Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical History
2.2. Collection and Culture of Breast Tumor-Derived Cells
2.3. Cell Lines Culture
2.4. Mycoplasma Assay
2.5. Scanning Electron Microscopy (SEM)
2.6. Transmission Electron Microscopy (TEM)
2.7. Cell Growth Curve
2.8. Wound Healing Scratch Assay
2.9. Immunocytochemistry (ICC)
2.10. Colony Formation Assay
2.11. Western Blot Analysis
2.12. Human Proteome Profiler Arrays
2.13. Sample Preparation and Analysis of Real-Time qPCR
2.14. MTT Cell Proliferation Assay
2.15. Flow Cytometry Analysis
2.16. Karyotyping
2.17. Histopathology
2.18. Mice Model
3. Results
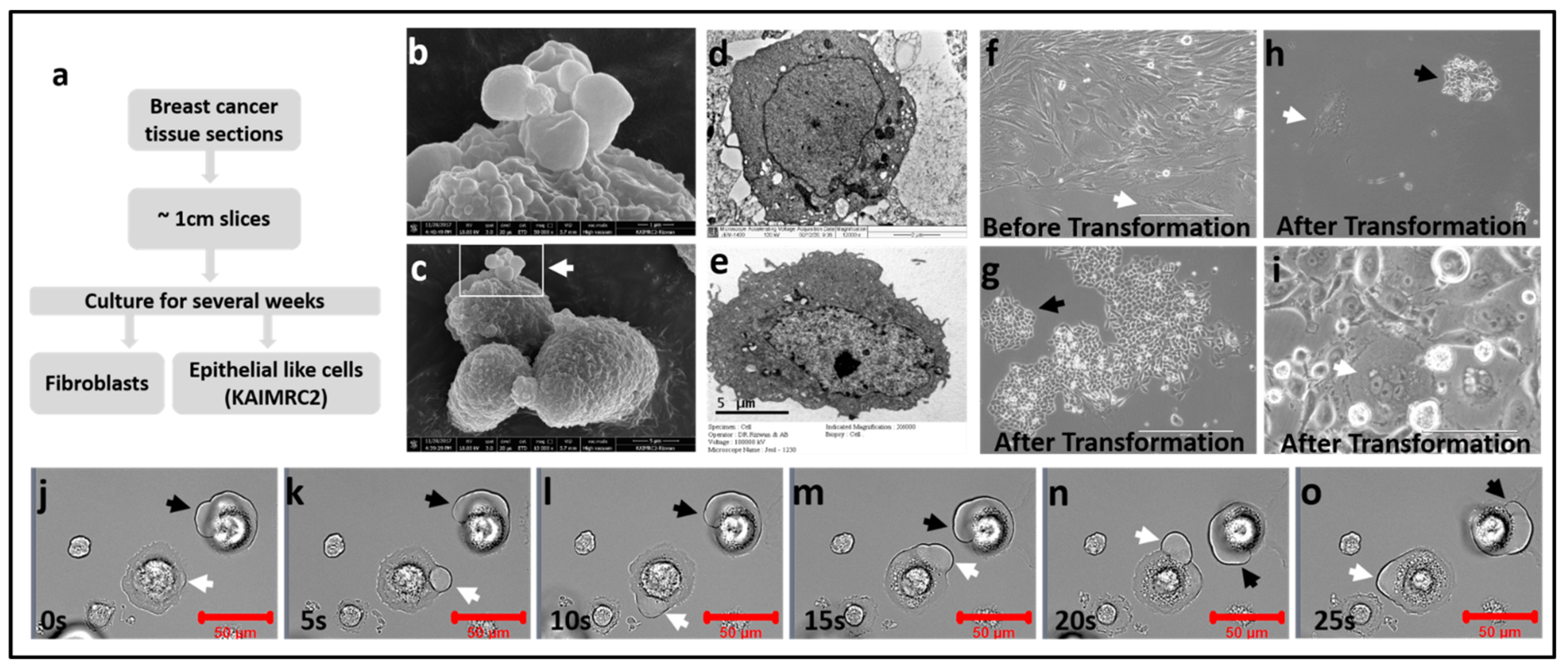
3.1. Isolation and Establishment of KAIMRC2 Cell Line
3.2. Growth Characteristics
3.3. Wound-Healing Assay
3.4. Colony Formation Assay
3.5. Chromosome Analysis
3.6. Immunophenotyping
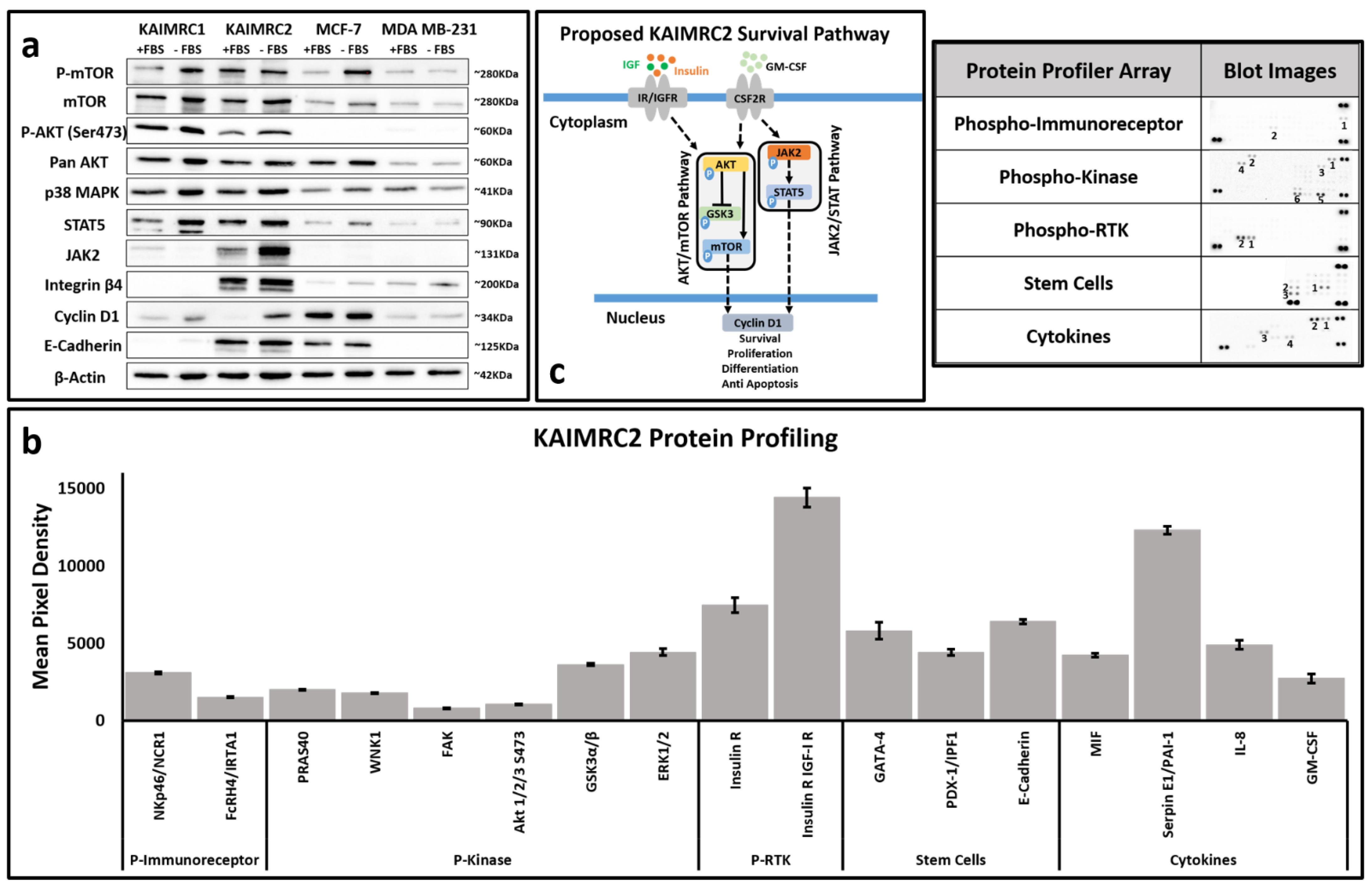
3.7. Molecular Characterization of KAIMRC2 Cell Line
3.8. Phospho-Protein Profiling
3.9. Drug Sensitivity
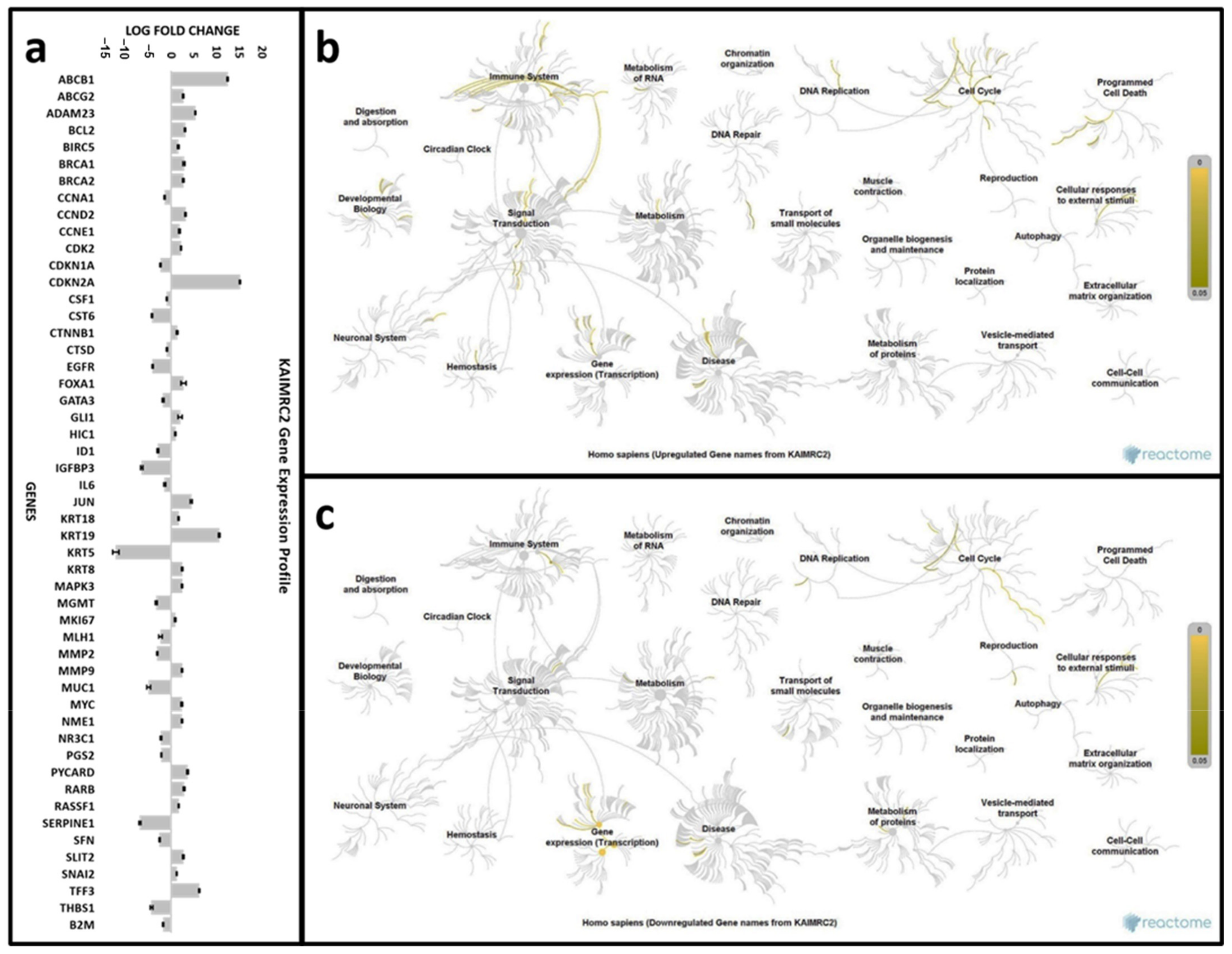
3.10. Gene Expression Analysis
3.11. In Vivo Tumorigenicity and Stemness
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Global Cancer Facts & Figures, 4th ed.; American Cancer Society: Atlanta, GA, USA, 2018; pp. 1–76. [Google Scholar]
- Mcpherson, K.; Steel, C.M.; Dixon, J.M. Breast cancer—Epidemiology, risk factors, and genetics Risk factors for breast cancer. BMJ 2000, 321, 624–628. [Google Scholar] [CrossRef]
- Chopra, S.; Davies, E.L. Breast cancer. Medicine 2020, 48, 113–118. [Google Scholar] [CrossRef]
- Saudi Cancer Registry. Saudi Cancer Registry Cancer Incidence Report Saudi Arabia, 2014; Saudi Health Council: Riyadh, Saudi Arabia, 2017. [Google Scholar]
