Transcriptionally Active Chromatin—Lessons Learned from the Chicken Erythrocyte Chromatin Fractionation
Abstract
1. Introduction
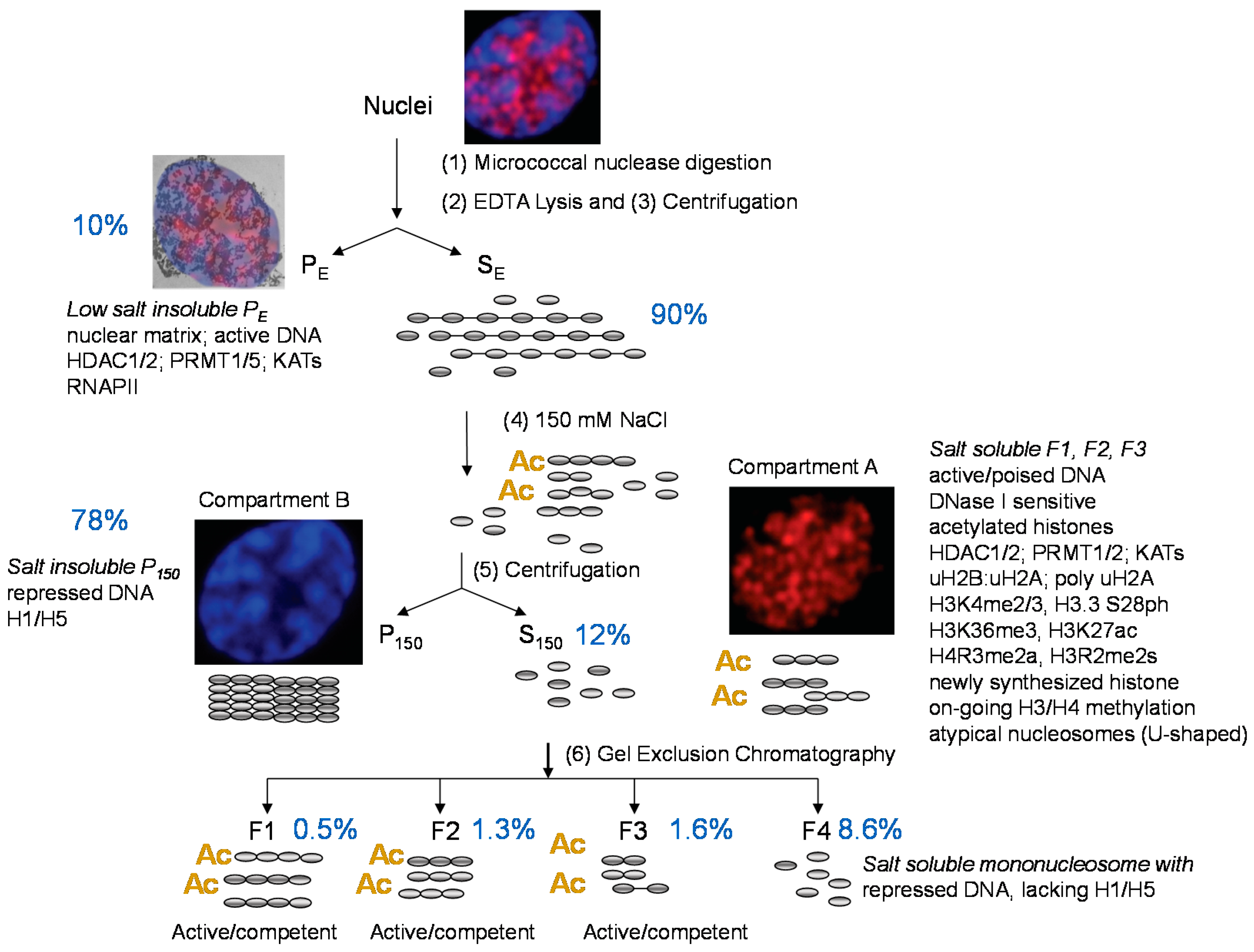
2. Enrichment of Transcriptionally Active/Competent Chromatin
3. Dynamically Acetylated Histones, DNase I Sensitivity and Salt Solubility
4. Phase Separation
5. F1 DNA Sequences and the Polychromatic Erythrocyte Transcriptome
6. Structure of Erythroid Transcriptionally Active Chromatin
7. Histone Post-Translational Modifications and Variants
8. Accessible Chromatin and Regulatory Regions
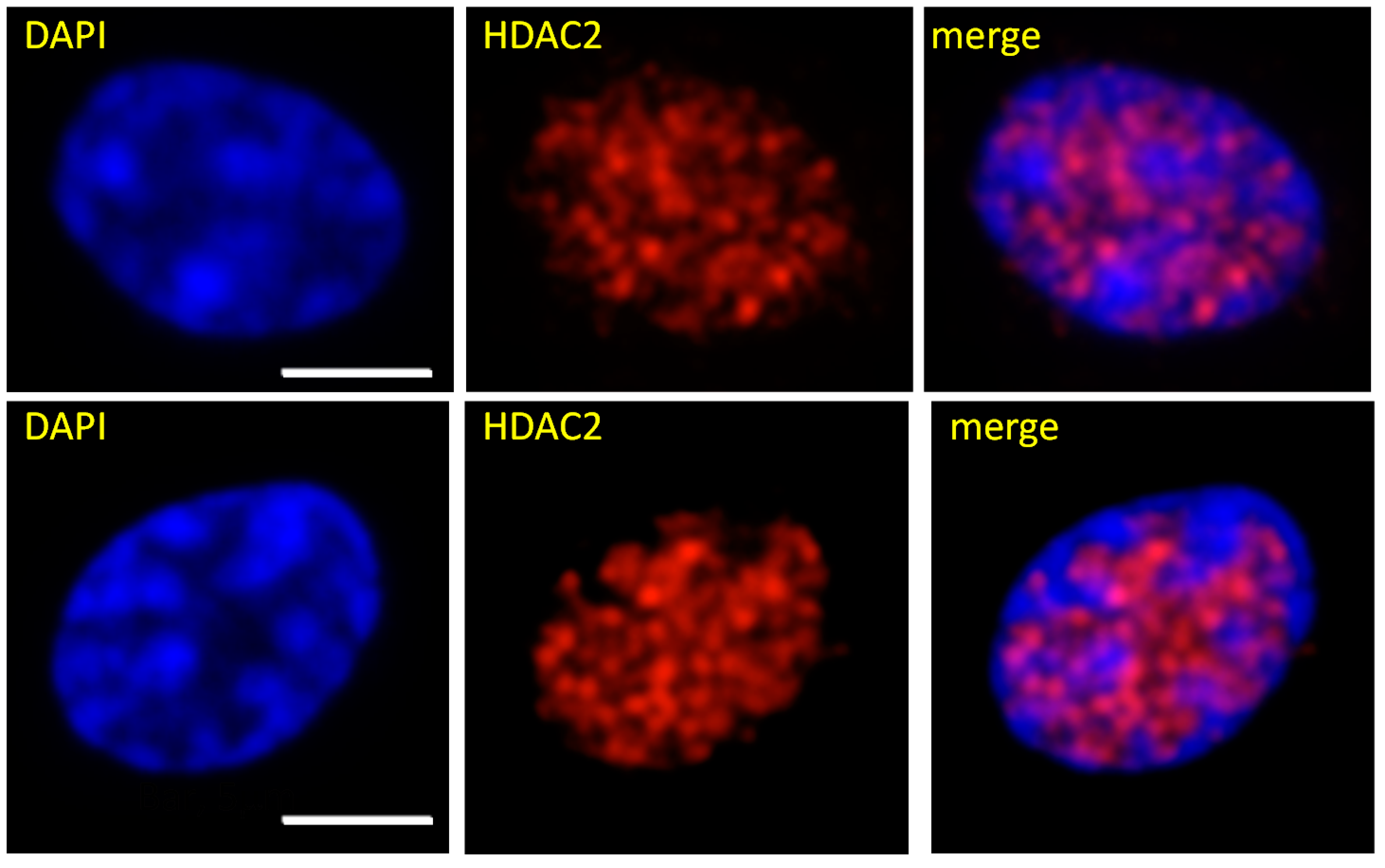
9. Chromatin Modifying Enzymes and Nuclear Location
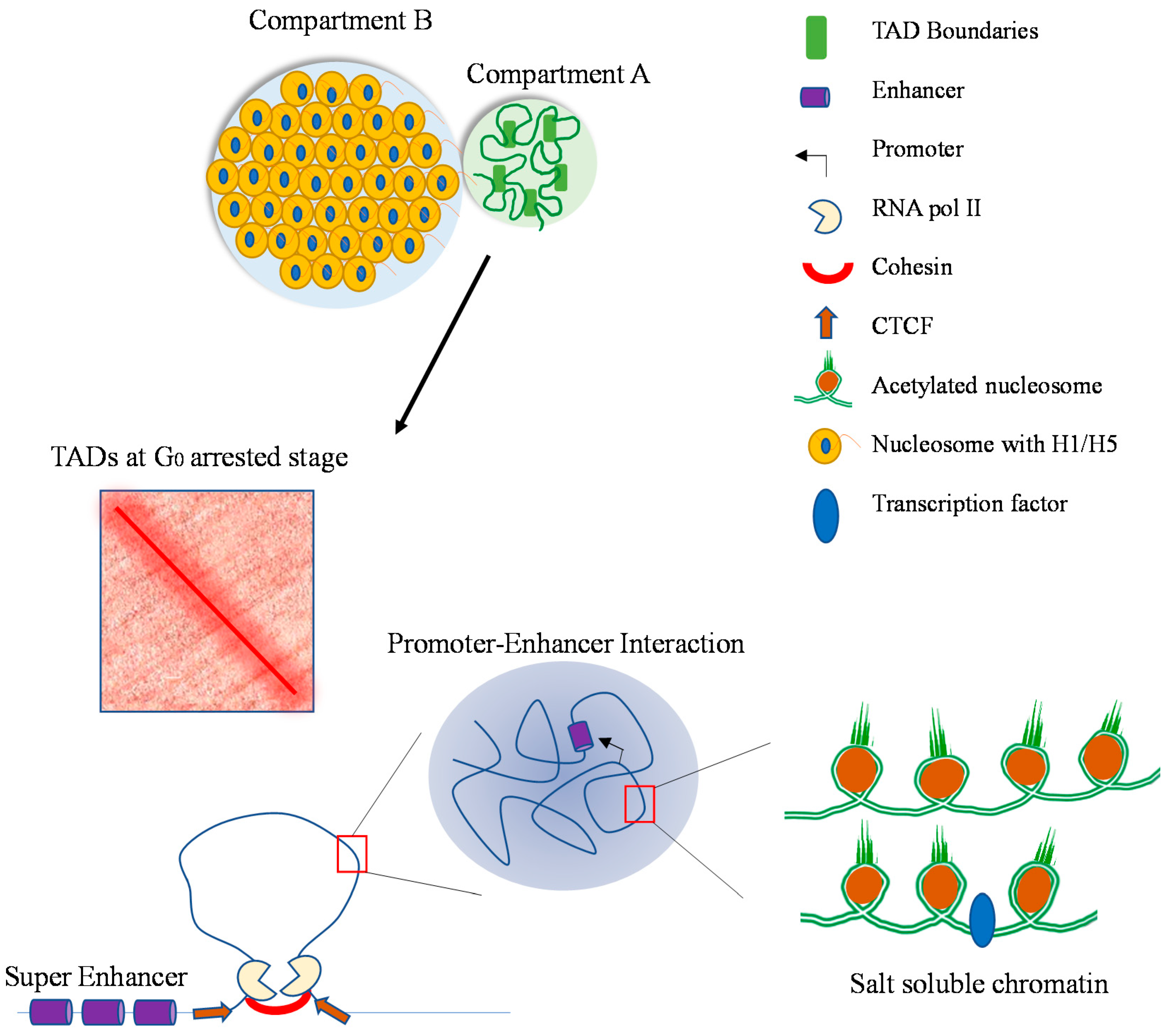
10. Compartment A and B Location in the Erythroid Nucleus
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beacon, M.T.H.; Davie, J.R. The chicken model organism for epigenomic research. Genome 2021, 64, 476–489. [Google Scholar] [CrossRef]
- Jahan, S.; Xu, W.; He, S.; González, C.; Delcuve, G.P.; Davie, J.R. The chicken erythrocyte epigenome. Epigenetics Chromatin 2016, 9, 19. [Google Scholar] [CrossRef]
- Weintraub, H.; Groudine, M. Chromosomal subunits in active genes have an altered conformation. Science 1976, 193, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Lawson, G.; Knoll, B.; March, C.; Woo, S.; Tsai, M.; O’Malley, B. Definition of 5′ and 3′ structural boundaries of the chromatin domain containing the ovalbumin multigene family. J. Biol. Chem. 1982, 257, 1501–1507. [Google Scholar] [CrossRef]
- Stalder, J.; Larsen, A.; Engel, J.D.; Dolan, M.; Groudine, M.; Weintraub, H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 1980, 20, 451–460. [Google Scholar] [CrossRef]
- Villeponteau, B.; Lundell, M.; Martinson, H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell 1984, 39, 469–478. [Google Scholar] [CrossRef]
- Villeponteau, B.; Martinson, H.G. Gamma rays and bleomycin nick DNA and reverse the DNase I sensitivity of beta-globin gene chromatin in vivo. Mol. Cell. Biol. 1987, 7, 1917–1924. [Google Scholar] [CrossRef]
- Hebbes, T.; Clayton, A.; Thorne, A.; Crane-Robinson, C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994, 13, 1823–1830. [Google Scholar] [CrossRef]
