Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Species, Mitotic Chromosome Preparations, C-Banding, and CMA3 Staining
2.2. FISH with rDNA and Repetitive Motifs
2.3. Microdissection and Whole Chromosome Painting
2.4. Comparative Genomic Hybridization
2.5. Microscopic Analyses and Image Processing
3. Results
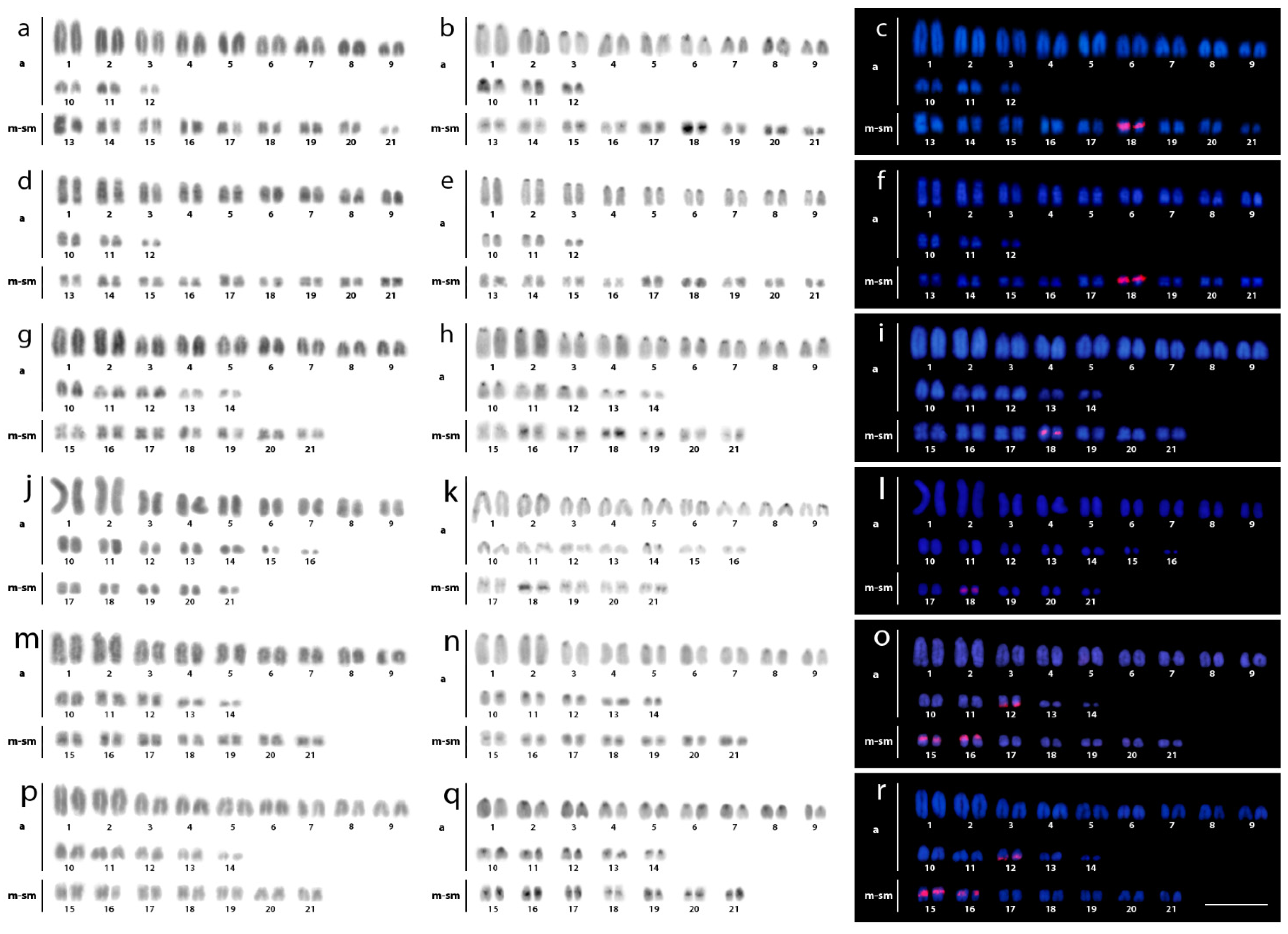
3.1. Karyotypes, C-Banding, and Chromomycin A3-Staining
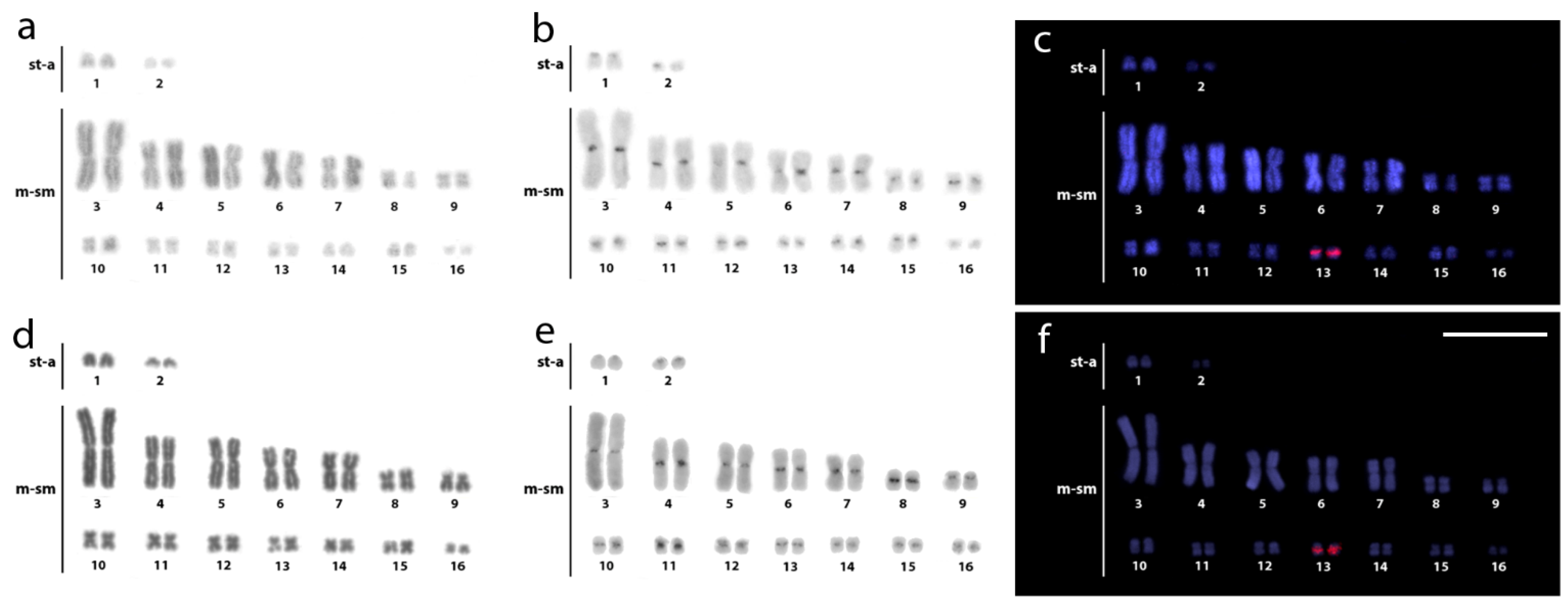
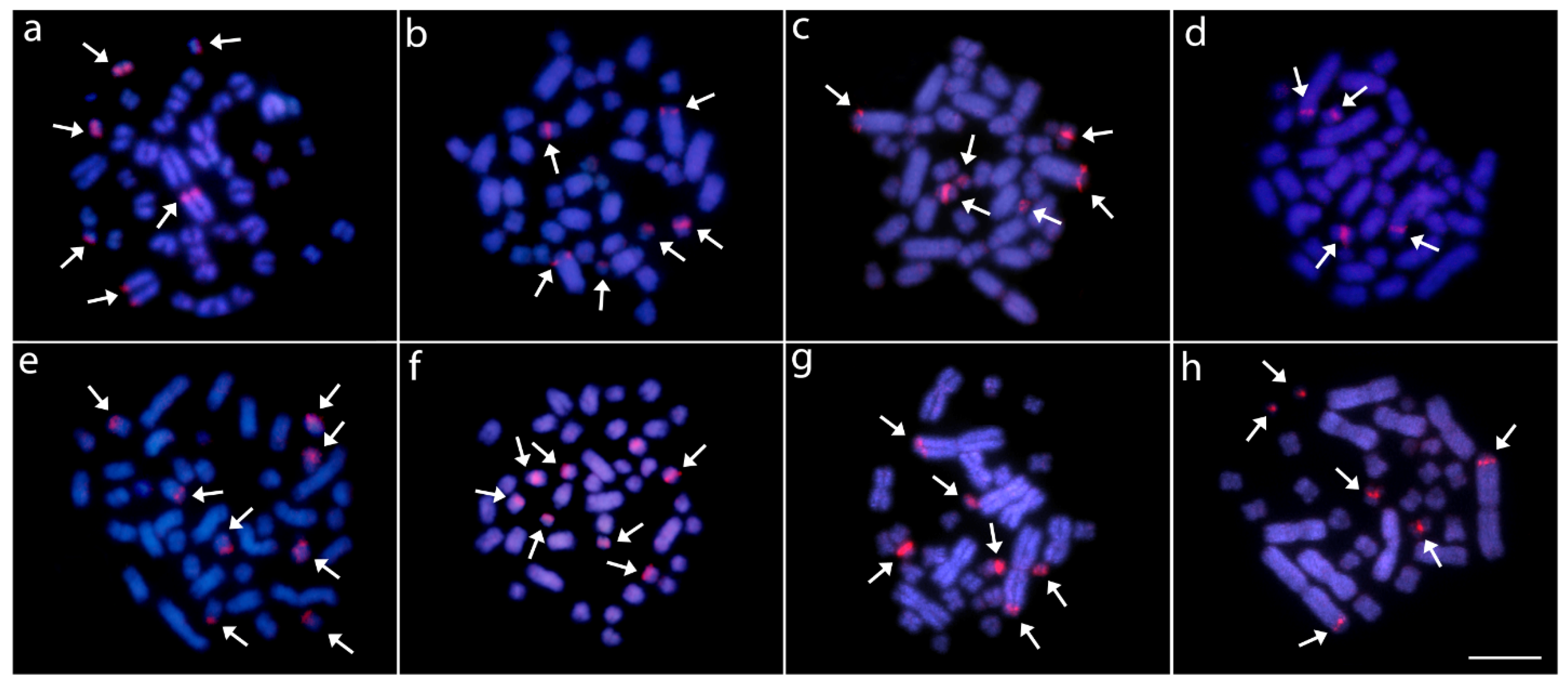
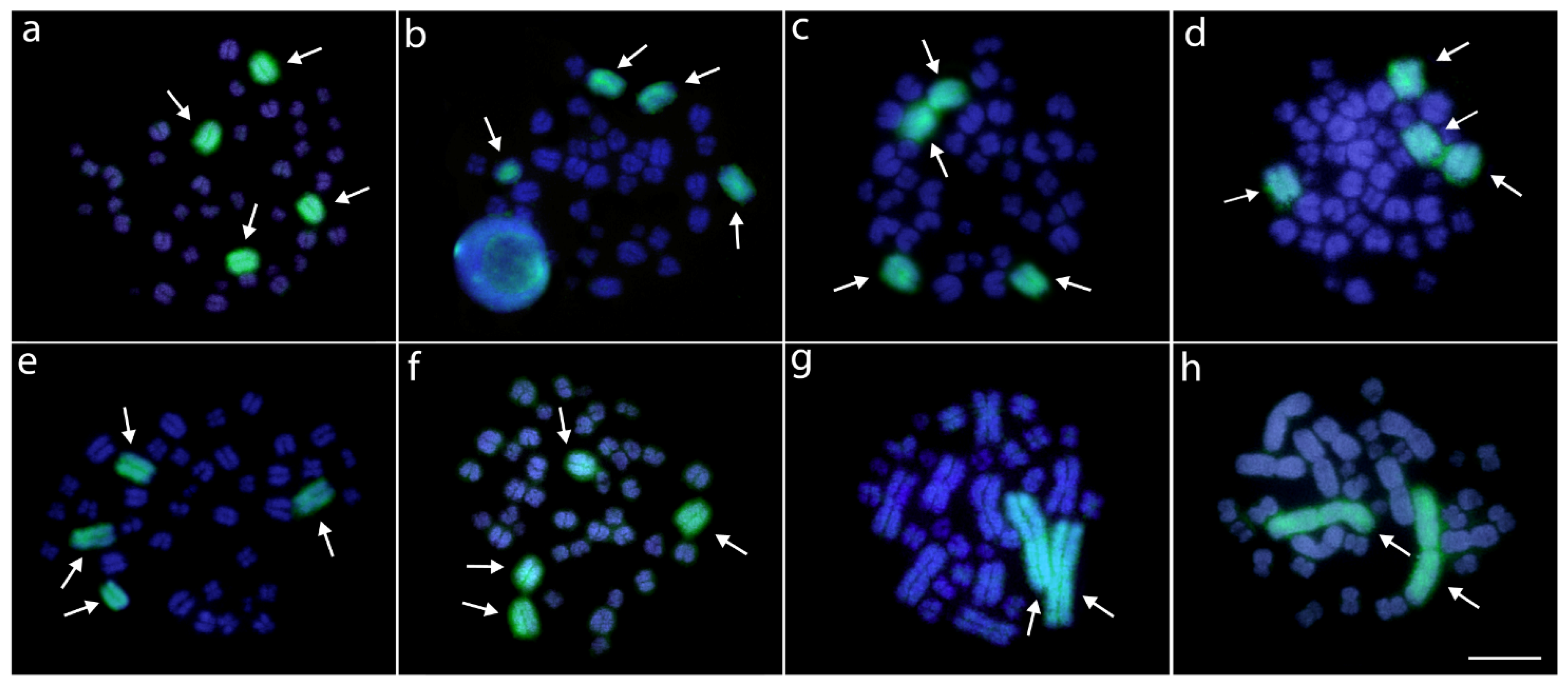
3.2. Fluorescence In Situ Hybridization (FISH) Mapping of Repetitive DNAs
3.3. WCP of AMI-1 Probe
3.4. Comparative Genomic Hybridization (CGH)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grigg, G.; Seebacher, F.; Franklin, C.E. (Eds.) Crocodilian Biology and Evolution, 1st ed.; Surrey Beatty: Chipping Norton, Australia, 2001; p. 446. [Google Scholar]
- Brochu, C.A. Phylogenetic approaches toward crocodylian history. Annu. Rev. Earth and Planet. Sci. 2003, 31, 357–397. [Google Scholar] [CrossRef]
- Bronzati, M.; Montefeltro, F.C.; Langer, M.C. Diversification events and the effects of mass extinction on Crocodyliformes evolutionary history. R. Soc. Open Sci. 2015, 2, 140385. [Google Scholar] [CrossRef]
- Stubbs, T.L.; Pierce, S.E.; Elsler, A.; Anderson, P.S.L.; Rayfield, E.J.; Benton, M.J. Ecological opportunity and the rise and fall of crocodylomorph evolutionary innovation. Proc. R. Soc. B 2021, 288, 20210069. [Google Scholar] [CrossRef]
- Janke, A.; Arnason, U. The complete mitochondrial genome of Alligator mississippiensis and the separation between recent Archosauria (birds and crocodiles). Mol. Biol. Evol. 1997, 14, 1266–1272. [Google Scholar] [CrossRef]
