All-Trans Retinoic Acid Increases DRP1 Levels and Promotes Mitochondrial Fission
Abstract
1. Introduction
2. Methods
2.1. Cell Culture
2.2. Animals
2.3. Western Blot
2.4. Isolation and Purification of Mitochondria from the Mouse Heart
2.5. Confocal Microscopy
2.6. Image Analysis
2.7. Modeling Mitochondrial Network
2.8. Transmission Electron Microscopy (TEM)
2.9. Statistics
3. Results
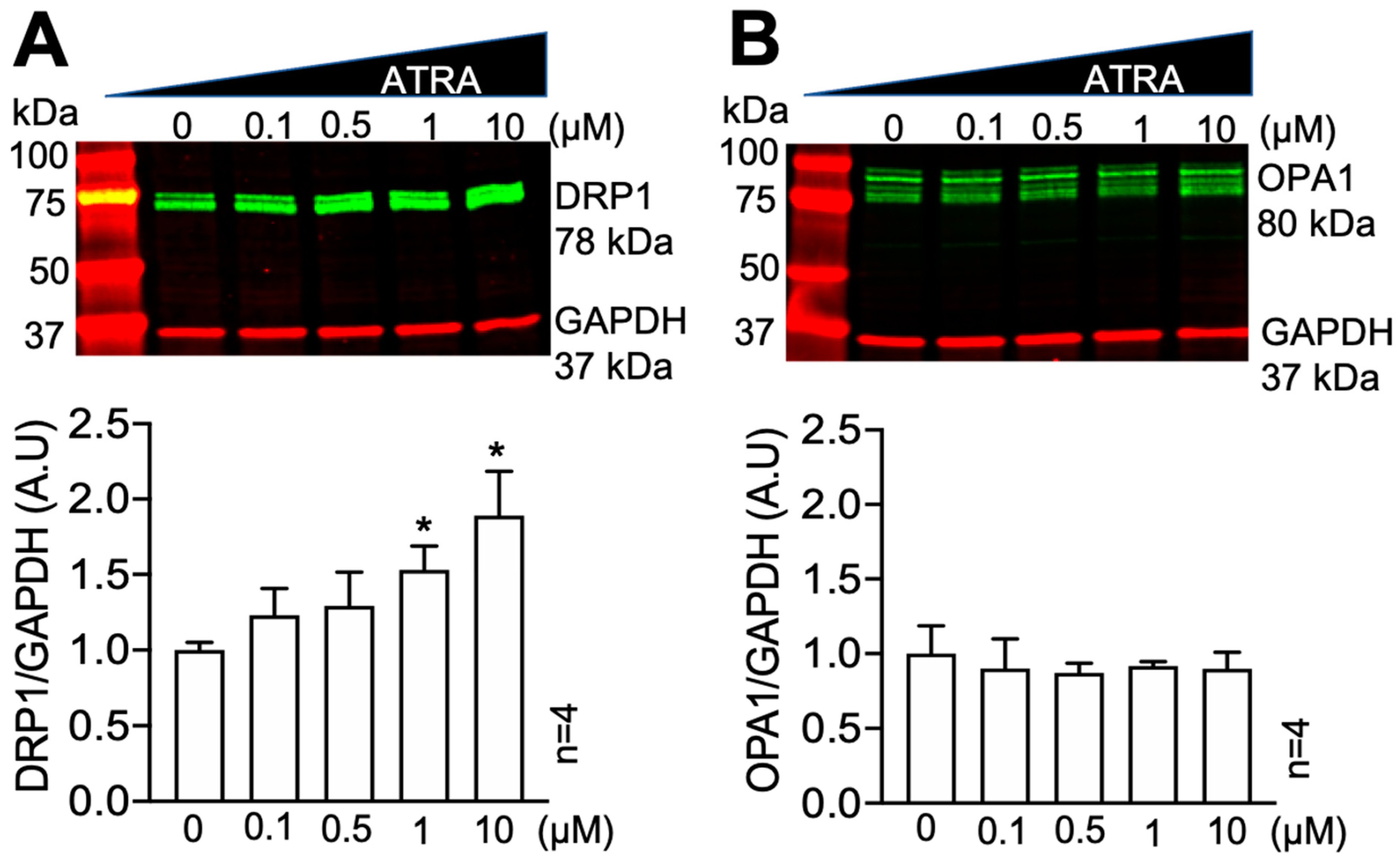
3.1. All-Trans Retinoic Acid (ATRA) Upregulates the Fission Protein DRP1
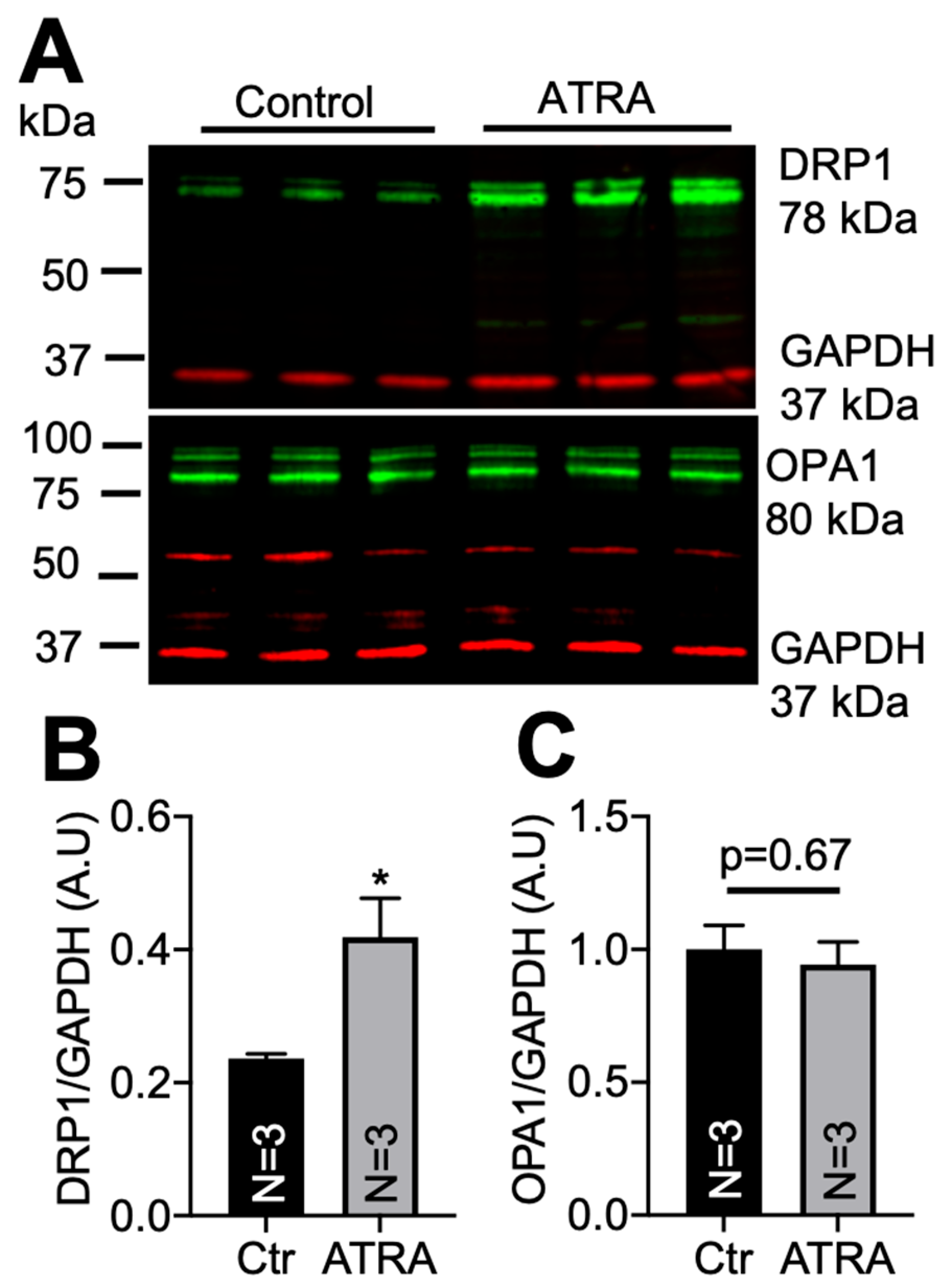
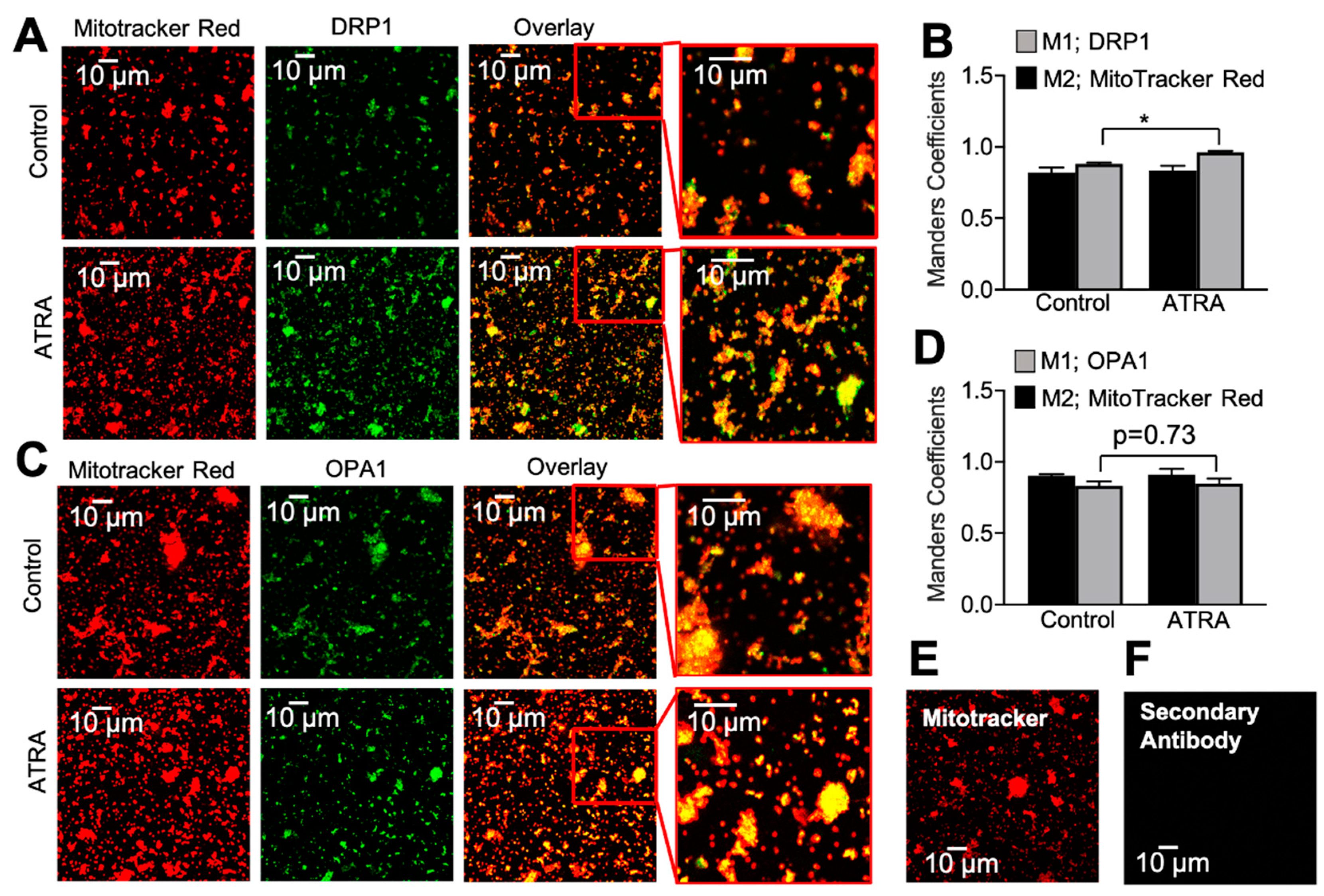
3.2. Localization of DRP1 and OPA1 Levels in Purified Mitochondria from ATRA-Treated Mouse Hearts
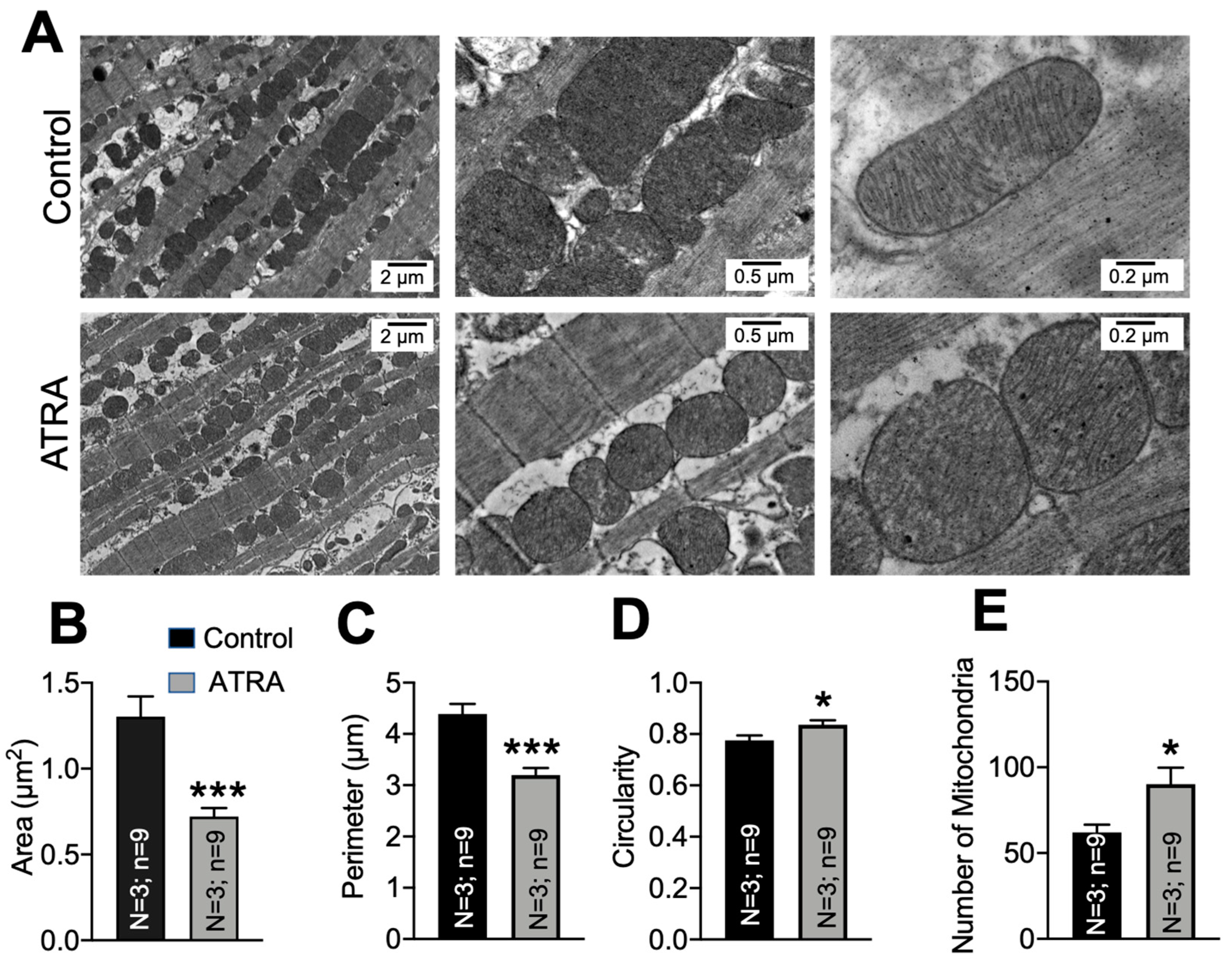
3.3. Mitochondrial Ultrastructural Changes in ATRA-Treated Mice
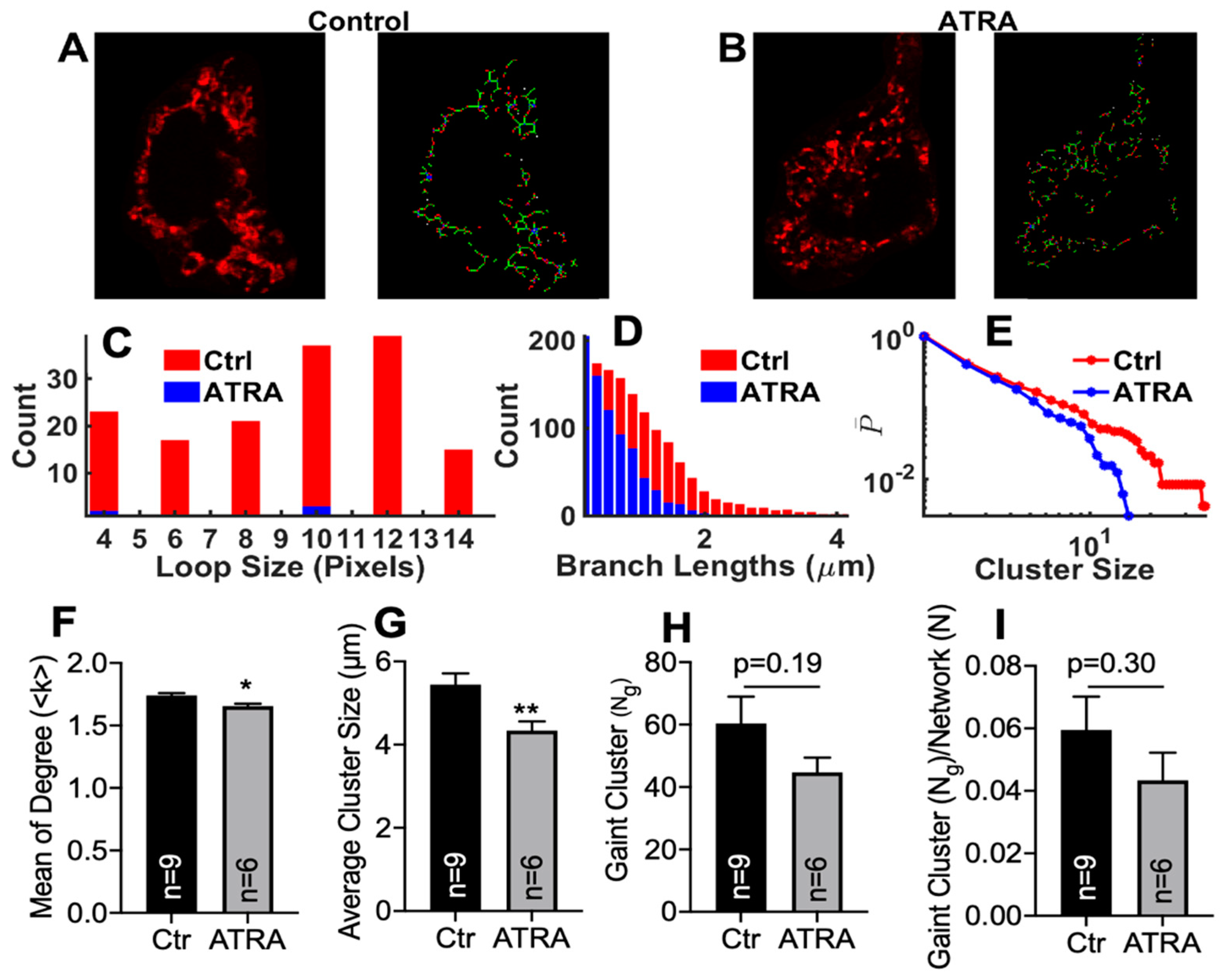
3.4. Modeling Mitochondrial Networks in ATRA-Treated HL-1 Cells
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 711–723. [Google Scholar] [CrossRef]
- Carreira, R.S.; Billups, P.; Gottlieb, R.A. Mitochondrial therapeutics for cardioprotection. Curr. Pharm. Des. 2011, 17, 2017–2035. [Google Scholar] [CrossRef]
- Chen, H.; Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic devel-opment. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Griparic, L.; Shurland, D.-L.; Van Der Bliek, A.M. Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Labrousse, A.M.; Zappaterra, M.D.; Rube, D.A.; van der Bliek, A.M. C. elegans Dynamin-Related Protein DRP-1 Controls Severing of the Mitochondrial Outer Membrane. Mol. Cell 1999, 4, 815–826. [Google Scholar] [CrossRef]
- Lackner, L.L.; Nunnari, J.M. The molecular mechanism and cellular functions of mitochondrial division. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2009, 1792, 1138–1144. [Google Scholar] [CrossRef]
- Detmer, S.A.; Chan, D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007, 8, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Youle, R.J. Mitochondrial fission and fusion. Essays Biochem. 2010, 47, 85–98. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Dorn, G.W. Mitochondrial Fusion is Essential for Organelle Function and Cardiac Homeostasis. Circ. Res. 2011, 109, 1327–1331. [Google Scholar] [CrossRef]
- Chen, Y.; Csordás, G.; Jowdy, C.; Schneider, T.G.; Csordás, N.; Wang, W.; Liu, Y.; Kohlhaas, M.; Meiser, M.; Bergem, S.; et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic re-sponses via interorganelle Ca2+ crosstalk. Circ. Res. 2012, 111, 863–875. [Google Scholar] [CrossRef]
- He, L.; Zhou, Q.; Huang, Z.; Xu, J.; Zhou, H.; Lv, D.; Lu, L.; Huang, S.; Tang, M.; Zhong, J.; et al. PINK1/Parkin-mediated mitophagy promotes apelin-13-induced vascular smooth muscle cell proliferation by AMPKalpha and exacerbates atherosclerotic lesions. J. Cell Physiol. 2019, 234, 8668–8682. [Google Scholar] [CrossRef]