- Hebbes, T.R.; Thorne, A.W.; Crane-Robinson, C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988, 7, 1395–1402. [Google Scholar] [CrossRef]
- Bates, D.L.; Thomas, J.O. Histories H1 and H5: One or two molecules per nucleosome? Nucleic Acids Res. 1981, 9, 5883–5894. [Google Scholar] [CrossRef]
- Ridsdale, J.A.; Hendzel, M.J.; Delcuve, G.P.; Davie, J.R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J. Biol. Chem. 1990, 265, 5150–5156. [Google Scholar] [CrossRef]
- Misteli, T. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell 2020, 183, 28–45. [Google Scholar] [CrossRef]
- Fishman, V.; Battulin, N.; Nuriddinov, M.; Maslova, A.; Zlotina, A.; Strunov, A.; Chervyakova, D.; Korablev, A.; Serov, O.; Krasikova, A. 3D organization of chicken genome demonstrates evolutionary conservation of topologically associated domains and highlights unique architecture of erythrocytes’ chromatin. Nucleic Acids Res. 2019, 47, 648–665. [Google Scholar] [CrossRef] [PubMed]
- Ridsdale, J.A.; Davie, J.R. Selective solubilization of. beta.-globin oligonucleosomes at low ionic strength. Biochemistry 1987, 26, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Morera, D.; MacKenzie, S.A. Is there a direct role for erythrocytes in the immune response? Vet. Res. 2011, 42, 89. [Google Scholar] [CrossRef]
- St Paul, M.; Paolucci, S.; Barjesteh, N.; Wood, R.D.; Sharif, S. Chicken erythrocytes respond to Toll-like receptor ligands by up-regulating cytokine transcripts. Res. Vet. Sci. 2013, 95, 87–91. [Google Scholar] [CrossRef]
- Williams, A.F. Dna Synthesis in Purified Populations of Avian Erythroid Cells. J. Cell Sci. 1972, 10, 27–46. [Google Scholar] [CrossRef]
- Rocha, E.; Davie, J.R.; van Holde, K.E.; Weintraub, H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J. Biol. Chem. 1984, 259, 8558–8563. [Google Scholar] [CrossRef]
- Davie, J.; Saunders, C. Chemical composition of nucleosomes among domains of calf thymus chromatin differing in micrococcal nuclease accessibility and solubility properties. J. Biol. Chem. 1981, 256, 12574–12580. [Google Scholar] [CrossRef]
- Ridsdale, J.; Davie, J. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched hi β-globin gene sequences. Nucleic Acids Res. 1987, 15, 1081–1096. [Google Scholar] [CrossRef]
