Rodrigo Zeni dos Santos, Leonardo Moura Giglio, Fausto Foresti, Claudio Oliveira, Manolo Penitente, Fábio Porto-Foresti, and Ricardo Utsunomia. 2021. "The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs" Cells 10, no. 6: 1527. https://doi.org/10.3390/cells10061527
APA StyleStornioli, J. H. F., Goes, C. A. G., Calegari, R. M., dos Santos, R. Z., Giglio, L. M., Foresti, F., Oliveira, C., Penitente, M., Porto-Foresti, F., & Utsunomia, R. (2021). The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs. Cells, 10(6), 1527. https://doi.org/10.3390/cells10061527






