Fibroblast Growth Factor Receptors (FGFRs) and Noncanonical Partners in Cancer Signaling
Abstract
1. Introduction
2. The FGFR Family
3. FGFR Activation and Signaling
4. Regulation of FGFR Signaling and FGFR Trafficking
5. FGF Signaling Diversity in Cancer
5.1. The FGF1 Subfamily
5.2. The FGF4 Subfamily
5.3. The FGF7 Subfamily
5.4. The FGF8 Subfamily
5.5. FGF9 Subfamily
5.6. FGF15/19 Subfamily
5.7. Intracellular FGFs
6. FGFR Genomic Aberrations in Cancer
| FGFR Gene | Cancer Subtype | Associated References |
|---|---|---|
| FGFR1 | Breast cancer | [10,176,177] |
| Colorectal cancer | [178,179] | |
| Gastrointestinal | [180] | |
| Glioma | [181,182] | |
| Head and neck | [183] | |
| Non-small cell lung cancer | [184,185,186,187,188] | |
| Ovarian cancer | [189] | |
| Pancreatic ductal adenocarcinoma | [190] | |
| Prostate cancer | [191,192,193,194] | |
| Small cell lung cancer | [186,195,196,197] | |
| Urothelial cancer | [189] | |
| FGFR2 | Breast cancer | [10,198] |
| Cholangiocarcinoma | [199,200,201] | |
| Endometrial cancer | [202] | |
| Gastric cancer | [9,203,204] | |
| Non-small cell lung cancer | [205] | |
| FGFR3 | Bladder cancer | [189,206,207,208,209,210,211,212,213,214] |
| Cervical cancer | [209,215,216] | |
| Glioma | [181,189] | |
| Non-small cell lung cancer | [189,205,217,218] | |
| FGFR4 | Adrenocortical cancers | [219] |
| Breast cancer | [189] |
6.1. FGFR1
6.2. FGFR2
6.3. FGFR3
6.4. FGFR4
6.5. FGFR5
7. The Role of Canonical FGFR Cofactors in Cancer
7.1. Heparan Sulfate Proteoglycans
7.2. Klotho
8. Noncanonical Regulators of FGFR Signaling in Cancer
8.1. ECM-Associated Signaling Partners
8.2. Cell Adhesion Molecules
8.3. Other Transmembrane Proteins
8.4. Other Receptor Tyrosine Kinases
8.5. Serine Threonine Kinases
9. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aFGF | Acidic FGF |
| AHCYL1 | S-adenosylhomocysteine hydrolase-like protein 1 |
| ANOS1 | Anosmin-1 or KAL1 |
| bFGF | Basic FGF |
| BICC1 | BicC family RNA binding protein 1 |
| BMP | Bone morphogenic protein |
| CAFs | Cancer associated fibroblasts |
| CAMs | Cell adhesion molecules |
| CBL | E3-ubiquitin ligase CBL |
| CCDC6 | Coiled-coil domain containing protein 6 |
| CRC | Colorectal cancer |
| CRK | Chicken tumor virus no. 10 regulator of kinase |
| CRKL | Closely related CRK-like |
| DAG | Diacylglycerol |
| DUSP6 | Dual specificity phosphatase 6 |
| ECM | Extracellular matrix |
| EGF | Epidermal growth factor |
| EMT | Epithelial-mesenchymal transition |
| EPHA1/2/4 | Ephrin type A receptor 1/2/4 |
| EPHB1 | Ephrin type B receptor 1 |
| ER | Endoplasmic reticulum |
| ER | Estrogen receptor |
| ERK1/2 | Extracellular signal-regulated protein kinase 1/2 |
| ESRP1/2 | Epithelial splicing regulatory protein 1/2 |
| FAK | Focal adhesion kinase |
| FGF | Fibroblast Growth Factor |
| FGFR | Fibroblast Growth Factor Receptor |
| FGFRL1 | FGFR like 1 |
| FHFs | Fibroblast homologous factors |
| FRS2 | FGFR substrate 2 |
| GALNT14 | Polypeptide N-acetyl galactosaminyltransferase |
| GIST | Gastrointestinal stromal tumors |
| GPCR | G protein-coupled receptors |
| GRB2/14 | Growth factor receptor bound 2/14 |
| HCC | Hepatocellular carcinoma |
| HER2 | Human epidermal growth factor receptor 2 |
| HSPGs | Heparan Sulfate Proteoglycans |
| ID1 | Inhibitor of DNA binding 1 transcription factor |
| iFGF | Intracellular FGF |
| IgI-IgIII | Immunoglobulin-like looped domains I-III |
| IgSF | Immunoglobulin superfamily |
| IP3 | Inositol trisphosphate |
| KGF | Keratinocyte growth factor |
| KIAA1217 | Sickle tail protein homolog |
| KIF5B | Kinesin family member 5B |
| LGALS1/3 | Galectin-1/3 |
| MAPK | Mitogen activated protein kinase |
| miRNA | Micro RNA |
| MM | Multiple myeloma |
| MMP9 | Matrix metallopeptidase 9 |