- Jahan, S.; Sun, J.-M.; He, S.; Davie, J.R. Transcription-dependent association of HDAC2 with active chromatin. J. Cell. Physiol. 2018, 233, 1650–1657. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Davie, J.R. Chromatin structure of erythroid-specific genes of immature and mature chicken erythrocytes. Biochem. J. 1989, 263, 179–186. [Google Scholar] [CrossRef]
- Ridsdale, J.A.; Rattner, J.B.; Davie, J.R. Erythroid-specific gene chromatin has an altered association with linker histones. Nucleic Acids Res. 1988, 16, 5915–5926. [Google Scholar] [CrossRef]
- Jahan, S. Characterization of Transcriptionally Active Chicken Erythrocyte Chromatin. Available online: https://mspace.lib.umanitoba.ca/handle/1993/32700 (accessed on 19 June 2020).
- Teves, S.S.; Henikoff, S. Salt Fractionation of Nucleosomes for Genome-Wide Profiling. Methods Mol. Biol. 2012, 833, 421–432. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.P.; Turner, B.M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995, 14, 3946–3957. [Google Scholar] [CrossRef]
- Zhang, D.E.; Nelson, D.A. Histone acetylation in chicken erythrocytes. Rates of deacetylation in immature and mature red blood cells. Biochem. J. 1988, 250, 241–245. [Google Scholar] [CrossRef]
- Zhang, D.E.; Nelson, D.A. Histone acetylation in chicken erythrocytes. Rates of acetylation and evidence that histones in both active and potentially active chromatin are rapidly modified. Biochem. J. 1988, 250, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, S.; Landsman, D.; Panchenko, A.R. Histone tails as signaling antennas of chromatin. Curr. Opin. Struct. Biol. 2021, 67, 153–160. [Google Scholar] [CrossRef]
- Peng, Z.; Mizianty, M.J.; Xue, B.; Kurgan, L.; Uversky, V.N. More than just tails: Intrinsic disorder in histone proteins. Mol. BioSyst. 2012, 8, 1886–1901. [Google Scholar] [CrossRef]
- Wang, X.; Moore, S.C.; Laszckzak, M.; Ausió, J. Acetylation Increases the α-Helical Content of the Histone Tails of the Nucleosome. J. Biol. Chem. 2000, 275, 35013–35020. [Google Scholar] [CrossRef] [PubMed]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Ebralidse, K.K.; Grachev, S.A.; Mirzabekov, A.D. A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nat. Cell Biol. 1988, 331, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.T. Structure of chromatin containing extensively acetylated H3 and H4. Cell 1978, 13, 691–699. [Google Scholar] [CrossRef]