- Iwabe, N.; Hara, Y.; Kumazawa, Y.; Shibamoto, K.; Saito, Y.; Miyata, T.; Katoh, K. Sister group relationship of turtles to the bird-crocodilian clade revealed by nuclear DNA-coded proteins. Mol. Biol. Evol. 2005, 22, 810–813. [Google Scholar] [CrossRef]
- Green, R.E.; Braun, E.L.; Armstrong, J.; Earl, D.; Nguyen, N.; Hickey, G.; Vandewege, M.W.; St. John, J.A.; Capella-Gutiérrez, S.; Castoe, T.A. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 2014, 346, 1254449. [Google Scholar] [CrossRef]
- Pan, T.; Miao, J.-S.; Zhang, H.-B.; Yan, P.; Lee, P.-S.; Jiang, X.-Y.; Ouyang, J.-H.; Deng, Y.-P.; Zhang, B.-W.; Wu, X.-B. Near-complete phylogeny of extant Crocodylia (Reptilia) using mitogenome-based data. Zool. J. Linn. Soc. 2021, 191, 1075–1089. [Google Scholar] [CrossRef]
- Espinosa, E.; Godshalk, R.; Hall, P.; Thorbjarnarson, J.; Tucker, A.; Verdade, L. Species Accounts. In Status Survey and Conservation Action Plan: Revised Action Plan for Crocodiles, 2nd ed.; Ross, P., Ed.; IUCN/SSC Crocodile Specialist Group: Gland, Switzerland; Cambridge, UK, 1998; Volume 1, pp. 3–73. [Google Scholar]
- Rueda-Almonacid, J.V.; Carr, J.L.; Mittermeier, R.A.; Rodríguez-Mahecha, J.V.; Mast, R.B.; Vogt, R.C.; Rhodin, A.G.J.; Ossa-Velásquez, J.O.; Rueda, J.N.; Mittermeier, C.G. Orden Crocodylia. In Conservación Internacional. Serie de Guías Tropicales de Campo N° 6. Las Tortugas y lós Crocodilianos de los Países Andinos del Trópico, 1st ed.; Mittermeier, R.A., Rylands, A., Eds.; Editorial Panamericana, Formas e Impresos: Bogotá, Colombia, 2007; Volume 1, pp. 387–432. [Google Scholar]
- Meredith, R.W.; Hekkala, E.R.; Amato, G.; Gatesy, J. A phylogenetic hypothesis for Crocodylus (Crocodylia) based on mitochondrial DNA: Evidence for a trans-Atlantic voyage from Africa to the New World. Mol. Phylogenet. Evol. 2011, 60, 183–191. [Google Scholar] [CrossRef]