- Suen, D.-F.; Narendra, D.P.; Tanaka, A.; Manfredi, G.; Youle, R.J. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11835–11840. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.; Tanaka, A.; Suen, D.-F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Sivitz, W.I.; Yorek, M.A. Mitochondrial Dysfunction in Diabetes: From Molecular Mechanisms to Functional Significance and Therapeutic Opportunities. Antioxid. Redox Signal. 2010, 12, 537–577. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, M.K.; Turner, N. Mitochondrial dysfunction and insulin resistance: An update. Endocr. Connect. 2015, 4, R1–R15. [Google Scholar] [CrossRef]
- Rovira-Llopis, S.; Bañuls, C.; Diaz-Morales, N.; Hernandez-Mijares, A.; Rocha, M.; Victor, V.M. Mitochondrial dynamics in type 2 diabetes: Pathophysiological implications. Redox Biol. 2017, 11, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Barot, M.; Gokulgandhi, M.R.; Mitra, A.K. Mitochondrial Dysfunction in Retinal Diseases. Curr. Eye Res. 2011, 36, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Dong, Z. Mitochondria in Kidney Injury: When the Power Plant Fails. J. Am. Soc. Nephrol. 2016, 27, 1869–1872. [Google Scholar] [CrossRef]
- Lane, M.; Boczonadi, V.; Bachtari, S.; Gomez-Duran, A.; Langer, T.; Griffiths, A.; Kleinle, S.; Dineiger, C.; Abicht, A.; Holinski-Feder, E.; et al. Mitochondrial dysfunction in liver failure requiring transplantation. J. Inherit. Metab. Dis. 2016, 39, 427–436. [Google Scholar] [CrossRef]
- Billups, B.; Forsythe, I.D. Presynaptic Mitochondrial Calcium Sequestration Influences Transmission at Mammalian Central Synapses. J. Neurosci. 2002, 22, 5840–5847. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Faas, G.C.; Saggau, P.; Craigen, W.J.; Sweatt, J.D. Mitochondrial Regulation of Synaptic Plasticity in the Hippocampus. J. Biol. Chem. 2003, 278, 17727–17734. [Google Scholar] [CrossRef]
- Liu, J.; Head, E.; Gharib, A.M.; Yuan, W.; Ingersoll, R.T.; Hagen, T.M.; Cotman, C.W.; Ames, B.N. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: Partial reversal by feeding acetyl-L-carnitine and/or R- -lipoic acid. Proc. Natl. Acad. Sci. USA 2002, 99, 2356–2361. [Google Scholar] [CrossRef]
- Verstreken, P.; Ly, C.V.; Venken, K.J.; Koh, T.-W.; Zhou, Y.; Bellen, H.J. Synaptic Mitochondria Are Critical for Mobilization of Reserve Pool Vesicles at Drosophila Neuromuscular Junctions. Neuron 2005, 47, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-Y.; Lu, M.-H.; Yuan, D.-J.; Xu, D.-E.; Yao, P.-P.; Ji, W.-L.; Chen, H.; Liu, W.-L.; Yan, C.-X.; Xia, Y.-Y.; et al. Mitochondrial Dysfunction in Neural Injury. Front. Neurosci. 2019, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Austin, G.L.; Young, L.E.A.; Johnson, L.A.; Sun, R. Mitochondrial Metabolism in Major Neurological Diseases. Cells 2018, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.R. Vitamin A and Retinoids as Mitochondrial Toxicants. Oxidative Med. Cell. Longev. 2015, 2015, 140267. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.A. Recommendations for Vitamin A Supplementation. J. Nutr. 2002, 132, 2902S–2906S. [Google Scholar] [CrossRef]