- Hao, F.; Murphy, K.J.; Kujirai, T.; Kamo, N.; Kato, J.; Koyama, M.; Okamato, A.; Hayashi, G.; Kurumizaka, H.; Hayes, J.J. Acetylation-modulated communication between the H3 N-terminal tail domain and the intrinsically disordered H1 C-terminal domain. Nucleic Acids Res. 2020, 48, 11510–11520. [Google Scholar] [CrossRef] [PubMed]
- Delcuve, G.P.; Moyer, R.; Bailey, G.; Davie, J.R. Gene-specific differences in the aflatoxin B1 adduction of chicken erythrocyte chromatin. Cancer Res. 1988, 48, 7146–7149. [Google Scholar]
- Locklear, L.; Ridsdale, A.J.; Bazett-Jones, D.P.; Davie, J.R. Ultrastructure of transcriptionally competent chromatin. Nucleic Acids Res. 1990, 18, 7015–7024. [Google Scholar] [CrossRef] [PubMed]
- Allegra, P.; Sterner, R.; Clayton, D.F.; Allfrey, V.G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J. Mol. Biol. 1987, 196, 379–388. [Google Scholar] [CrossRef]
- Chen-Cleland, T.; Boffa, L.; Carpaneto, E.; Mariani, M.; Valentin, E.; Mendez, E.; Allfrey, V. Recovery of transcriptionally active chromatin restriction fragments by binding to organomercurial-agarose magnetic beads. A rapid and sensitive method for monitoring changes in higher order chromatin structure during gene activation and repression. J. Biol. Chem. 1993, 268, 23409–23416. [Google Scholar] [CrossRef]
- Walia, H.; Chen, H.Y.; Sun, J.-M.; Holth, L.T.; Davie, J.R. Histone Acetylation Is Required to Maintain the Unfolded Nucleosome Structure Associated with Transcribing DNA. J. Biol. Chem. 1998, 273, 14516–14522. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Davie, J.R. Nucleosomal histones of transcriptionally active/competent chromatin preferentially exchange with newly synthesized histones in quiescent chicken erythrocytes. Biochem. J. 1990, 271, 67–73. [Google Scholar] [CrossRef]
- Li, W.; Nagaraja, S.; Delcuve, G.P.; Hendzel, M.J.; Davie, J.R. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. Biochem. J. 1993, 296, 737–744. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Davie, J.R. Western blotting and immunochemical detection of histones electrophoretically resolved on acid-urea-Triton- and sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 1992, 200, 339–341. [Google Scholar] [CrossRef]