- Althubiti, M.A.; Eldein, M.M.N. Trends in the incidence and mortality of cancer in Saudi Arabia. Saudi Med. J. 2018, 39, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Dutil, J.; Chen, Z.; Monteiro, A.N.; Teer, J.K.; Eschrich, S.A. An Interactive Resource to Probe Genetic Diversity and Estimated Ancestry in Cancer Cell Lines. Cancer Res. 2019, 79, 1263–1273. [Google Scholar] [CrossRef]
- Kondo, T.; Setoguchi, T.; Taga, T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc. Natl. Acad. Sci. USA 2004, 101, 781–786. [Google Scholar] [CrossRef]
- Phi, L.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 5416923. [Google Scholar] [CrossRef]
- Bighetti-Trevisan, R.L.; Sousa, L.O.; Castilho, R.M.; Almeida, L.O. Cancer Stem Cells: Powerful Targets to Improve Current Anticancer Therapeutics. Stem Cells Int. 2019, 2019, 9618065. [Google Scholar] [CrossRef]
- Kim, W.T.; Ryu, C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017, 50, 285–298. [Google Scholar] [CrossRef]
- Ali, R.; Samman, N.; Al Zahrani, H.; Nehdi, A.; Rahman, S.; Khan, A.L.; Al Balwi, M.; Alriyees, L.A.; Alzaid, M.; Al Askar, A.; et al. Isolation and characterization of a new naturally immortalized human breast carcinoma cell line, KAIMRC1. BMC Cancer 2017, 17, 803. [Google Scholar] [CrossRef]
- Kodack, D.P.; Farago, A.F.; Dastur, A.; Held, M.A.; Dardaei, L.; Friboulet, L.; Von Flotow, F.; Damon, L.J.; Lee, D.; Parks, M.; et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017, 21, 3298–3309. [Google Scholar] [CrossRef]
- Howe, B.; Umrigar, A.; Tsien, F. Chromosome Preparation from Cultured Cells. J. Vis. Exp. 2014, 3–7, e50203. [Google Scholar] [CrossRef]