- Tanumihardjo, S.A. Assessing Vitamin A Status: Past, Present and Future. J. Nutr. 2004, 134, 290S–293S. [Google Scholar] [CrossRef]
- Allen, L.H.; Haskell, M. Estimating the Potential for Vitamin A Toxicity in Women and Young Children. J. Nutr. 2002, 132, 2907S–2919S. [Google Scholar] [CrossRef]
- O’Reilly, K.; Bailey, S.J.; Lane, M.A. Retinoid-Mediated Regulation of Mood: Possible Cellular Mechanisms. Exp. Biol. Med. 2008, 233, 251–258. [Google Scholar] [CrossRef]
- National Institute of Health Office of Diary Supplements. Dietary Supplement Fact Sheet—Vitamin A. Vitam. Vitamin A-Health Professional. 2020. Available online: https://ods.od.nih.gov/factsheets/VitaminA-HealthProfessional/ (accessed on 14 February 2020).
- Kam, R.K.T.; Deng, Y.; Chen, Y.; Zhao, H. Retinoic acid synthesis and functions in early embryonic development. Cell Biosci. 2012, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Davies, W.L.; Hankins, M.W.; Foster, R.G. Vertebrate ancient opsin and melanopsin: Divergent irradiance detectors. Photochem. Photobiol. Sci. 2010, 9, 1444–1457. [Google Scholar] [CrossRef]
- Rosselot, C.; Spraggon, L.; Chia, I.; Batourina, E.; Riccio, P.; Lu, B.; Niederreither, K.; Dolle, P.; Duester, G.; Chambon, P.; et al. Non-cell-autonomous retinoid signaling is crucial for renal development. Development 2010, 137, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Mallipattu, S.K.; He, J.C. The Beneficial Role of Retinoids in Glomerular Disease. Front. Med. 2015, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Rhinn, M.; Dollé, P. Retinoic acid signaling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef]
- Duester, G. Retinoic Acid Synthesis and Signaling during Early Organogenesis. Cell 2008, 134, 921–931. [Google Scholar] [CrossRef]
- Okumura, L.M.; Okumura, P.C.B.; Veroneze, C. Administration of all-trans retinoic acid through enteral tubes in acute promyelocytic leukemia: The handling of cytotoxic agents and clinical benefits. Rev. Bras. Hematol. Hemoter. 2017, 39, 86–88. [Google Scholar] [CrossRef]
- Schmidt, N.; Gans, E.H. Tretinoin: A Review of Its Anti-inflammatory Properties in the Treatment of Acne. J. Clin. Aesthetic Dermatol. 2011, 4, 22–29. [Google Scholar]
- Mukherjee, S.; Date, A.; Patravale, V.; Korting, H.C.; Roeder, A.; Weindl, G. Retinoids in the treatment of skin aging: An overview of clinical efficacy and safety. Clin. Interv. Aging 2006, 1, 327–348. [Google Scholar] [CrossRef]
- Tourniaire, F.; Musinovic, H.; Gouranton, E.; Astier, J.; Marcotorchino, J.; Arreguin, A.; Bernot, D.; Palou, A.; Bonet, M.L.; Ribot, J.; et al. All-trans retinoic acid induces oxidative phosphorylation and mitochondria biogenesis in adipocytes. J. Lipid Res. 2015, 56, 1100–1109. [Google Scholar] [CrossRef]
- Vantaggiato, C.; Castelli, M.; Giovarelli, M.; Orso, G.; Bassi, M.T.; Clementi, E.; De Palma, C. The Fine Tuning of Drp1-Dependent Mitochondrial Remodeling and Autophagy Controls Neuronal Differentiation. Front. Cell. Neurosci. 2019, 13, 120. [Google Scholar] [CrossRef]