- Nickel, B.E.; Allis, C.D.; Davie, J.R. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry 1989, 28, 958–963. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Espino, P.S.; Davie, J.R. Phosphorylated serine 28 of histone H3 is associated with destabilized nucleosomes in transcribed chromatin. Nucleic Acids Res. 2007, 35, 6640–6647. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Davie, J.R. Dynamically acetylated histones of chicken erythrocytes are selectively methylated. Biochem. J. 1991, 273, 753–758. [Google Scholar] [CrossRef]
- Beacon, T.H.; Xu, W.; Davie, J.R. Genomic landscape of transcriptionally active histone arginine methylation marks, H3R2me2s and H4R3me2a, relative to nucleosome depleted regions. Gene 2020, 742, 144593. [Google Scholar] [CrossRef]
- Davie, J.R.; Xu, W.; Delcuve, G.P. Histone H3K4 trimethylation: Dynamic interplay with pre-mRNA splicing. Biochem. Cell Biol. 2016, 94, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bieberstein, N.I.; Oesterreich, F.C.; Straube, K.; Neugebauer, K.M. First Exon Length Controls Active Chromatin Signatures and Transcription. Cell Rep. 2012, 2, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Pollina, E.A.; Ucar, D.; Mahmoudi, S.; Karra, K.; Wong, E.D.; Devarajan, K.; Daugherty, A.C.; Kundaje, A.; Mancini, E.; et al. H3K4me3 Breadth Is Linked to Cell Identity and Transcriptional Consistency. Cell 2014, 158, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, Z.; Wu, D.; Zhang, L.; Lin, X.; Su, J.; Rodriguez, B.; Xi, Y.; Xia, Z.; Chen, X.; et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 2015, 47, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Fang, Y.; Tan, H.K.; Goh, Y.; Choy, J.Y.H.; Koh, B.T.H.; Tan, J.H.; Bertin, N.; Ramadass, A.; Hunter, E.; et al. Super-Enhancers and Broad H3K4me3 Domains Form Complex Gene Regulatory Circuits Involving Chromatin Interactions. Sci. Rep. 2017, 7, 2186. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, A.; Márquez, E.J.; Shin, D.-G.; Vera-Licona, P.; Ucar, D. Chromatin interaction networks revealed unique connectivity patterns of broad H3K4me3 domains and super enhancers in 3D chromatin. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Beacon, T.H.; Xu, W.; Davie, J.R. Atypical chromatin structure of immune-related genes expressed in chicken erythrocytes. Biochem. Cell Biol. 2020, 98, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.R.; Murphy, L.C. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 1990, 29, 4752–4757. [Google Scholar] [CrossRef]