| mTOR | Mammalian target of rapamycin |
| NCAM | Neural cell adhesion molecule |
| NFκB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NRPs | Neuropilins |
| NSCLC | Nonsmall cell lung cancer |
| PDAC | Pancreatic ductal adenocarcinoma |
| PDGFRA | Platelet derived growth factor α |
| PDPK1 | 3-phosphoinositide-dependent protein kinase 1 |
| PI3K | Phosphatidylinositide 3-kinase |
| PI3KR1/2 | PI3K subunit alpha/beta or p85 PI3K subunit |
| PIP2 | Phosphatidylinositol 4,5-bisphosphate |
| PIP3 | Phosphatidylinositol 3,4,5-trisphosphate |
| PKC | Protein kinase C |
| PLCγ | Phospholipase C gamma |
| PP2A | Protein phosphatase 2A |
| PTPRG | Protein tyrosine phosphatase 6 |
| ptdFGFR4 | Pituitary-derived FGFR4 |
| PTPN11 | Protein tyrosine phosphatase nonreceptor type 11 |
| PYK2 | Nonreceptor protein tyrosine kinase 2 |
| RET | Rearranged during transfection |
| RSK2 | Ribosomal S6 kinase 2 |
| RTK | Receptor Tyrosine Kinase |
| SEF | Similar expression to FGF |
| SFK | SRC-family kinases |
| SH2 | SRC homology 2 |
| SH3BP4 | SH3-binding protein 4 |
| SHC | SRC-homology-2 containing transforming protein |
| SHIP2 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 |
| SNP | Single nucleotide polymorphism |
| SPRY1/2/4 | Sprouty 1/2/4 |
| STAT1/3/5 | Signal transducer and activator of transcription 1/3/5 |
| TAB1 | TAK1-binding protein |
| TACC1/3 | Transforming acidic coiled coil 1/3 |
| TAK1 | TGFα-activated kinase 1 |
| TGFBR1/3 | TGFβ receptor 1/3 |
| TGFβ | Transforming growth factor beta |
| TNBC | Triple negative breast cancer |
| VEGFR | Vascular endothelial growth factor receptor |
| Y | Tyrosine |
References
- Wintheiser, G.A.; Silberstein, P. Physiology, tyrosine kinase receptors. Statpearls 2020, internet. [Google Scholar]
- Bennasroune, A.; Gardin, A.; Aunis, D.; Cremel, G.; Hubert, P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit. Rev. Oncol. Hematol. 2004, 50, 23–38. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Fan, C.M.; Flores-Villanueva, P.O.; Generali, D.; Li, Y. The Fibroblast Growth Factor Receptors in Breast Cancer: From Oncogenesis to Better Treatments. Int. J. Mol. Sci. 2020, 21, 2011. [Google Scholar] [CrossRef]
- Gartside, M.G.; Chen, H.B.; Ibrahimi, O.A.; Byron, S.A.; Curtis, A.V.; Wellens, C.L.; Bengston, A.; Yudt, L.M.; Eliseenkova, A.V.; Ma, J.H.; et al. Loss-of-Function Fibroblast Growth Factor Receptor-2 Mutations in Melanoma. Mol. Cancer Res. 2009, 7, 41–54. [Google Scholar] [CrossRef]
- Ricol, D.; Cappellen, D.; El Marjou, A.; Gil-Diez-de-Medina, S.; Girault, J.M.; Yoshida, T.; Ferry, G.; Tucker, G.; Poupon, M.F.; Chopin, D.; et al. Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer. Oncogene 1999, 18, 7234–7243. [Google Scholar] [CrossRef] [PubMed]
- Amann, T.; Bataille, F.; Spruss, T.; Dettmer, K.; Wild, P.; Liedtke, C.; Muhlbauer, M.; Kiefer, P.; Oefner, P.J.; Trautwein, C.; et al. Reduced Expression of Fibroblast Growth Factor Receptor 2IIIb in Hepatocellular Carcinoma Induces a More Aggressive Growth. Am. J. Pathol. 2010, 176, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, J.H.; Jang, H.J.; Han, B.; Zang, D.Y. Pathological and Prognostic Impacts of FGFR2 Overexpression in Gastric Cancer: A Meta-Analysis. J. Cancer 2019, 10, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Arao, T.; Hamaguchi, T.; Shimada, Y.; Kato, K.; Oda, I.; Taniguchi, H.; Koizumi, F.; Yanagihara, K.; Sasaki, H.; et al. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br. J. Cancer 2012, 106, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Lambros, M.B.; Horlings, H.M.; Pearson, A.; Sharpe, R.; Natrajan, R.; Geyer, F.C.; van Kouwenhove, M.; Kreike, B.; Mackay, A.; et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010, 29, 2013–2023. [Google Scholar] [CrossRef]