- Nan, J.L.; Zhu, W.; Rahman, M.S.; Liu, M.; Li, D.; Su, S.; Zhang, N.; Hu, X.; Yu, H.; Gupta, M.P. Molecular regulation of mitochondrial dynamics in cardiac disease. Biochim. Biophys. Acta-Mol. Cell Res. 2017, 1864, 1260–1273. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chan, D.C. Physiological functions of mitochondrial fusion. Ann. N. Y. Acad. Sci. 2010, 1201, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gong, Q.; Stice, J.P.; Knowlton, A.A. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc. Res. 2009, 84, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Papanicolaou, K.N.; Khairallah, R.J.; Ngoh, G.A.; Chikando, A.; Luptak, I.; O’Shea, K.M.; Riley, D.D.; Lugus, J.J.; Colucci, W.S.; Lederer, W.J.; et al. Mitofusin-2 Maintains Mitochondrial Structure and Contributes to Stress-Induced Permeability Transition in Cardiac Myocytes. Mol. Cell. Biol. 2011, 31, 1309–1328. [Google Scholar] [CrossRef] [PubMed]
- Tandler, B.; Hoppel, C.L. Possible division of cardiac mitochondria. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 1972, 173, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Marín-García, J.; Akhmedov, A.T. Mitochondrial dynamics and cell death in heart failure. Heart Fail. Rev. 2016, 21, 123–136. [Google Scholar] [CrossRef]
- Song, M.; Franco, A.; Fleischer, J.A.; Zhang, L.; Dorn, G.W. Abrogating Mitochondrial Dynamics in Mouse Hearts Accelerates Mitochondrial Senescence. Cell Metab. 2017, 26, 872–883.e5. [Google Scholar] [CrossRef]
- Kuzmicic, J.; del Campo, A.; López-Crisosto, C.; Morales, P.E.; Pennanen, C.; Bravo-Sagua, R.; Hechenleitner, J.; Zepeda, R.; Castro, P.F.; Verdejo, H.E.; et al. Mitochondrial Dynamics: A Potential New Therapeutic Target for Heart Failure. Rev. Española Cardiol. 2011, 64, 916–923. [Google Scholar] [CrossRef]
- Claycomb, W.C.; Lanson, N.A.; Stallworth, B.S.; Egeland, D.B.; Delcarpio, J.B.; Bahinski, A.; Izzo, N.J. HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 1998, 95, 2979–2984. [Google Scholar] [CrossRef]
- White, S.M.; Constantin, P.E.; Claycomb, W.C. Cardiac physiology at the cellular level: Use of cultured HL-1 cardio-myocytes for studies of cardiac muscle cell structure and function. Am. J. Physiol. Heart Circ. Physiol. 2004, 286, H823–H829. [Google Scholar] [CrossRef]
- Singh, H.; Lu, R.; Rodríguez, P.F.G.; Wu, Y.; Bopassa, J.C.; Stefani, E.; Toro, L. Visualization and quantification of cardiac mitochondrial protein clusters with STED microscopy. Mitochondrion 2012, 12, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.M. Purification of a Crude Mitochondrial Fraction by Density-Gradient Centrifugation. Curr. Protoc. Cell Biol. 1999, 4, 3.4.1–3.4.22. [Google Scholar] [CrossRef] [PubMed]
- Trigo, D.; Goncalves, M.B.; Corcoran, J.P.T. The regulation of mitochondrial dynamics in neurite outgrowth by retinoic acid receptor β signaling. FASEB J. 2019, 33, 7225–7235. [Google Scholar] [CrossRef] [PubMed]