- Oaks, J.R. A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 2011, 65, 3285–3297. [Google Scholar] [CrossRef]
- McAliley, L.R.; Willis, R.E.; Ray, D.A.; White, P.S.; Brochu, C.A.; Densmore, L.D. Are crocodiles really monophyletic?—Evidence for subdivisions from sequence and morphological data. Mol. Phylogenet. Evol. 2006, 39, 16–32. [Google Scholar] [CrossRef]
- Martin, S. Global diversity of crocodiles (Crocodilia, Reptilia) in freshwater. Hydrobiologia 2008, 595, 587–591. [Google Scholar] [CrossRef]
- Hekkala, E.; Shirley, M.H.; Amato, G.; Austin, J.D.; Charter, S.; Thorbjarnarson, J.; Vliet, K.A.; Houck, M.L.; Desalle, R.; Blum, M.J. An ancient icon reveals new mysteries: Mummy DNA resurrects a cryptic species within the Nile crocodile. Mol. Ecol. 2011, 20, 4199–4215. [Google Scholar] [CrossRef]
- Shirley, M.H.; Vliet, K.A.; Carr, A.N.; Austin, J.D. Rigorous approaches to species delimitation have significant implications for African crocodilian systematics and conservation. Proc. R. Soc. B 2014, 281, 20132483. [Google Scholar] [CrossRef]
- Srikulnath, K.; Thapana, W.; Muangmai, N. Role of chromosome changes in Crocodylus evolution and diversity. Genom. Inform. 2015, 13, 102–111. [Google Scholar] [CrossRef]
- Barreiros, J.P. Crocodylia: Uma Longa História de Sucesso Evolutivo, 1st ed.; Atlântida Revista de Cultura: Açores, Portugal, 2016; pp. 1–12. [Google Scholar]
- Muniz, F.L.; Ximenes, A.M.; Bittencourt, P.S.; Hernández-Rangel, S.M.; Campos, Z.; Hrbek, T.; Farias, I.P. Detecting population structure of Paleosuchus trigonatus (Alligatoridae: Caimaninae) through microsatellites markers developed by next generation sequencing. Mol. Biol. Rep. 2019, 46, 1–12. [Google Scholar] [CrossRef]
- Uetz, P.; Freed, P.; Aguilar, R.; Hošek, J. (Eds.) The Reptile Database. Available online: http://www.reptile-database.org (accessed on 31 August 2020).
