In Vivo Stimulation of α- and β-Adrenoceptors in Mice Differentially Alters Small RNA Content of Circulating Extracellular Vesicles
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Blood EVs in Mouse
2.2. Basal Characterization of Mouse Blood Serum EVs
2.2.1. Analysis of EV Morphology
2.2.2. Protein Immunoblot Analysis of EVs
2.2.3. Identification of miRNAs in EVs by Reverse Transcriptase-Quantitative Polymerase Chain
Reaction (RT-qPCR)
2.3. Sequencing of Small Noncoding RNA Isolated from Mouse Blood EVs with ISO and PE Infusion
2.3.1. Isoproterenol and Phenylephrine Infusion Mouse Model
2.3.2. Sequencing of Small Noncoding RNA Isolated from Mouse Blood EVs with ISO and PE Infusion
2.3.3. Echocardiography
2.4. Statistics
3. Results
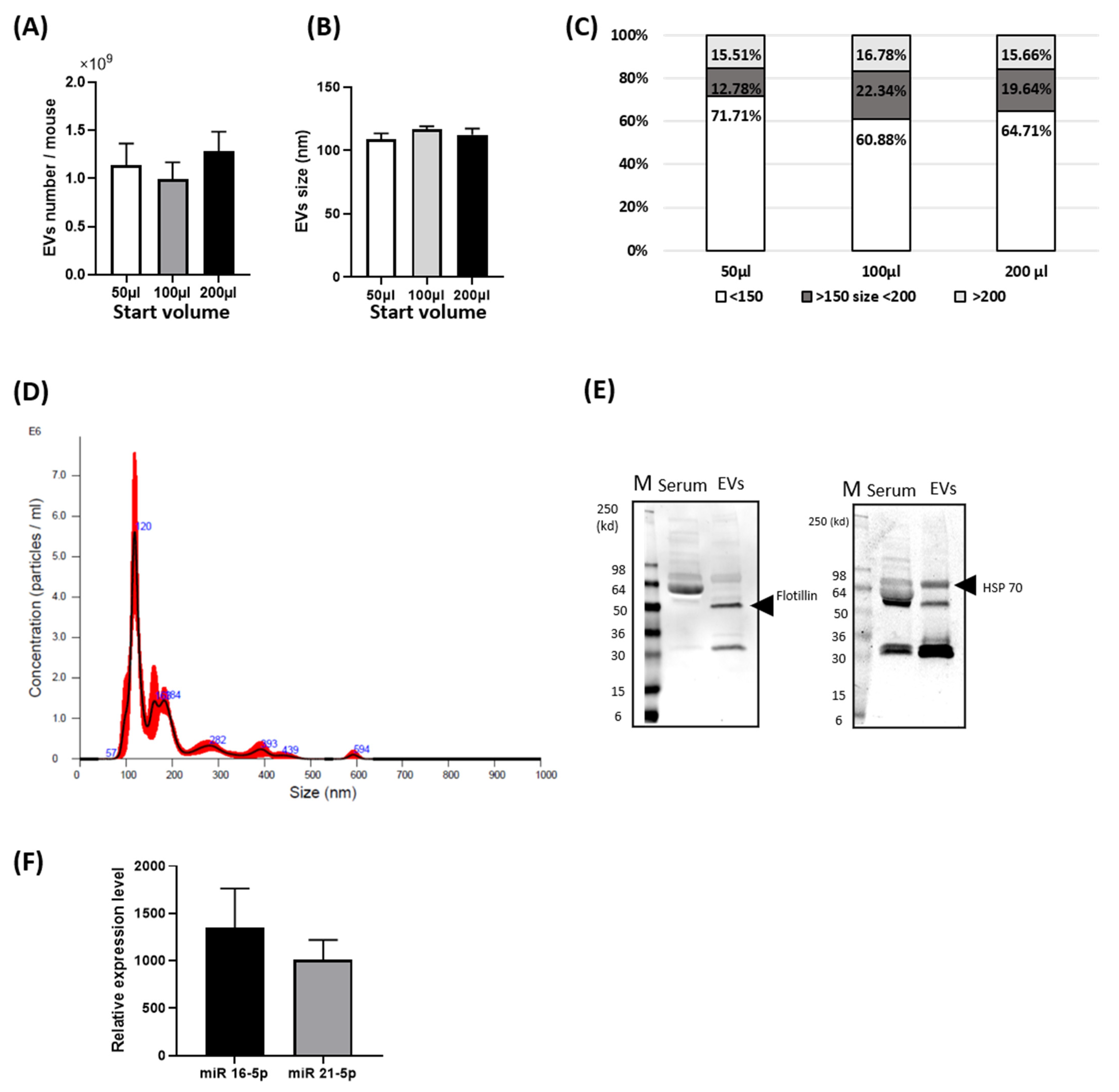
3.1. Basal Characterization of Circulating EVs from Mouse Blood
3.2. Phenotypic Characterization of Mice after Chronic Infusion of PE or ISO
3.3. Quality Evaluation of Total RNA of Serum EVs
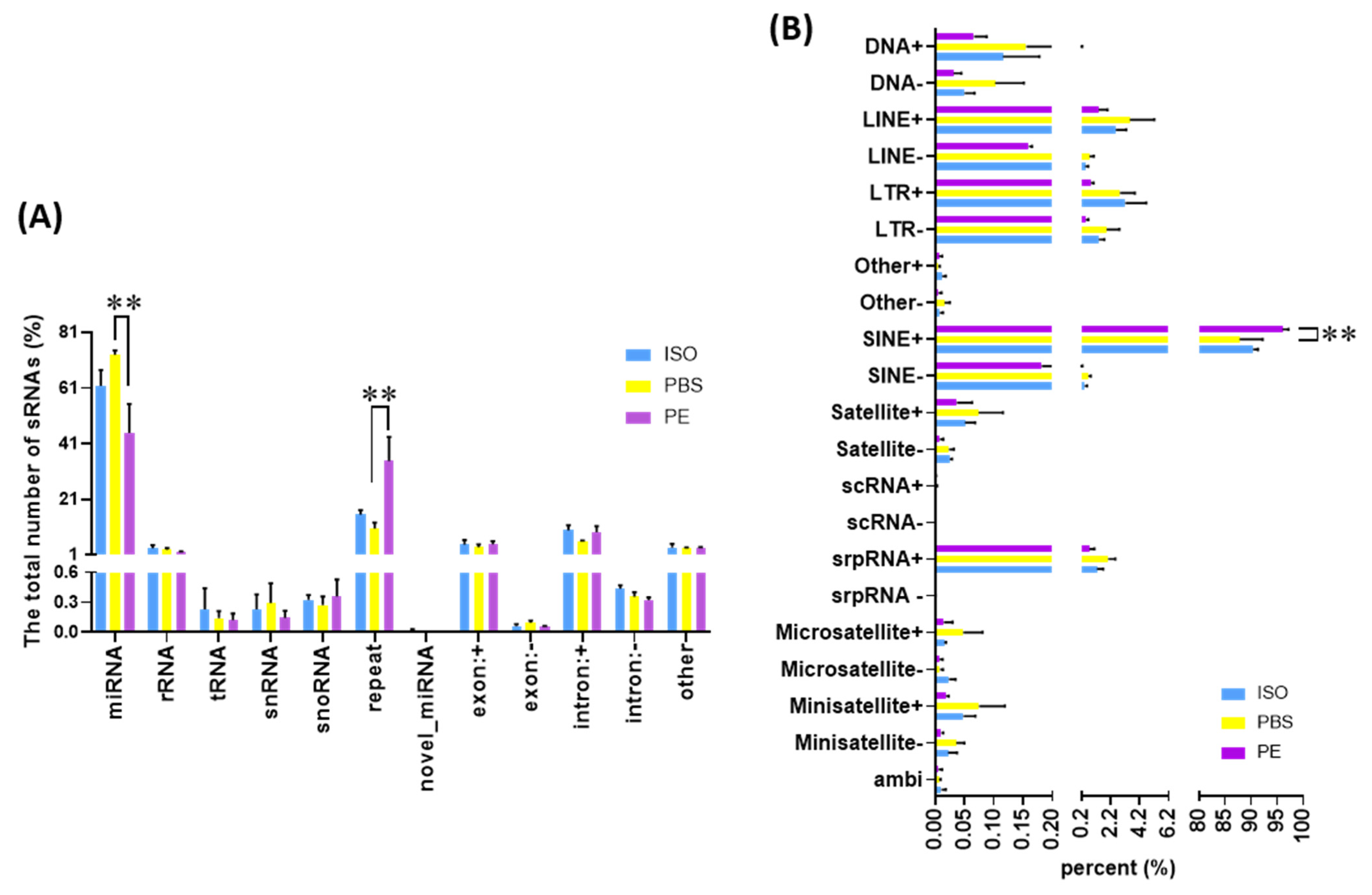
3.4. Comparison of Small ncRNA Content of Serum EVs
3.5. Differentially Expressed miRs in Different Serum EVs
3.6. Functional Pathway Analysis of Differentially Expressed Serum EV miRs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waldenstrom, A.; Ronquist, G. Role of exosomes in myocardial remodeling. Circ. Res. 2014, 114, 315–324. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Zhu, H.J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Invest. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Gozal, D. Exosomal miRNAs as potential biomarkers of cardiovascular risk in children. J. Transl Med. 2014, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.D.; French, K.M.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of Therapeutic Covariant MicroRNA Clusters in Hypoxia-Treated Cardiac Progenitor Cell Exosomes Using Systems Biology. Circ. Res. 2015, 116, 255–263. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Aghabozorgi, A.S.; Ahangari, N.; Eftekhaari, T.E.; Torbati, P.N.; Bahiraee, A.; Ebrahimi, R.; Pasdar, A. Circulating exosomal miRNAs in cardiovascular disease pathogenesis: New emerging hopes. J. Cell. Physiol. 2019, 234, 21796–21809. [Google Scholar] [CrossRef]