- Burkholder, T.; Foltz, C.; Karlsson, E.; Linton, C.G.; Smith, J.M. Health Evaluation of Experimental Laboratory Mice. Curr. Protoc. Mouse Biol. 2012, 2, 145–165. [Google Scholar] [CrossRef]
- Barhoumi, T.; Fraulob-Aquino, J.C.; Mian, M.O.R.; Ouerd, S.; Idris-Khodja, N.; Huo, K.-G.; Rehman, A.; Caillon, A.; Dancose-Giambattisto, B.; Ebrahimian, T.; et al. Matrix metalloproteinase-2 knockout prevents angiotensin II-induced vascular injury. Cardiovasc. Res. 2017, 113, 1753–1762. [Google Scholar] [CrossRef]
- Nitulescu, G.M.; Van De Venter, M.; Nitulescu, G.; Ungurianu, A.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Grădinaru, D.; Tsatsakis, A.; Tsoukalas, D.; et al. The Akt pathway in oncology therapy and beyond (Review). Int. J. Oncol. 2018, 53, 2319–2331. [Google Scholar] [CrossRef]
- Faes, S.; Dormond, O. PI3K and AKT: Unfaithful Partners in Cancer. Int. J. Mol. Sci. 2015, 16, 21138–21152. [Google Scholar] [CrossRef]
- Hiraga, T.; Ito, S.; Nakamura, H. EpCAM expression in breast cancer cells is associated with enhanced bone metastasis formation. Int. J. Cancer 2016, 138, 1698–1708. [Google Scholar] [CrossRef]
- Yao, J.; Jin, Q.; Wang, X.-D.; Zhu, H.-J.; Ni, Q.-C. Aldehyde dehydrogenase 1 expression is correlated with poor prognosis in breast cancer. Medicine 2017, 96, e7171. [Google Scholar] [CrossRef]
- Li, J.-Y.; Ou, Z.-L.; Yu, S.-J.; Gu, X.-L.; Yang, C.; Chen, A.-X.; Di, G.-H.; Shen, Z.-Z.; Shao, Z.-M. The chemokine receptor CCR4 promotes tumor growth and lung metastasis in breast cancer. Breast Cancer Res. Treat. 2011, 131, 837–848. [Google Scholar] [CrossRef]
- Cheng, X.; Wu, H.; Jin, Z.-J.; Ma, D.; Yuen, S.; Jing, X.-Q.; Shi, M.-M.; Shen, B.-Y.; Peng, C.-H.; Zhao, R.; et al. Up-regulation of chemokine receptor CCR4 is associated with Human Hepatocellular Carcinoma malignant behavior. Sci. Rep. 2017, 7, 12362. [Google Scholar] [CrossRef]