- Nicolai, M.P.J.; Matzke, N.J. Trait-based range expansion aided in the global radiation of Crocodylidae. Glob. Ecol. Biogeogr. 2019, 28, 1244–1258. [Google Scholar] [CrossRef]
- Shirley, M.H.; Carr, A.N.; Nestler, J.H.; Vliet, K.A.; Brochu, C.A. Systematic revision of the living African slender-snouted crocodiles (Mecistops Gray, 1844). Zootaxa 2018, 4504, 151–193. [Google Scholar] [CrossRef]
- Bezuijen, M.R.; Shwedick, B.; Simpson, B.K.; Staniewicz, A.; Stuebing, R. Tomistoma schlegelii. IUCN Red List Threat. Species 2014, 2014, E.T21981A2780499. [Google Scholar] [CrossRef]
- Lee, M.S.Y.; Yates, A.M. Tip-dating and homoplasy: Reconciling the shallow molecular divergences of modern gharials with their long fossil record. Proc. R. Soc. B. 2018, 285, 20181071. [Google Scholar] [CrossRef]
- Muniz, F.L.; Campos, Z.; Rangel, S.M.H.; Martínez, J.G.; Souza, B.C.; De Thoisy, B.; Botero-Arias, R.; Hrbek, T.; Farias, I.P. Delimitation of evolutionary units in Cuvier´s dwarf caiman, Paleosuchus palpebrosus (Cuvier, 1807): Insights from conservation of a broadly distributes species. Conserv. Genet. 2018, 19, 599–610. [Google Scholar] [CrossRef]
- Bittencourt, P.S.; Campos, Z.; Muniz, F.L.; Marioni, B.; Souza, B.C.; Da Silveira, R.; de Thoisy, B.; Hrbek, T.; Farias, I.P. Evidence of cryptic lineages within a small South American crocodilian: The Schneider´s dwarf caiman Paleosuchus trigonatus (Alligatoridae: Caimaninae). PeerJ 2019, 7, e6580. [Google Scholar] [CrossRef]
- Balaguera-Reina, S.A.; Velasco, A. Caiman crocodilus. IUCN Red List Threat. Species 2019, 2019, E.T46584A3009688. [Google Scholar] [CrossRef]
- Jiang, H.; Wu, X. Alligator sinensis. IUCN Red List Threat. Species 2018, 2018. [Google Scholar] [CrossRef]
- Thorbjarnarson, J.; Wang, X.; Ming, S.; He, L.; Ding, Y.; Wu, Y.; McMurry, S.T. Wild populations of the Chinese alligator approach extinction. Biol. Conserv. 2002, 103, 93–102. [Google Scholar] [CrossRef]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Ezaz, T. Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef]
- Straková, B.; Rovatsos, M.; Kubička, L.; Kratochvíl, L. Evolution of sex determination in amniotes: Did stress and sequential hermaphroditism produce environmental determination? BioEssays 2020, 42, e2000050. [Google Scholar] [CrossRef]
- Ezaz, T.; Quinn, A.E.; Miura, I.; Sarre, S.D.; Georges, A.; Graves, J.A.M. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005, 13, 763–776. [Google Scholar] [CrossRef]
- Ezaz, T.; Valenzuela, N.; Grützner, F.; Miura, I.; Georges, A.; Burke, R.L.; Graves, J.A.M. An XX/XY sex michrocromosome system in a freshwater turtle, Chelodina longicollis (Testudines: Chelidae) with genetic sex determination. Chromosome Res. 2006, 14, 139–150. [Google Scholar] [CrossRef]
- Kawai, A.; Nishida-Umehara, C.; Ishijima, J.; Tsuda, Y.; Ota, H.; Matsuda, Y. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet. Genome Res. 2007, 117, 92–102. [Google Scholar] [CrossRef]
- Martinez, P.A.; Ezaz, T.; Valenzuela, N.; Georges, A.; Graves, J.A.M. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii: A new piece in the puzzle of sex chromosome evolution in turtles. Chromosome Res. 2008, 16, 815–825. [Google Scholar] [CrossRef]
- Badenhorst, D.; Stanyon, R.; Engstrom, T.; Valenzuela, N. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 2013, 21, 137–147. [Google Scholar] [CrossRef]
- Koubová, M.; Pokorná, M.J.; Rovatsos, M.; Farkačová, K.; Altmanová, M.; Kratochvil, L. Sex determination in Madagascar geckos of the genus Paroedura (Squamata: Gekkonidae) are differentiated sex chromosomes indeed so evolutionary stable? Chromosome Res. 2014, 22, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Gamble, T.; Matsuda, Y.; Zarkower, D.; Sarre, S.D.; Georges, A.; Graves, J.A.M.; Ezaz, T. Non-homologous sex chromosomes in two geckos (Gekkonidae: Gekkota) with female heterogamety. Cytogenet. Genome Res. 2014, 143, 251–258. [Google Scholar] [CrossRef]
- Montiel, E.E.; Badenhorst, D.; Tamplin, J.; Burke, R.L.; Valenzuela, N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 2016, 126, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Viana, P.F.; Ezaz, T.; Cioffi, M.B.; Almeida, B.J.; Feldberg, E. Evolutionary Insights of the ZW Sex Chromosomes in Snakes: A New Chapter Added by the Amazonian Puffing Snakes of the Genus Spilotes. Genes 2019, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Viana, P.F.; Ezaz, T.; Cioffi, M.B.; Liehr, T.; Al-Rikabi, A.; Goll, L.G.; Rocha, A.M.; Feldberg, E. Landscape of snake’ sex chromosomes evolution spanning 85 MYR reveals ancestry of sequences despite distinct evolutionary trajectories. Sci. Rep. 2020, 10, 12499. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.M.; Clark, H.F. The somatic chromosomes of five crocodilian species. Cytogenetics 1967, 6, 193–203. [Google Scholar] [CrossRef]