- Mancuso, T.; Barone, A.; Salatino, A.; Molinaro, C.; Marino, F.; Scalise, M.; Torella, M.; Angelis, A.D.; Urbanek, K.; Torella, D.; et al. Unravelling the Biology of Adult Cardiac Stem Cell-Derived Exosomes to Foster Endogenous Cardiac Regeneration and Repair. Int. J. Mol. Sci. 2020, 21, 3725. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sucharov, J.; Stauffer, B.L.; Miyamoto, S.D.; Sucharov, C.C. Exosomes from pediatric dilated cardiomyopathy patients modulate a pathological response in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H818–H826. [Google Scholar] [CrossRef] [PubMed]
- Beninson, L.A.; Brown, P.N.; Loughridge, A.B.; Saludes, J.P.; Maslanik, T.; Hills, A.K.; Woodworth, T.; Craig, W.; Yin, H.; Fleshner, M. Acute stressor exposure modifies plasma exosome-associated heat shock protein 72 (Hsp72) and microRNA (miR-142-5p and miR-203). PLoS ONE 2014, 9, e108748. [Google Scholar]
- Bristow, M.R.; Hershberger, R.E.; Port, J.D.; Minobe, W.; Rasmussen, R. Beta-1-Adrenergic and Beta-2-Adrenergic Receptor-Mediated Adenylate-Cyclase Stimulation in Nonfailing and Failing Human Ventricular Myocardium. Mol. Pharmacol. 1989, 35, 295–303. [Google Scholar] [PubMed]
- Brodde, O.E.; Zerkowski, H.R.; Doetsch, N.; Motomura, S.; Khamssi, M.; Michel, M.C. Myocardial Beta-Adrenoceptor Changes in Heart-Failure—Concomitant Reduction in Beta-1-Adrenoceptor and Beta-2-Adrenoceptor Function Related to the Degree of Heart-Failure in Patients with Mitral-Valve Disease. J. Am. Coll. Cardiol. 1989, 14, 323–331. [Google Scholar] [CrossRef]
- Ding, S.Q.; Chen, J.; Wang, S.N.; Duan, F.X.; Chen, Y.Q.; Shi, Y.J.; Hu, J.G.; Lu, H.Z. Identification of serum exosomal microRNAs in acute spinal cord injured rats. Exp. Biol. Med. 2019, 244, 1149–1161. [Google Scholar] [CrossRef]