- Campbell, J.; Ryan, C.J.; Brough, R.; Bajrami, I.; Pemberton, H.N.; Chong, I.Y.; Costa-Cabral, S.; Frankum, J.; Gulati, A.; Holme, H.; et al. Large-Scale Profiling of Kinase Dependencies in Cancer Cell Lines. Cell Rep. 2016, 14, 2490–2501. [Google Scholar] [CrossRef]
- Vecchione, A.; Cooper, H.J.; Trim, K.J.; Akbarzadeh, S.; Heath, J.K.; Wheldon, L.M. Protein partners in the life history of activated fibroblast growth factor receptors. Proteomics 2007, 7, 4565–4578. [Google Scholar] [CrossRef] [PubMed]
- Balek, L.; Nemec, P.; Konik, P.; Bosakova, M.K.; Varecha, M.; Gudernova, I.; Medalova, J.; Krakow, D.; Krejci, P. Proteomic analyses of signalling complexes associated with receptor tyrosine kinase identify novel members of fibroblast growth factor receptor 3 interactome. Cell. Signal. 2018, 42, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Latko, M.; Czyrek, A.; Porebska, N.; Kucinska, M.; Otlewski, J.; Zakrzewska, M.; Opalinski, L. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 2019, 8, 455. [Google Scholar] [CrossRef]
- Lee, P.L.; Johnson, D.E.; Cousens, L.S.; Fried, V.A.; Williams, L.T. Purification and complementary-dna cloning of a receptor for basic fibroblast growth-factor. Science 1989, 245, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Ruta, M.; Howk, R.; Ricca, G.; Drohan, W.; Zabelshansky, M.; Laureys, G.; Barton, D.E.; Francke, U.; Schlessinger, J.; Givol, D. A novel protein tyrosine kinase gene whose expression is modulated during endothelial-cell differentiation. Oncogene 1988, 3, 9–15. [Google Scholar]
- Ruta, M.; Burgess, W.; Givol, D.; Epstein, J.; Neiger, N.; Kaplow, J.; Crumley, G.; Dionne, C.; Jaye, M.; Schlessinger, J. Receptor for Acidic Fibroblast Growth-Factor is Related to the Tyrosine Kinase Encoded by the Fms-Like Gene (Flg). Proc. Natl. Acad. Sci. USA 1989, 86, 8722–8726. [Google Scholar] [CrossRef] [PubMed]
- Kornbluth, S.; Paulson, K.E.; Hanafusa, H. Novel tyrosine kinase identified by phosphotyrosine antibody screening of cDNA libraries. Mol. Cell. Biol. 1988, 8, 5541–5544. [Google Scholar] [CrossRef]
- Keegan, K.; Johnson, D.E.; Williams, L.T.; Hayman, M.J. Isolation of an additional member of the fibroblast growth-factor receptor family, FGFR-3. Proc. Natl. Acad. Sci. USA 1991, 88, 1095–1099. [Google Scholar] [CrossRef]
- Partanen, J.; Makela, T.P.; Eerola, E.; Korhonen, J.; Hirvonen, H.; Claessonwelsh, L.; Alitalo, K. FGFR-4, a novel acidic fibroblast growth-factor receptor with a distinct expression pattern. EMBO J. 1991, 10, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, M.; Fraser, J.; McDonald, M.; Yuan, S.N.; White, D.; Grandison, P.; Kumble, K.; Watson, J.D.; Murison, J.G. Identification of a new fibroblast growth factor receptor, FGFR5. Gene 2001, 271, 171–182. [Google Scholar] [CrossRef]
- Wiedemann, M.; Trueb, B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics 2000, 69, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Regeenes, R.; Silva, P.N.; Chang, H.H.; Arany, E.J.; Shukalyuk, A.I.; Audet, J.; Kilkenny, D.M.; Rocheleau, J.V. Fibroblast growth factor receptor 5 (FGFR5) is a co-receptor for FGFR1 that is up-regulated in beta-cells by cytokine-induced inflammation. J. Biol. Chem. 2018, 293, 17218–17228. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.N.; Altamentova, S.M.; Kilkenny, D.M.; Rocheleau, J.V. Fibroblast Growth Factor Receptor Like-1 (FGFRL1) Interacts with SHP-1 Phosphatase at Insulin Secretory Granules and Induces Beta-cell ERK1/2 Protein Activation. J. Biol. Chem. 2013, 288, 17859–17870. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Lu, J.; Chen, H.; Werner, S.; Williams, L.T. The human fibroblast growth-factor receptor genes—a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their 3rd immunoglobulin domain. Mol. Cell. Biol. 1991, 11, 4627–4634. [Google Scholar] [CrossRef]