- Cohen, M.M.; Gans, C. The chromosomes of the Order Crocodilia. Cytogenetics 1970, 9, 81–105. [Google Scholar] [CrossRef]
- King, M.; Honeycutt, R.; Contreras, N. Chromosomal repatterning in crocodiles: C, G, and N-banding and the in situ hybridization of 18S and 26S rRNA cistrons. Genetica 1986, 70, 191–201. [Google Scholar] [CrossRef]
- Lui, J.F.; Valencia, E.F.T.; Boer, J.A. Karyotypic analysis and chromosome biometry of cell cultures of the yellow throated alligator (Caiman latirostris DAUDIN). Rev. Brasil. Genet. 1994, 17, 165–169. [Google Scholar]
- Valleley, E.M.A.; Harrison, C.J.; Cook, Y.; Ferguson, M.W.J.; Sharpe, P.T. The karyotype of Alligator mississippiensis, and chromosomal mapping of the ZFY/X homologue, Zfc. Chromosoma 1994, 103, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Kawagoshi, T.; Nishida, C.; Ota, H.; Kumazawa, Y.; Endo, H.; Matsuda, Y. Molecular structures of centromeric heterochromatin and karyotypic evolution in the Siamese crocodile (Crocodylus siamensis) (Crocodylidae, Crocodylia). Chromosome Res. 2008, 16, 1119–1132. [Google Scholar] [CrossRef]
- Uno, Y.; Nishida, C.; Tarui, H.; Ishishita, S.; Takagi, C.; Nishimura, O.; Ishijima, J.; Ota, H.; Kosaka, A.; Matsubara, K.; et al. Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS ONE 2012, 7, e53027. [Google Scholar] [CrossRef]
- Kasai, F.; O’Brien, P.C.M.; Martin, S.; Ferguson-Smith, M.A. Extensive homology of chicken macrochromosomes in the karyotypes of Trachemys scripta elegans and Crocodylus niloticus revealed by chromosome painting despite long divergence times. Cytogenet. Genome Res. 2012, 136, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.C.S.; Viana, P.F.; Gross, M.C.; Feldberg, E.; Da Silveira, R.; Cioffi, M.B.; Bertollo, L.A.C.; Schneider, C.H. Looking for genetic effects of polluted anthropized environments on Caiman crocodilus crocodilus (Reptilia, Crocodylia): A comparative genotoxic and chromosomal analysis. Ecotoxicol. Environ. Saf. 2021, 209, 111835. [Google Scholar] [CrossRef]
- Olmo, E.; Signorino, G.G. Chromorep: A Reptile Chromosomes Database. Available online: http://chromorep.univpm.it/?q=node/13 (accessed on 24 August 2020).
- Degrandi, T.M.; Gunski, R.J.; Garnero, A.V.; Oliveira, E.H.C.; Kretschmer, R.; Souza, M.S.; Barcellos, S.A.; Hass, I. The distribution of 45S rDNA sites in bird suggests multiple evolutionary histories. Genet. Mol. Biol. 2020, 43, e20180331. [Google Scholar] [CrossRef]
- Amavet, P.; Markariani, R.; Fenocchio, A. Comparative cytogenetic analysis of the South American alligators Caiman latirostris and Caiman yacare (Reptilia, Alligatoridae) from Argentina. Caryologia 2003, 56, 489–493. [Google Scholar] [CrossRef][Green Version]
- Valenzuela, N. Temperature dependent sex determination in reptiles. In Reptilian Incubation: Environment & Behaviour, 1st ed.; Deeming, D.C., Ed.; Nottinghan University Press: Nottingham, UK, 2004; Volume 1, pp. 65–80. [Google Scholar]
- González, E.J.; Martínez-López, M.; Morales-Garduza, M.A.; García-Morales, R.; Charruau, P.; Gallardo-Cruz, J.A. The sex-determination pattern in crocodilians: A systematic review of three decades of research. J. Anim. Ecol. 2019, 88, 1417–1427. [Google Scholar] [CrossRef]
- Viana, P.F.; Ribeiro, L.B.; Lima, T.; de Carvalho, V.T.; Vogt, R.C.; Gross, M.C.; Feldberg, E. An optimized protocol for obtaining mitotic chromosomes from cultured reptilian lymphocytes. Nucleus 2016, 59, 191–195. [Google Scholar] [CrossRef]
- Johnson Pokorná, M.; Altmanová, M.; Rovatsos, M.; Velenský, P.; Vodička, R.; Rehák, I.; Kratochvíl, L. First description of the karyotype and sex chromosomes in the Komodo dragon (Varanus komodoensis). Cytogenet. Genome Res. 2016, 148, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Schmid, M. Chromosome banding in Amphibia. IV. Differentiation of GC and AT-rich regions in Anura. Chromosoma 1980, 77, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Martins, C.; Bertollo, L.A.C. Comparative chromosome mapping of repetitive sequences. Implications for genomic evolution in the fish, Hoplias malabaricus. BMC Genet. 2009, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef]
- Kubat, Z.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 2008, 51, 350–356. [Google Scholar] [CrossRef]
- Yano, C.F.; Bertollo, L.A.C.; Cioffi, M.B. Fish-FISH: Molecular Cytogenetics in Fish Species. In Fluorescence In Situ Hybridization (FISH)—Application Guide, 2nd ed.; Liehr, T., Ed.; Springer: Berlin, Germany, 2017; Volume 1, pp. 429–444. [Google Scholar] [CrossRef]
- Kosyakova, N.; Liehr, T.; Al-Rikabi, A. FISH-microdissection. In Fluorescence In Situ Hybridization (FISH)—Application Guide, 2nd ed.; Liehr, T., Ed.; Springer: Berlin, Germany, 2017; Volume 1, pp. 81–99. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning, A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. 58–63. [Google Scholar]