- Zhao, F.; Cheng, L.; Shao, Q.; Chen, Z.; Lv, X.; Li, J.; He, L.; Sun, Y.; Ji, Q.; Lu, P.; et al. Characterization of serum small extracellular vesicles and their small RNA contents across humans, rats, and mice. Sci. Rep. 2020, 10, 4197. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Yeri, A.; Cheah, P.S.; Chung, A.; Danielson, K.; De Hoff, P.; Filant, J.; Laurent, C.D.; Laurent, L.D.; Magee, R.; et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 2019, 177, 446–462.e16. [Google Scholar] [CrossRef]
- McDonald, M.K.; Capasso, K.E.; Ajit, S.K. Purification and microRNA profiling of exosomes derived from blood and culture media. J. Vis. Exp. 2013, 76, e50294. [Google Scholar] [CrossRef]
- Kwon, J.S.; Schumacher, S.M.; Gao, E.; Chuprun, J.K.; Ibetti, J.; Roy, R.; Khan, M.; Kishore, R.; Koch, W.J. Characterization of betaARKct engineered extracellular vesicles and model specific cardioprotection. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H1276–H1289. [Google Scholar] [CrossRef]
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010, 27, 796–810. [Google Scholar] [CrossRef]
- Schumacher, S.M.; Gao, E.; Cohen, M.; Lieu, M.; Chuprun, J.K.; Koch, W.J. A peptide of the RGS domain of GRK2 binds and inhibits Galpha(q) to suppress pathological cardiac hypertrophy and dysfunction. Sci. Signal. 2016, 9, ra30. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Miller, J.W.; Urbinati, C.R.; Teng-Umnuay, P.; Stenberg, M.G.; Byrne, B.J.; Thornton, C.A.; Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000, 19, 4439–4448. [Google Scholar] [CrossRef]
- Swinnen, B.; Robberecht, W.; Van Den Bosch, L. RNA toxicity in non-coding repeat expansion disorders. EMBO J. 2020, 39, e101112. [Google Scholar] [CrossRef] [PubMed]
- Zu, T.; Gibbens, B.; Doty, N.S.; Gomes-Pereira, M.; Huguet, A.; Stone, M.D.; Margolis, J.; Peterson, M.; Markowski, T.W.; Ingram, M.A.; et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA 2011, 108, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, A.; Wang, H.; Klein, J.D.; Tan, L.; Wang, Z.M.; Du, J.; Naqvi, N.; Liu, B.C.; Wang, X.H. miR-26a Limits Muscle Wasting and Cardiac Fibrosis through