- Aliper, A.M.; Frieden-Korovkina, V.P.; Buzdin, A.; Roumiantsev, S.A.; Zhavoronkov, A. A role for G-CSF and GM-CSF in nonmyeloid cancers. Cancer Med. 2014, 3, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Anggorowati, N.; Kurniasari, C.R.; Damayanti, K.; Cahyanti, T.; Widodo, I.; Ghozali, A.; Romi, M.M.; Sari, D.C.R.; Arfian, N. Histochemical and Immunohistochemical Study of α-SMA, Collagen, and PCNA in Epithelial Ovarian Neoplasm. Asian Pac. J. Cancer Prev. 2017, 18, 667–671. [Google Scholar] [PubMed]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 13856. [Google Scholar] [CrossRef] [PubMed]
- Desai, B.; Ma, T.; Zhu, J.; Chellaiah, M. Characterization of the Expression of Variant and Standard CD44 in Prostate Cancer Cells: Identification of the Possible Molecular Mechanism of CD44/MMP9 Complex Formation on the Cell Surface. J. Cell. Biochem. 2009, 108, 272–284. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Waghray, M.; Yalamanchili, M.; Dziubinski, M.; Zeinali, M.; Erkkinen, M.; Yang, H.; Schradle, K.A.; Urs, S.; Di Magliano, M.P.; Welling, T.H.; et al. GM-CSF Mediates Mesenchymal–Epithelial Cross-Talk in Pancreatic Cancer. Cancer Discov. 2016, 6, 886–899. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]
- Vadlakonda, L.; Dash, A.; Pasupuleti, M.; Kumar, K.A.; Reddanna, P. The paradox of Akt-mTOR interactions. Front. Oncol. 2013, 3, 165. [Google Scholar] [CrossRef]
- Zhu, Y.; Pereira, R.O.; O’Neill, B.T.; Riehle, C.; Ilkun, O.; Wende, A.R.; Rawlings, T.A.; Zhang, Y.C.; Zhang, Q.; Klip, A.; et al. Cardiac PI3K-Akt Impairs Insulin-Stimulated Glucose Uptake Independent of mTORC1 and GLUT4 Translocation. Mol. Endocrinol. 2013, 27, 172–184. [Google Scholar] [CrossRef]