- Zwick, M.S.; Hanson, R.E.; McKnight, T.D.; Islam-Faridi, M.N.; Stelly, D.M.; Wing, R.A.; Price, H.J. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome 1997, 40, 138–142. [Google Scholar] [CrossRef]
- Symonová, R.; Sember, A.; Majtánová, Z.; Ráb, P. Characterization of fish genomes by GISH and CGH. In Fish Cytogenetic Techniques Ray-Fin Fishes Chondrichthyans, 1st ed.; Ozouf-Costaz, C., Pisano, E., Foresti, F., Toledo, L.F.A., Eds.; CCR Press: Boca Raton, FL, USA, 2015; pp. 118–131. [Google Scholar]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Matthey, R.; van Brink, J.M. Sex chromosomes in Amniota. Evolution 1957, 11, 163–165. [Google Scholar] [CrossRef]
- Ohno, S. Sex Chromosomes and Sex-linked Genes. In Monographs on Endocrinology, 1st ed.; Labhart, A., Mann, T., Samuels, L.T., Zander, J., Eds.; Springer: Berlin, Germany; New York, NY, USA, 1967; Volume 1, pp. 33–46. [Google Scholar] [CrossRef]
- Beçak, W.; Beçak, M.L. Order: CROCODILIA, Suborder: EUSUCHIA, Family: CROCODYLIDAE, Caiman crocodilus (Linnaeus). Folio R-15 1971, 1, 1–3. [Google Scholar]
- Olmo, E.A. Reptilia. In Animal Cytogenetics. Chordata 3, 1st ed.; John, B., Ed.; Gebrueder Borntraeger: Berlin, Germany; Stuttgart, Germany, 1986; Volume 4, pp. 1–100. [Google Scholar]
- Amavet, P.; Siroski, P.; Donayo, P.; Medina, M. Karyotype of Caiman latirostris and Caiman yacare (Reptilia, Alligatoridae). In Proceedings of the 15th Working Meeting of the Crocodile Specialist Group, Varadero, Cuba, 17–20 January 2000; pp. 135–138. [Google Scholar]
- Alfaro, M.E.; Santini, F.; Brock, C.; Alamillo, H.; Dornburg, A.; Rabosky, D.L.; Carnevale, G.; Harmon, L.J. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl. Acad. Sci. USA 2009, 106, 13410–13414. [Google Scholar] [CrossRef]
- Jetz, W.; Thomas, G.H.; Joy, J.B.; Hartmann, K.; Mooers, A.O. The global diversity of birds in space and time. Nature 2012, 491, 444–448. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef]
- Bista, B.; Valenzuela, N. Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 2020, 11, 416. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Altmanová, M.; Johnson Pokorná, M.; Velenský, P.; Baca, A.S.; Kratochvíl, L. Evolution of karyotypes in chameleons. Genes 2017, 8, 382. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Reisz, R.R. Four well-constrained calibration points from the vertebrate fossil record for molecular clock estimates. BioEssays 2005, 27, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Slijepcevic, P. Telomere length and telomere-centromere relationships? Mutat. Res. 1998, 404, 215–220. [Google Scholar] [CrossRef]
- Porter, C.A.; Haiduk, M.W.; de Queiroz, K. Evolution and phylogenetic significance of ribosomal gene location in chromosomes of squamate reptiles. Copeia 1994, 1994, 302–313. [Google Scholar] [CrossRef]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Camper, J.D.; Hanks, B. Variation in the nucleolus organizer region among new world snakes. J. Herpetol. 1995, 29, 468–471. [Google Scholar] [CrossRef]
- O’Meally, S.; Patel, H.R.; Stiglec, R.; Sarre, S.D.; Georges, A.; Graves, J.A.M.; Ezaz, T. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 2010, 18, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Altmanová, M.; Augstenová, B.; Mazzoleni, S.; Velenský, P.; Kratochvíl, L. ZZ/ZW sex determination with multiple neo-sex chromosomes is common in Madagascan chameleons of the genus Furcifer (Reptilia: Chamaeleonidae). Genes 2019, 10, 1020. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 2020, 10, 4276. [Google Scholar] [CrossRef]
- Perry, B.W.; Schield, D.R.; Adams, R.H.; Castoe, T.A. Microchromosomes exhibit distinct features of vertebrate chromosome structure and function with underappreciated ramifications for genome evolution. Mol. Biol. Evol. 2021, 38, 904–910. [Google Scholar] [CrossRef]
- Burt, D.W. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 2002, 96, 97–112. [Google Scholar] [CrossRef]
- Norris, T.B.; Rickards, G.K.; Daugherty, C.H. Chromosomes of tuatara, Sphenodon, a chromosome heteromorphism and an archaic reptilian karyotype. Cytogenet. Genome Res. 2004, 105, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Auer, H.; Mayr, B.; Lambrou, M.; Schleger, W. An extended chicken karyotype, including the NOR chromosome. Cytogenet. Cell Genet. 1987, 45, 218–221. [Google Scholar] [CrossRef] [PubMed]
- McQueen, H.A.; Siriaco, G.; Bird, A.P. Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res. 1998, 8, 621–630. [Google Scholar] [CrossRef]