- Valentijn, J.A.; van Weeren, L.; Ultee, A.; Koster, A.J. Novel localization of Rab3D in rat intestinal goblet cells and Brunner’s gland acinar cells suggests a role in early Golgi trafficking. Am. J. Physiol.-Gastrointest. Liver Physiol. 2007, 293, G165–G177. [Google Scholar] [CrossRef]
- Ranson, N.; Veldhuis, M.; Mitchell, B.; Fanning, S.; Cook, A.L.; Kunde, D.; Eri, R. NLRP3-Dependent and -Independent Processing of Interleukin (IL)-1β in Active Ulcerative Colitis. Int. J. Mol. Sci. 2018, 20, 57. [Google Scholar] [CrossRef]
- Manders, E.M.M.; Verbeek, F.J.; Aten, J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993, 169, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Zamponi, N.; Zamponi, E.; Cannas, S.A.; Billoni, O.V.; Helguera, P.R.; Chialvo, D.R. Mitochondrial network complexity emerges from fission/fusion dynamics. Sci. Rep. 2018, 8, 363. [Google Scholar] [CrossRef]
- Shah, S.I.; Paine, J.G.; Perez, C.; Ullah, G. Mitochondrial fragmentation and network architecture in degenerative diseases. PLoS ONE 2019, 14, e0223014. [Google Scholar] [CrossRef]
- Sukhorukov, V.M.; Dikov, D.; Reichert, A.S.; Meyer-Hermann, M. Emergence of the Mitochondrial Reticulum from Fission and Fusion Dynamics. PLoS Comput. Biol. 2012, 8, e1002745. [Google Scholar] [CrossRef]
- Graham, L.; Orenstein, J.M. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2007, 2, 2439–2450. [Google Scholar] [CrossRef]
- Demeter-Haludka, V.; Kovács, M.; Petrus, A.; Patai, R.; Muntean, D.M.; Siklós, L.; Végh, Á. Examination of the Role of Mitochondrial Morphology and Function in the Cardioprotective Effect of Sodium Nitrite Administered 24 h Before Ischemia/Reperfusion Injury. Front. Pharmacol. 2018, 9, 286. [Google Scholar] [CrossRef]
- Kalkhoran, S.B.; Munro, P.; Qiao, F.; Ong, S.-B.; Hall, A.R.; Cabrera-Fuentes, H.; Chakraborty, B.; Boisvert, W.A.; Yellon, D.M.; Hausenloy, D.J. Unique morphological characteristics of mitochondrial subtypes in the heart: The effect of ischemia and ischemic preconditioning. Discoveries 2017, 5, e71. [Google Scholar] [CrossRef]
- Verma, A.K.; Rice, H.M.; Shapas, B.G.; Boutwell, R.K. Inhibition of 12-O-Tetradecanoylphorbol-13-acetate-induced Ornithine Decarboxylase Activity in Mouse Epidermis by Vitamin A Analogs (Retinoids). Cancer Res. 1978, 38, 793. [Google Scholar]
- Jansz, A.; Flad, H.D.; Koffler, D.; Miescher, P.A. The Effect of Vitamin A on Experimental Immune Thyroiditis. Int. Arch. Allergy 1967, 31, 69–77. [Google Scholar] [CrossRef]
- Strack, S.; Cribbs, J.T. Allosteric Modulation of Drp1 Mechanoenzyme Assembly and Mitochondrial Fission by the Variable Domain*. J. Biol. Chem. 2012, 287, 10990–11001. [Google Scholar] [CrossRef]
- Szalkai, B.; Varga, B.; Grolmusz, V. Comparing advanced graph-theoretical parameters of the connectomes of the lobes of the human brain. Cogn. Neurodyn. 2018, 12, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Schnakenberg, J. Network theory of microscopic and macroscopic behavior of master equation systems. Rev. Mod. Phys. 1976, 48, 571–585. [Google Scholar] [CrossRef]