Exosome-Mediated microRNA Transfer in Chronic Kidney Disease. Theranostics 2019, 9, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Urabe, F.; Kosaka, N.; Sawa, Y.; Yamamoto, Y.; Ito, K.; Yamamoto, T.; Kimura, T.; Egawa, S.; Ochiya, T. miR-26a regulates extracellular vesicle secretion from prostate cancer cells via targeting SHC4, PFDN4, and CHORDC1. Sci. Adv. 2020, 6, eaay3051. [Google Scholar] [CrossRef]
- Sun, L.P.; Yu, Y.; Niu, B.F.; Wang, D.Q. Red Blood Cells as Potential Repositories of MicroRNAs in the Circulatory System. Front. Genet. 2020, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.S.; Li, L.; Li, L.; Chu, S.; Shiang, K.D.; Li, M.; Sun, H.Y.; Xu, J.; Xiao, F.J.; Sun, G.; et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood 2015, 125, 1302–1313. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Korkola, J.E.; Sadelain, M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2010, 28, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Palmer, L.E.; Lechauve, C.; Zhao, G.; Yao, Y.; Luan, J.; Vourekas, A.; Tan, H.; Peng, J.; Schuetz, J.D.; et al. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood 2019, 133, 2518–2528. [Google Scholar] [CrossRef]
- Hasan, S.; Mosier, M.J.; Szilagyi, A.; Gamelli, R.L.; Muthumalaiappan, K. Discrete beta-adrenergic mechanisms regulate early and late erythropoiesis in erythropoietin-resistant anemia. Surgery 2017, 162, 901–916. [Google Scholar] [CrossRef]
- Fonseca, R.B.; Mohr, A.M.; Wang, L.; Sifri, Z.C.; Rameshwar, P.; Livingston, D.H. The impact of a hypercatecholamine state on erythropoiesis following severe injury and the role of IL-6. J. Trauma Acute Care Surg. 2005, 59, 884–889, discussion 889–890. [Google Scholar] [CrossRef]
- O’Dell, T.J.; Connor, S.A.; Guglietta, R.; Nguyen, P.V. β-Adrenergic receptor signaling and modulation of long-term potentiation in the mammalian hippocampus. Learn. Mem. 2015, 22, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Vicencio, J.M.; Yellon, D.M.; Sivaraman, V.; Das, D.; Boi-Doku, C.; Arjun, S.; Zheng, Y.; Riquelme, J.A.; Kearney, J.; Sharma, V.; et al. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J. Am. Coll. Cardiol. 2015, 65, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Q.; Zhao, T.; Liu, X.; Bai, G.; Xin, Y.; Shen, H.; Wei, B. Expression profile of serum-related exosomal miRNAs from parathyroid tumor. Endocrine 2021, 72, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Cheng, J.; Yao, Y.; Lou, C.; Wang, L.; Huang, X.; Zhang, Y. Combination of Four Serum Exosomal MiRNAs as Novel Diagnostic Biomarkers for Early-Stage Gastric Cancer. Front. Genet. 2020, 11, 237. [Google Scholar] [CrossRef]
- May, S.M.; Abbott, T.E.F.; Del Arroyo, A.G.; Reyes, A.; Martir, G.; Stephens, R.C.M.; Brealey, D.; Cuthbertson, B.H.; Wijeysundera, D.N. MicroRNA signatures of perioperative myocardial injury after elective noncardiac surgery: A prospective observational mechanistic cohort study. Br. J. Anaesth. 2020, 125, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef]


| miRNA | PE (Read Count) | PBS (Read Count) | log2FoldChange |
|---|---|---|---|
| mmu-miR-3057-5p | 27.55 | 0.19 | 4.16 |
| mmu-miR-872-3p | 33.03 | 2.29 | 2.80 |
| mmu-miR-1843b-3p | 115.24 | 19.78 | 2.26 |
| mmu-miR-3068-5p | 182.56 | 30.11 | 2.25 |
| mmu-miR-1981-5p | 795.19 | 309.24 | 1.30 |
| mmu-miR-340-3p | 191.34 | 76.10 | 1.28 |
| mmu-miR-30e-3p | 781.70 | 339.02 | 1.16 |
| mmu-miR-26a-5p | 41,301.67 | 24,470.54 | 0.74 |
| RNA | PE (Read Count) | PBS (Read Count) | log2FoldChange |
|---|---|---|---|
| mmu-miR-365-3p | 0.90 | 52.51 | −3.85 |
| mmu-miR-365-1-5p | 0.00 | 29.88 | −3.77 |
| mmu-miR-214-5p | 0.52 | 43.74 | −3.70 |
| mmu-miR-200a-5p | 8.86 | 189.11 | −3.64 |
| mmu-miR-205-5p | 421.15 | 8177.68 | −3.56 |
| mmu-miR-31-3p | 0.00 | 22.30 | −3.53 |
| mmu-miR-323-3p | 1.79 | 45.17 | −3.50 |
| mmu-miR-135b-5p | 1.79 | 34.68 | −3.18 |
| mmu-miR-654-3p | 2.69 | 53.94 | −3.13 |
| mmu-miR-494-3p | 2.83 | 46.99 | −3.05 |
| mmu-miR-34a-5p | 7.50 | 91.43 | −2.93 |
| mmu-miR-532-3p | 2.16 | 43.68 | −2.81 |
| mmu-miR-196a-5p | 7.01 | 81.23 | −2.75 |