- Davie, J.; Murphy, L.C. Inhibition of Transcription Selectively Reduces the Level of Ubiquitinated Histone H2B in Chromatin. Biochem. Biophys. Res. Commun. 1994, 203, 344–350. [Google Scholar] [CrossRef]
- Minsky, N.; Shema, E.; Field, Y.; Schuster, M.; Segal, E.; Oren, M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008, 10, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Beacon, T.H.; He, S.; Gonzalez, C.; Xu, W.; Delcuve, G.P.; Jia, S.; Hu, P.; Davie, J.R. Chromatin organization of transcribed genes in chicken polychromatic erythrocytes. Gene 2019, 699, 80–87. [Google Scholar] [CrossRef]
- Sun, J.-M.; Ferraiuolo, R.; Davie, J.R. In situ footprinting of chicken histone H5 gene in mature and immature erythrocytes reveals common factor-binding sites. Chromosoma 1996, 104, 504–510. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Davie, J.R. Nuclear factor 1 is a component of the nuclear matrix. J. Cell. Biochem. 1994, 55, 252–263. [Google Scholar] [CrossRef]
- Hendzel, M.; Delcuve, G.; Davie, J. Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem. 1991, 266, 21936–21942. [Google Scholar] [CrossRef]
- Spencer, V.A.; Davie, J.R. Dynamically Acetylated Histone Association with Transcriptionally Active and Competent Genes in the Avian Adult β-Globin Gene Domain. J. Biol. Chem. 2001, 276, 34810–34815. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Davie, J.R. Nuclear distribution of histone deacetylase: A marker enzyme for the internal nuclear matrix. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1992, 1130, 307–313. [Google Scholar] [CrossRef]
- Hendzel, M.; Sun, J.; Chen, H.; Rattner, J.; Davie, J. Histone acetyltransferase is associated with the nuclear matrix. J. Biol. Chem. 1994, 269, 22894–22901. [Google Scholar] [CrossRef]
- Li, W.; Chen, H.Y.; Davie, J.R. Properties of chicken erythrocyte histone deacetylase associated with the nuclear matrix. Biochem. J. 1996, 314, 631–637. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Moniwa, M.; Samuel, S.; Davie, J.R. Purification and Characterization of Chicken Erythrocyte Histone Deacetylase 1. Biochemistry 1999, 38, 5939–5947. [Google Scholar] [CrossRef] [PubMed]