- Hoffecker, I.T.; Yang, Y.; Bernardinelli, G.; Orponen, P.; Högberg, B. A computational framework for DNA sequencing microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 19282–19287. [Google Scholar] [CrossRef]
- Rohani, A.; Kashatus, J.A.; Sessions, D.T.; Sharmin, S.; Kashatus, D.F. Mito Hacker: A set of tools to enable high-throughput analysis of mitochondrial network morphology. Sci. Rep. 2020, 10, 18941. [Google Scholar] [CrossRef]
- Valente, A.J.; Maddalena, L.A.; Robb, E.L.; Moradi, F.; Stuart, J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017, 119, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Shirihai, O.S. Mitochondrial Dynamics in the Regulation of Nutrient Utilization and Energy Expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Burke, N.; Dongworth, R.K.; Hausenloy, D.J. Mitochondrial fusion and fission proteins: Novel therapeutic targets for combating cardiovascular disease. Br. J. Pharmacol. 2014, 171, 1890–1906. [Google Scholar] [CrossRef]
- Suárez-Rivero, J.M.; Villanueva-Paz, M.; De La Cruz-Ojeda, P.; De La Mata, M.; Cotán, D.; Oropesa-Ávila, M.; De Lavera, I.; Álvarez-Córdoba, M.; Luzón-Hidalgo, R.; Sánchez-Alcázar, J.A. Mitochondrial Dynamics in Mitochondrial Diseases. Diseases 2016, 5, 1. [Google Scholar] [CrossRef]
- Sabouny, R.; Shutt, T.E. Reciprocal Regulation of Mitochondrial Fission and Fusion. Trends Biochem. Sci. 2020, 45, 564–577. [Google Scholar] [CrossRef]
- Martin, O.J.; Lai, L.; Soundarapandian, M.M.; Leone, T.C.; Zorzano, A.; Keller, M.P.; Attie, A.D.; Muoio, D.M.; Kelly, D.P. A role for peroxisome proliferator-activated receptor γ coactivator-1 in the control of mitochondrial dy-namics during postnatal cardiac growth. Circ. Res. 2014, 114, 626–636. [Google Scholar] [CrossRef]
- Choudhary, V.; Kaddour-Djebbar, I.; Lakshmikanthan, V.; Ghazaly, T.; Thangjam, G.S.; Sreekumar, A.; Lewis, R.W.; Mills, I.G.; Bollag, W.B.; Kumar, M.V. Novel Role of Androgens in Mitochondrial Fission and Apoptosis. Mol. Cancer Res. 2011, 9, 1067–1077. [Google Scholar] [CrossRef]
- Richter, V.; Singh, A.P.; Kvansakul, M.; Ryan, M.T.; Osellame, L.D. Splitting up the powerhouse: Structural insights into the mechanism of mitochondrial fission. Cell. Mol. Life Sci. 2015, 72, 3695–3707. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, Y.; Zhang, Z.; Sesaki, H. Mitochondrial division: Molecular machinery and physiological functions. Curr. Opin. Cell Biol. 2011, 23, 427–434. [Google Scholar] [CrossRef][Green Version]
- Hoppins, S.; Nunnari, J. Mitochondrial Dynamics and Apoptosis—The ER Connection. Science 2012, 337, 1052–1054. [Google Scholar] [CrossRef]
- Twig, G.; Elorza, A.; Molina, A.A.J.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.N.; Zhang, Y.; Ren, J. Mitophagy, Mitochondrial Dynamics, and Homeostasis in Cardiovascular Aging. Oxidative Med. Cell. Longev. 2019, 2019, 9825061. [Google Scholar] [CrossRef]