- Kohn, A.D.; Kovacina, K.S.; Roth, R.A. Insulin stimulates the kinase activity of RAC-PK, a pleckstrin homology domain containing ser/thr kinase. EMBO J. 1995, 14, 4288–4295. [Google Scholar] [CrossRef]
- Hou, J.; Wu, J.; Dombkowski, A.; Zhang, K.; Holowatyj, A.; Boerner, J.L.; Yang, Z.-Q. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am. J. Transl. Res. 2012, 4, 247–256. [Google Scholar]
- Mitsui, E.; Yoshida, S.; Shinoda, Y.; Matsumori, Y.; Tsujii, H.; Tsuchida, M.; Wada, S.; Hasegawa, M.; Ito, A.; Mino, K.; et al. Identification of ryuvidine as a KDM5A inhibitor. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]



| Description | Markers | Cell Lines | ||||
|---|---|---|---|---|---|---|
| MCF-7 | MDA-MB231 | MCF10A | KAIMRC1 | KAIMRC2 | ||
| Epithelial, Fibroblasts and Tumor Cell Lines Marker | CD47 | *** | *** | *** | *** | *** |
| Monocytes, Macrophages and Fibroblasts Marker | CD116 | * | - | ** | - | ** |
| Monocytes and Granulocytes Marker | SSEA-1 | ** | - | - | - | * |
| Surface Marker of Some Breast Cancers | HER-2 | - | - | - | - | - |
| Epithelial Marker | CD49c | ** | *** | *** | - | *** |
| Breast and Colon Epithelial Marker | CD227 | ** | *** | *** | - | ** |
| Messenchymal and Fibroblast Cells Makrer | Vimentin | - | - | - | - | ** |
| Marker for Breast Cancer Stem Cells | CD24 | *** | - | *** | - | ** |
| Tumor Epithelial Cells | EpCAM-EBA1 | ** | - | ** | - | ** |
| Epithelial Cells and Cancer Stem Cells Marker | CD44 | ** | *** | *** | ** | *** |
| Stem Cells and Epithelial Adherent Junctions Marker | E-cadherin | ** | - | *** | *** | *** |
| Cancer Stem Cells and Breast Cancer Cells Marker | ALDH-1A1 | *** | *** | *** | ** | *** |