| mmu-miR-96-5p | 21.56 | 206.15 | −2.69 |
| mmu-miR-493-5p | 4.19 | 56.35 | −2.68 |
| mmu-miR-495-3p | 4.44 | 49.01 | −2.63 |
| mmu-miR-148a-5p | 7.88 | 67.58 | −2.62 |
| mmu-miR-29c-5p | 3.73 | 45.12 | −2.61 |
| mmu-miR-200c-3p | 576.38 | 3452.40 | −2.41 |
| mmu-miR-31-5p | 43.08 | 322.11 | −2.40 |
| mmu-miR-183-5p | 274.95 | 1734.91 | −2.38 |
| mmu-miR-218-5p | 70.60 | 459.62 | −2.38 |
| mmu-miR-145a-5p | 98.67 | 692.59 | −2.30 |
| mmu-miR-200b-3p | 768.80 | 3522.19 | −2.06 |
| mmu-miR-363-3p | 355.44 | 1637.29 | −1.98 |
| mmu-miR-203-3p | 886.93 | 3145.32 | −1.74 |
| mmu-miR-182-5p | 205.97 | 706.32 | −1.68 |
| mmu-miR-100-5p | 2259.26 | 7238.42 | −1.59 |
| mmu-miR-23b-3p | 216.79 | 646.32 | −1.49 |
| mmu-miR-200a-3p | 576.49 | 1708.26 | −1.49 |
| mmu-miR-342-3p | 74.86 | 185.36 | −1.24 |
| mmu-miR-27b-3p | 4002.02 | 7246.92 | −0.84 |
| miRNA | ISO | PBS | log2FoldChange |
|---|---|---|---|
| mmu-miR-340-3p | 320.51 | 85.06 | 1.79 |
| mmu-miR-3103-3p | 0.00 | 20.73 | −3.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, J.-S.; Barr, E.W.; Chuprun, J.K.; Koch, W.J. In Vivo Stimulation of α- and β-Adrenoceptors in Mice Differentially Alters Small RNA Content of Circulating Extracellular Vesicles. Cells 2021, 10, 1211. https://doi.org/10.3390/cells10051211
Kwon J-S, Barr EW, Chuprun JK, Koch WJ. In Vivo Stimulation of α- and β-Adrenoceptors in Mice Differentially Alters Small RNA Content of Circulating Extracellular Vesicles. Cells. 2021; 10(5):1211. https://doi.org/10.3390/cells10051211
Chicago/Turabian StyleKwon, Jin-Sook, Eric W. Barr, J. Kurt Chuprun, and Walter J. Koch. 2021. "In Vivo Stimulation of α- and β-Adrenoceptors in Mice Differentially Alters Small RNA Content of Circulating Extracellular Vesicles" Cells 10, no. 5: 1211. https://doi.org/10.3390/cells10051211
APA StyleKwon, J.-S., Barr, E. W., Chuprun, J. K., & Koch, W. J. (2021). In Vivo Stimulation of α- and β-Adrenoceptors in Mice Differentially Alters Small RNA Content of Circulating Extracellular Vesicles. Cells, 10(5), 1211. https://doi.org/10.3390/cells10051211