- Ferreira-da-Silva, A.; Valacca, C.; Rios, E.; Pópulo, H.; Soares, P.; Sobrinho-Simões, M.; Scorrano, L.; Máximo, V.; Campello, S. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS ONE 2015, 10, e0122308. [Google Scholar] [CrossRef] [PubMed]
- Giovarelli, M.; Zecchini, S.; Martini, E.; Garrè, M.; Barozzi, S.; Ripolone, M.; Napoli, L.; Coazzoli, M.; Vantaggiato, C.; Roux-Biejat, P.; et al. Drp1 overexpression induces desmin disassembling and drives kinesin-1 activation promoting mitochonrial trafficking in skeletal muscle. Cell Death Differ. 2020, 27, 2383–2401. [Google Scholar] [CrossRef]
- Touvier, T.; De Palma, C.; Rigamonti, E.; Scagliola, A.; Incerti, E.; Mazelin, L.; Thomas, J.-L.; D’Antonio, M.; Politi, L.S.; Schaeffer, L.; et al. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 2015, 6, e1663. [Google Scholar] [CrossRef] [PubMed]
- Szabadkai, G.; Simoni, A.M.; Chami, M.; Wieckowski, M.R.; Youle, R.J.; Rizzuto, R. Drp-1-Dependent Division of the Mitochondrial Network Blocks Intraorganellar Ca2+ Waves and Pro-tects against Ca2+-Mediated Apoptosis. Mol. Cell 2004, 16, 59–68. [Google Scholar] [CrossRef]
- Benard, G.; Bellance, N.; James, D.; Parrone, P.; Fernandez, H.; Letellier, T.; Rossignol, R. Mitochondrial bioenergetics and structural network organization. J. Cell Sci. 2007, 120, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Shirakabe, A.; Maejima, Y.; Zhai, P.; Sciarretta, S.; Toli, J.; Nomura, M.; Mihara, K.; Egashira, K.; Ohishi, M.; et al. Endogenous Drp1 Mediates Mitochondrial Autophagy and Protects the Heart against Energy Stress. Circ. Res. 2015, 116, 264–278. [Google Scholar] [CrossRef]
- Fan, H.; He, Z.; Huang, H.; Zhuang, H.; Liu, H.; Liu, X.; Yang, S.; He, P.; Yang, H.; Feng, D. Mitochondrial Quality Control in Cardiomyocytes: A Critical Role in the Progression of Cardiovascular Diseases. Front. Physiol. 2020, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.-B.; Subrayan, S.; Lim, S.Y.; Yellon, D.M.; Davidson, S.M.; Hausenloy, D.J. Inhibiting Mitochondrial Fission Protects the Heart against Ischemia/Reperfusion Injury. Circulation 2010, 121, 2012–2022. [Google Scholar] [CrossRef]
- Ashrafian, H.; Docherty, L.; Leo, V.; Towlson, C.; Neilan, M.; Steeples, V.; Lygate, C.A.; Hough, T.; Townsend, S.; Williams, D.; et al. A Mutation in the Mitochondrial Fission Gene Dnm1l Leads to Cardiomyopathy. PLoS Genet. 2010, 6, e1001000. [Google Scholar] [CrossRef]
- Tang, Y.; Mi, C.; Liu, J.; Gao, F.; Long, J. Compromised mitochondrial remodeling in compensatory hypertrophied myocardium of spontaneously hy-pertensive rat. Cardiovasc. Pathol. 2014, 23, 101–106. [Google Scholar] [CrossRef]
- Pennanen, C.; Parra, V.; López-Crisosto, C.; Morales, P.E.; Del Campo, A.; Gutierrez, T.; Rivera-Mejías, P.; Kuzmicic, J.; Chiong, M.; Zorzano, A.; et al. Mitochondrial fission is required for cardiomyocyte hypertrophy mediated by a Ca2+-calcineurin signal-ing pathway. J. Cell Sci. 2014, 127 Pt 12, 2659–2671. [Google Scholar] [CrossRef]