- Smith, J.; Bruley, C.K.; Paton, I.R.; Dunn, I.; Jones, C.T.; Windsor, D.; Morrice, D.R.; Law, A.S.; Masabanda, J.; Sazanov, A.; et al. Differences in gene density on chicken macrochromosomes and microchromosomes. Anim. Genet. 2000, 31, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Andreozzi, L.; Federico, C.; Motta, S.; Saccone, S.; Sazanova, A.L.; Sazanov, A.A.; Smirnov, A.F.; Galkina, S.A.; Lukina, N.A.; Rodionov, A.V.; et al. Compositional mapping of chicken chromosomes and identification of the gene-richest regions. Chromosome Res. 2001, 9, 521–532. [Google Scholar] [CrossRef]
- Kuraku, S.; Ishijima, J.; Nishida-Umehara, C.; Agata, K.; Kuratani, S.; Matsuda, Y. cDNA-based gene mapping and GC3 profiling in the soft-shelled turtle suggest a chromosomal size-dependent GC bias shared by sauropsids. Chromosome Res. 2006, 14, 187–202. [Google Scholar] [CrossRef]
- Olmo, E. Rate of chromosome changes and speciation in reptiles. Genetica 2005, 125, 185–203. [Google Scholar] [CrossRef]
- Shedlock, A.M.; Botka, C.W.; Zhao, S.; Shetty, J.; Zhang, T.; Liu, J.S.; Deschavanne, P.J.; Edwards, S.V. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc. Natl. Acad. Sci. USA 2007, 104, 2767–2772. [Google Scholar] [CrossRef]
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.H.; Blackmon, H.; Reyes-Velasco, J.; Schield, D.R.; Card, D.C.; Andrew, A.L.; Waynewood, N.; Castoe, T.A. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 2016, 59, 295–310. [Google Scholar] [CrossRef]
- Kapusta, A.; Suh, A.; Feschotte, C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA 2017, 114, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Balaresque, P.; King, T.E.; Parkin, E.J.; Heyer, E.; Carvalho-Silva, D.; Kraaijenbrink, T.; de Knijff, P.; Tyler-Smith, C.; Jobling, M.A. Gene conversion violates the stepwise mutation model for microsatellites in Y-chromosomal palindromic repeats. Hum. Mutat. 2014, 35, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, L.; Macaulay, M.; Cardle, L.; Morgante, M.; Ivanissevich, S.d.; Maestri, E.; Powell, W.; Waugh, R. Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant. J. 1999, 17, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposon on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. Genome evolution in reptilia, the sister group of mammals. Annu. Rev. Genom. Hum. Genet. 2010, 11, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Figliuolo, V.S.P.; Goll, L.; Viana, P.F.; Feldberg, E.; Gross, M.C. First record on sex chromosomes in a species of the family Cynodontidae: Cynodon gibbus (Agassiz, 1829). Cytogenet Genome Res. 2020, 160, 29–37. [Google Scholar] [CrossRef]
- Pasquesi, G.I.M.; Adams, R.H.; Card, D.C.; Schield, D.R.; Corbin, A.B.; Perry, B.W.; Reyes-Velasco, J.; Ruggiero, R.P.; Vandewege, M.W.; Shortt, J.A.; et al. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat. Commun. 2018, 9, 2774. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Suntronpong, A.; Panthum, T.; Malaivijitnond, M.; Srikulnath, K. Dark matter of primate genomes: Satellite DNA repeats and their evolutionary dynamics. Cells 2020, 9, 2714. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Kovarik, A.; Matyasek, R.; Chase, M.W.; Clarkson, J.J.; Grandbastien, M.A.; Leitch, A.R. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 2007, 175, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Majka, J.; Majka, M.; Kwiatek, M.; Wiśniewska, H. Similarities and differences in the nuclear genome organization within Pooideae species revealed by comparative genomic in situ hybridization (GISH). J. Appl. Genet. 2017, 58, 151–161. [Google Scholar] [CrossRef]
- Barby, F.F.; Bertollo, L.A.C.; de Oliveira, E.A.; Yano, C.F.; Hatanaka, T.; Ráb, P.; Sember, A.; Ezaz, T.; Artoni, R.F.; Liehr, T.; et al. Emerging patterns of genome organization in Notopteridae species (Teleostei, Osteoglossiformes) as revealed by Zoo-FISH and Comparative Genomic Hybridization (CGH). Sci. Rep. 2019, 9, 1112. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.P.; Gharrett, A.J.; Wilmot, R.L.; Smoker, W.W. Genetic linkage mapping of allozyme loci in even- and odd-year pink salmon (Oncorhynchus gorbuscha). J. Hered. 2004, 95, 421–429. [Google Scholar] [CrossRef][Green Version]
- Mandáková, T.; Joly, S.; Krzywinski, M.; Mummenhoff, K.; Lysak, M.A. Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant. Cell 2010, 22, 2277–2290. [Google Scholar] [CrossRef] [PubMed]
- Bomfleur, B.; Decombeix, A.-L.; Schwendemann, A.B.; Escapa, I.H.; Taylor, E.L.; Taylor, T.N.; McLoughlin, S. Habit and ecology of the petriellales, an unusual group of seed plants from the Triassic of Gondwana. Int. J. Plant. Sci. 2014, 175, 1062–1075. [Google Scholar] [CrossRef]
- Samad, M.S.; Biswas, A.; Bakken, L.R.; Clough, T.J.; de Klein, C.A.M.; Richards, K.G.; Lanigan, G.J.; Morales, S.E. Phylogenetic and functional potential links pH and N2O emission in pasture soils. Sci Rep. 2016, 6, 35990. [Google Scholar] [CrossRef]