- Heinzel, T.; Lavinsky, R.M.; Mullen, T.-M.; Söderström, M.; Laherty, C.D.; Torchia, J.A.; Yang, W.-M.; Brard, G.; Ngo, S.D.; Davie, J.R.; et al. A complex containing N-CoR, mSln3 and histone deacetylase mediates transcriptional repression. Nature 1997, 387, 43–48. [Google Scholar] [CrossRef]
- Laherty, C.D.; Yang, W.-M.; Sun, J.-M.; Davie, J.R.; Seto, E.; Eisenman, R.N. Histone Deacetylases Associated with the mSin3 Corepressor Mediate Mad Transcriptional Repression. Cell 1997, 89, 349–356. [Google Scholar] [CrossRef]
- Laherty, C.D.; Billin, A.N.; Lavinsky, R.M.; Yochum, G.S.; Bush, A.C.; Sun, J.-M.; Mullen, T.-M.; Davie, J.R.; Rose, D.W.; Glass, C.K.; et al. SAP30, a Component of the mSin3 Corepressor Complex Involved in N-CoR-Mediated Repression by Specific Transcription Factors. Mol. Cell 1998, 2, 33–42. [Google Scholar] [CrossRef]
- Khan, D.H.; Gonzalez, C.; Cooper, C.; Sun, J.-M.; Chen, H.Y.; Healy, S.; Xu, W.; Smith, K.T.; Workman, J.L.; Leygue, E.; et al. RNA-dependent dynamic histone acetylation regulates MCL1 alternative splicing. Nucleic Acids Res. 2013, 42, 1656–1670. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Moniwa, M.; Litchfield, D.W.; Seto, E.; Davie, J.R. The Transcriptional Repressor Sp3 Is Associated with CK2-phosphorylated Histone Deacetylase 2. J. Biol. Chem. 2002, 277, 35783–35786. [Google Scholar] [CrossRef]
- Tsai, S.-C.; Seto, E. Regulation of Histone Deacetylase 2 by Protein Kinase CK2. J. Biol. Chem. 2002, 277, 31826–31833. [Google Scholar] [CrossRef]
- Pflum, M.K.H.; Tong, J.K.; Lane, W.S.; Schreiber, S.L. Histone Deacetylase 1 Phosphorylation Promotes Enzymatic Activity and Complex Formation. J. Biol. Chem. 2001, 276, 47733–47741. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Davie, J.R. Differential Distribution of Unmodified and Phosphorylated Histone Deacetylase 2 in Chromatin. J. Biol. Chem. 2007, 282, 33227–33236. [Google Scholar] [CrossRef]
- Spencer, V.A. Chromatin immunoprecipitation: A tool for studying histone acetylation and transcription factor binding. Methods 2003, 31, 67–75. [Google Scholar] [CrossRef]