| Compounds | Target | Biological Description | IC50 | IC50 Range | R2 |
|---|---|---|---|---|---|
| BIO-acetoxime | Glycogen Synthase Kinase 3 | Selective GSK-3α/β inhibitor | 10.52 | 9.057 to 11.96 | 0.9240 |
| CD 437 | Retinoic Acid Receptors | Enhances reprogramming efficiency of MEFs | 5.540 | 4.327 to 7.052 | 0.9528 |
| TCS 359 | FLT3 | Potent inhibitor of FLT3 receptor tyrosine kinase | 3.092 | 2.602 to 3.664 | 0.9629 |
| Ryuvidine | cdk | Cyclin-dependent kinase (cdk) 4 inhibitor | 0.8339 | 0.6558 to 1.084 | 0.9243 |
| IKK 16 | IKK | Selective inhibitor of IκB kinase (IKK). Inhibits TNF-α-stimulated IκB degradation and expression of adhesion molecules E-selectin, ICAM and VCAM | 7.176 | 6.024 to 8.406 | 0.9293 |
| Purvalanol A | cdk | Cyclin-dependent kinase inhibitor | 12.61 | 10.28 to 15.45 | 0.9176 |
| Ro 31-8220 mesylate | Broad Spectrum Inhibitor | Protein kinase C inhibitor, with activity at other protein. Also inhibits voltage-dependent Na+ channels in the micromolar range. | 7.025 | 5.955 to 8.298 | 0.9539 |
| NH 125 | CaM Kinase III | Histidine protein kinase and eukaryotic elongation factor 2 (eEF-2) kinase (CaMK III) inhibitor. Blocks G1/S cell cycle progression | 1.147 | 0.9333 to 1.409 | 0.9520 |
| ER 27319 maleate | Syk | Selective inhibitor of Syk kinase | ~5.947 | Very wide | 0.908 |
| M 344 | pan-HDAC | Histone deacetylase inhibitor | 0.6241 | 0.3621 to 1.040 | 0.8225 |
| SGI 1027 | DNMT1 | DNA methyltransferase inhibitor | 3.843 | 3.279 to 4.513 | 0.9669 |
| BIX 01294 | G9a/GLP | G9a-like protein and G9a histone lysine methyltransferase inhibitor | 8.220 | 7.711 to 8.766 | 0.9900 |
| TC-E 5003 | PRMT1 | Selective PRMT1 arginine methyltransferase inhibitor | 7.446 | 6.006 to 9.228 | 0.9341 |