- Sessions, S.K.; Kezer, J. Evolutionary cytogenetics of Bolitoglossine salamanders (Family Plethodontidae). In Amphibian Cytogenetics and Evolution, 1st ed.; Green, D.M., Sessions, S.K., Eds.; San Diego Academic Press: San Diego, CA, USA, 1991; pp. 89–130. [Google Scholar] [CrossRef]
- Aprea, G.; Andreone, G.; Capriglione, T.; Odierna, V.; Vences, M. Evidence for a remarkable stasis of chromosome evolution in Malagasy tree-frogs (Boophis: Mantellidae). Ital. J. Zool. 2004, 2, 237–243. [Google Scholar] [CrossRef]
- Ellegren, H. Evolutionary stasis: The stable chromosome of birds. Trends Ecol. Evol. 2010, 25, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Molina, W.F. Chromosomal changes and stasis in marine fish groups. In Fish Cytogenetics, 1st ed.; Pisano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 69–110. [Google Scholar] [CrossRef]
- Gaffaroglu, M.; Majtánová, Z.; Symonová, R.; Pelikánová, S.; Unal, S.; Lajbner, Z.; Ráb, P. Present and future salmonid cytogenetics. Genes 2020, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
- Barby, F.F.; Ráb, P.; Lavoué, S.; Ezaz, T.; Bertollo, L.A.C.; Kilian, A.; Maruyama, S.R.; de Oliveira, E.A.; Artoni, R.A.; Santos, M.H. From chromosomes to genome: Insights into the evolutionary relationships and biogeography of Old World knifefishes (Notopteridae; Osteoglossiformes). Genes 2018, 9, 306. [Google Scholar] [CrossRef]
- Wake, D.B.; Roth, G.; Wake, M.H. On the problem of stasis in organismal evolution. J. Theor. Biol. 1983, 101, 211–224. [Google Scholar] [CrossRef]
- Stockdale, M.T.; Benton, M.J. Environmental drivers of body size evolution in crocodile-line archosaurs. Commun. Biol. 2021, 4, 38. [Google Scholar] [CrossRef]
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; Cambridge University Press: Cambridge, UK, 1973; p. 468. [Google Scholar]
- King, M. Chromosome change and speciation in lizards. In Essays on Evolution and Speciation in Honour of M. J. D., 1st ed.; Atchley, W.R., Woodruff, D.S., Eds.; Cambridge University Press: London, UK, 1981; pp. 262–285. [Google Scholar]
- White, M.J.D. Modes of Speciation, 1st ed.; W.R. Freeman and Company: San Francisco, CA, USA, 1978. [Google Scholar]
- King, M. Species Evolution: The Role of Chromosome Change, 3rd ed.; Cambridge University Press: Cambridge, UK, 1995; p. 322. [Google Scholar]
- Potter, S.; Bragg, J.G.; Blom, M.P.K.; Deakin, J.E.; Kirkpatrick, M.; Eldridge, M.D.B.; Moritz, C. Chromosomal speciation in the genomics era: Disentangling phylogenetic evolution of rock-wallabies. Front. Genet. 2017, 8, 10. [Google Scholar] [CrossRef]
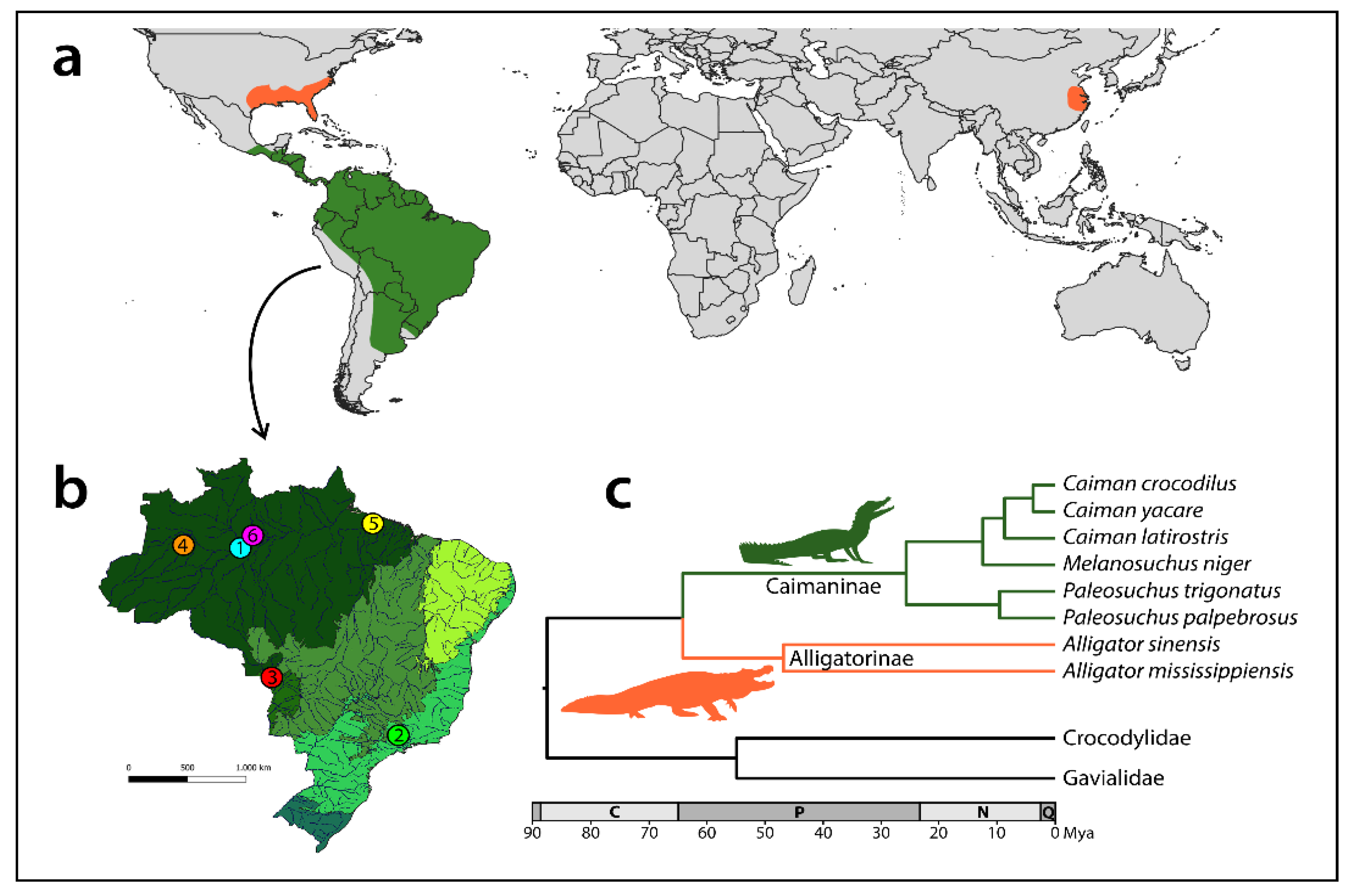


| Species | N | Locality/Origin of Samples | |
|---|---|---|---|
| ① Caiman crocodilus (Spectacled caiman) | 2♀, 2♂ | Amazonas (BR) (Amazon Basin) | 3°22′34.7″ S 60°19′20.7″ W |
| ② Caiman latirostris (Broad-snouted caiman) | 4♀, 6♂ | São Paulo (BR) (Cerrado) | 22°33′53.1″ S 48°00′35.2″ W |
| ③ Caiman yacare (Yacare caiman) | 2♀, 8♂ | Mato Grosso (BR) (Pantanal) | 16°19′32.0″ S 57°46′35.7″ W |
| ④ Melanosuchus niger (Black caiman) | 2♀, 2♂ | Amazonas (BR) (Amazon Basin) | 3°25′50.4″ S 66°02′35.0″ W |
| ⑤ Paleosuchus palpebrosus (Cuvier’s dwarf caiman) | 3♀, 3♂ | Pará (BR) (Amazon Basin) | 1°18′19.7″ S 48°19′05.0″ W |
| ⑥ Paleosuchus trigonatus (Schneider’s smooth-fronted caiman) | 3♀, 4♂ | Amazonas (BR) (Amazon Basin) | 3°06′52.0″ S 60°01′58.0″ W |
| Alligator mississippiensis (American alligator) | 2♀, 2♂ | Canberra University collection (Australia) | |
| Alligator sinensis (Chinese alligator) | 4♀, 1♂, 1 unsexed | Private collections (Germany) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, V.C.S.; Altmanová, M.; Viana, P.F.; Ezaz, T.; Bertollo, L.A.C.; Ráb, P.; Liehr, T.; Al-Rikabi, A.; Feldberg, E.; Hatanaka, T.; et al. Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles. Cells 2021, 10, 1397. https://doi.org/10.3390/cells10061397
Oliveira VCS, Altmanová M, Viana PF, Ezaz T, Bertollo LAC, Ráb P, Liehr T, Al-Rikabi A, Feldberg E, Hatanaka T, et al. Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles. Cells. 2021; 10(6):1397. https://doi.org/10.3390/cells10061397
Chicago/Turabian StyleOliveira, Vanessa C. S., Marie Altmanová, Patrik F. Viana, Tariq Ezaz, Luiz A. C. Bertollo, Petr Ráb, Thomas Liehr, Ahmed Al-Rikabi, Eliana Feldberg, Terumi Hatanaka, and et al. 2021. "Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles" Cells 10, no. 6: 1397. https://doi.org/10.3390/cells10061397
APA StyleOliveira, V. C. S., Altmanová, M., Viana, P. F., Ezaz, T., Bertollo, L. A. C., Ráb, P., Liehr, T., Al-Rikabi, A., Feldberg, E., Hatanaka, T., Scholz, S., Meurer, A., & de Bello Cioffi, M. (2021). Revisiting the Karyotypes of Alligators and Caimans (Crocodylia, Alligatoridae) after a Half-Century Delay: Bridging the Gap in the Chromosomal Evolution of Reptiles. Cells, 10(6), 1397. https://doi.org/10.3390/cells10061397