- Khan, D.H.; Jahan, S.; Davie, J.R. Pre-mRNA Splicing: Role of Epigenetics and Implications in Disease; Elsevier Ltd.: Amsterdam, The Netherlands, 2012; Volume 52, pp. 377–388. [Google Scholar]
- Liao, Y.; Lupiani, B.; Ai-Mahmood, M.; Reddy, S.M. Marek’s disease virus US3 protein kinase phosphorylates chicken HDAC 1 and 2 and regulates viral replication and pathogenesis. PLoS Pathog. 2021, 17, e1009307. [Google Scholar] [CrossRef]
- Li, X.; Hu, X.; Patel, B.; Zhou, Z.; Liang, S.; Ybarra, R.; Qiu, Y.; Felsenfeld, G.; Bungert, J.; Huang, S. H4R3 methylation facilitates β-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 2010, 115, 2028–2037. [Google Scholar] [CrossRef]
- Huang, S.; Litt, M.; Felsenfeld, G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005, 19, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, N.; Weintraub, H. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell 1985, 43, 471–482. [Google Scholar] [CrossRef]
- Kantidze, O.; Iarovaia, O.V.; Philonenko, E.S.; Yakutenko, I.I.; Razin, S.V. Unusual compartmentalization of CTCF and other transcription factors in the course of terminal erythroid differentiation. Biochim. Biophys. Acta (BBA) Bioenerg. 2007, 1773, 924–933. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, J.; Wiaderkiewicz, R.; Ruiz-Carrillo, A. Histone H5 in the control of DNA synthesis and cell proliferation. Science 1989, 245, 68–71. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beacon, T.H.; Davie, J.R. Transcriptionally Active Chromatin—Lessons Learned from the Chicken Erythrocyte Chromatin Fractionation. Cells 2021, 10, 1354. https://doi.org/10.3390/cells10061354
Beacon TH, Davie JR. Transcriptionally Active Chromatin—Lessons Learned from the Chicken Erythrocyte Chromatin Fractionation. Cells. 2021; 10(6):1354. https://doi.org/10.3390/cells10061354
Chicago/Turabian StyleBeacon, Tasnim H., and James R. Davie. 2021. "Transcriptionally Active Chromatin—Lessons Learned from the Chicken Erythrocyte Chromatin Fractionation" Cells 10, no. 6: 1354. https://doi.org/10.3390/cells10061354
APA StyleBeacon, T. H., & Davie, J. R. (2021). Transcriptionally Active Chromatin—Lessons Learned from the Chicken Erythrocyte Chromatin Fractionation. Cells, 10(6), 1354. https://doi.org/10.3390/cells10061354