| GSK J4 | JMJD3/UTX | Histone demethylase JMJD3/UTX inhibitor | 8.234 | 6.407 to 10.61 | 0.9156 |
| JIB 04 | pan-JMJD | Pan Jumonji histone demethylase inhibitor | 0.4134 | 0.264 to 0.654 | 0.8575 |
| Mice | Size in Length and Width (mm) after 4 Weeks | Size in Length and Width (mm) after 8 Weeks | Tumor Weight (g) after 8 Weeks | Body Weight (g) after 8 Weeks |
|---|---|---|---|---|
| 1 | 5.5–5.6 | 6.7–6.3 | 1.7 | 24 |
| 2 | 9–7 | 13–11 | 1.75 | 26 |
| 3 | 7–8 | 9.4–10.3 | 2.5 | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, R.; Al Zahrani, H.; Barhoumi, T.; Alhallaj, A.; Mashhour, A.; Alshammari, M.A.; Alshawakir, Y.A.; Baz, O.; Alanazi, A.H.; Khan, A.L.; et al. Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2. Cells 2021, 10, 1303. https://doi.org/10.3390/cells10061303
Ali R, Al Zahrani H, Barhoumi T, Alhallaj A, Mashhour A, Alshammari MA, Alshawakir YA, Baz O, Alanazi AH, Khan AL, et al. Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2. Cells. 2021; 10(6):1303. https://doi.org/10.3390/cells10061303
Chicago/Turabian StyleAli, Rizwan, Hajar Al Zahrani, Tlili Barhoumi, Alshaimaa Alhallaj, Abdullah Mashhour, Musaad A. Alshammari, Yasser A. Alshawakir, Omar Baz, Abdullah H. Alanazi, Abdul Latif Khan, and et al. 2021. "Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2" Cells 10, no. 6: 1303. https://doi.org/10.3390/cells10061303
APA StyleAli, R., Al Zahrani, H., Barhoumi, T., Alhallaj, A., Mashhour, A., Alshammari, M. A., Alshawakir, Y. A., Baz, O., Alanazi, A. H., Khan, A. L., Al Nikhli, H., Al Balwi, M. A., Al Riyees, L., & Boudjelal, M. (2021). Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2. Cells, 10(6), 1303. https://doi.org/10.3390/cells10061303