- Kageyama, Y.; Hoshijima, M.; Seo, K.; Bedja, D.; Sysa-Shah, P.; Andrabi, S.A.; Chen, W.; Höke, A.; Dawson, V.L.; Dawson, T.M.; et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014, 33, 2798–2813. [Google Scholar] [CrossRef]
- Sharp, W.W.; Fang, Y.H.; Han, M.; Zhang, H.J.; Hong, Z.; Banathy, A.; Morrow, E.; Ryan, J.J.; Archer, S.L. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: Therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J. 2014, 28, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Mihara, K.; Chen, Y.; Scorrano, L.; Dorn, G.W., II. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cul-tured fibroblasts. Cell Metab. 2015, 21, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Huang, Y.; Chen, G.; Chen, Z.; Tweardy, D.J.; Dong, S. Aberrant Chromatin Remodeling by Retinoic Acid Receptor α Fusion Proteins Assessed at the Single-Cell Level. Mol. Biol. Cell 2007, 18, 3941–3951. [Google Scholar] [CrossRef] [PubMed]
- Frezza, C.; Cipolat, S.; De Brito, O.M.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 Controls Apoptotic Cristae Remodeling Independently from Mitochondrial Fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Olichon, A.; Baricault, L.; Gas, N.; Guillou, E.; Valette, A.; Belenguer, P.; Lenaers, G. Loss of OPA1 Perturbates the Mitochondrial Inner Membrane Structure and Integrity, Leading to Cytochrome c Release and Apoptosis. J. Biol. Chem. 2003, 278, 7743–7746. [Google Scholar] [CrossRef] [PubMed]

| Condition | Mean Degree <k> | Ng/N | C1 | C2 | ||
|---|---|---|---|---|---|---|
| Exp | Theory | Exp | Theory | |||
| Ctrl | 1.68 | 1.68 | 0.060 | 0.062 | 5.5 × 10−3 | 2.0 × 10−4 |
| ATRA | 1.58 | 1.59 | 0.047 | 0.045 | 3.0 × 10−3 | 2.0 × 10−4 |
| Condition | X1 | X2 | X3 |
|---|---|---|---|
| Ctrl | 0.436 | 0.541 | 0.023 |
| ATRA | 0.349 | 0.631 | 0.020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chidipi, B.; Shah, S.I.; Reiser, M.; Kanithi, M.; Garces, A.; Cha, B.J.; Ullah, G.; Noujaim, S.F. All-Trans Retinoic Acid Increases DRP1 Levels and Promotes Mitochondrial Fission. Cells 2021, 10, 1202. https://doi.org/10.3390/cells10051202
Chidipi B, Shah SI, Reiser M, Kanithi M, Garces A, Cha BJ, Ullah G, Noujaim SF. All-Trans Retinoic Acid Increases DRP1 Levels and Promotes Mitochondrial Fission. Cells. 2021; 10(5):1202. https://doi.org/10.3390/cells10051202
Chicago/Turabian StyleChidipi, Bojjibabu, Syed Islamuddin Shah, Michelle Reiser, Manasa Kanithi, Amanda Garces, Byeong J. Cha, Ghanim Ullah, and Sami F. Noujaim. 2021. "All-Trans Retinoic Acid Increases DRP1 Levels and Promotes Mitochondrial Fission" Cells 10, no. 5: 1202. https://doi.org/10.3390/cells10051202
APA StyleChidipi, B., Shah, S. I., Reiser, M., Kanithi, M., Garces, A., Cha, B. J., Ullah, G., & Noujaim, S. F. (2021). All-Trans Retinoic Acid Increases DRP1 Levels and Promotes Mitochondrial Fission. Cells, 10(5), 1202. https://doi.org/10.3390/cells10051202