- Werner, S.; Duan, D.S.R.; Devries, C.; Peters, K.G.; Johnson, D.E.; Williams, L.T. Differential splicing in the extracellular region of fibroblast growth-factor receptor-1 generates receptor variants with different ligand-binding specificities. Mol. Cell. Biol. 1992, 12, 82–88. [Google Scholar] [CrossRef]
- Miki, T.; Bottaro, D.P.; Fleming, T.P.; Smith, C.L.; Burgess, W.H.; Chan, A.M.L.; Aaronson, S.A. Determination of ligand-binding specificity by alternative splicing—2 distinct growth-factor receptors encoded by a single gene. Proc. Natl. Acad. Sci. USA 1992, 89, 246–250. [Google Scholar] [CrossRef]
- Yayon, A.; Zimmer, Y.; Shen, G.H.; Avivi, A.; Yarden, Y.; Givol, D. A Confined variable region confers ligand specificity on fibroblast growth-factor receptors—implications for the origin of the immunoglobulin fold. EMBO J. 1992, 11, 1885–1890. [Google Scholar] [CrossRef]
- Orrurtreger, A.; Bedford, M.T.; Burakova, T.; Arman, E.; Zimmer, Y.; Yayon, A.; Givol, D.; Lonai, P. Developmental localization of the splicing alternatives of fibroblast growth-factor receptor-2 (fgfr2). Dev. Biol. 1993, 158, 475–486. [Google Scholar] [CrossRef]
- Warzecha, C.C.; Sato, T.K.; Nabet, B.; Hogenesch, J.B.; Carstens, R.P. ESRP1 and ESRP2 Are Epithelial Cell-Type-Specific Regulators of FGFR2 Splicing. Mol. Cell 2009, 33, 591–601. [Google Scholar] [CrossRef]
- Johnson, D.E.; Lee, P.L.; Lu, J.; Williams, L.T. Diverse forms of a receptor for acidic and basic fibroblast growth-factors. Mol. Cell. Biol. 1990, 10, 4728–4736. [Google Scholar] [CrossRef]
- Hou, J.Z.; Kan, M.; McKeehan, K.; McBride, G.; Adams, P.; McKeehan, W.L. Fibroblast growth-factor receptors from liver vary in 3 structural domains. Science 1991, 251, 665–668. [Google Scholar] [CrossRef]
- Shi, E.; Kan, M.; Xu, J.M.; Wang, F.; Hou, J.Z.; McKeehan, W.L. Control of fibroblast growth-factor receptor kinase signal-transduction by heterodimerization of combinatorial splice variants. Mol. Cell. Biol. 1993, 13, 3907–3918. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kalinina, J.; Dutta, K.; Ilghari, D.; Beenken, A.; Goetz, R.; Eliseenkova, A.V.; Cowburn, D.; Mohammadi, M. The Alternatively Spliced Acid Box Region Plays a Key Role in FGF Receptor Autoinhibition. Structure 2012, 20, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Odagiri, H.; Nakatani, H.; Miyagawa, K.; Naito, K.; Sakamoto, H.; Katoh, O.; Yoshida, T.; Sugimura, T.; Terada, M. K-SAM, an amplified gene in stomach-cancer, is a member of the heparin-binding growth-factor receptor genes. Proc. Natl. Acad. Sci. USA 1990, 87, 5983–5987. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, A.; Tada, K.; Shukunami, C.; Hiraki, Y.; Kurokawa, T.; Magane, N.; Kurokawa-Seo, M. A novel alternatively spliced fibroblast growth factor receptor 3 isoform lacking the acid box domain is expressed during chondrogenic differentiation of ATDC5 cells. J. Biol. Chem. 2001, 276, 11031–11040. [Google Scholar] [CrossRef]
- Miki, T.; Fleming, T.P.; Bottaro, D.P.; Rubin, J.S.; Ron, D.; Aaronson, S.A. Expression cDNA cloning of the KGF receptor by creation of a transforming autocrine loop. Science 1991, 251, 72–75. [Google Scholar] [CrossRef]
- Bae, J.H.; Boggon, T.J.; Tome, F.; Mandiyan, V.; Lax, I.; Schlessinger, J. Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 2866–2871. [Google Scholar] [CrossRef]
- Peng, W.C.; Lin, X.; Torres, J. The strong dimerization of the transmembrane domain of the fibroblast growth factor receptor (FGFR) is modulated by C-terminal juxtamembrane residues. Protein Sci. 2009, 18, 450–459. [Google Scholar] [CrossRef]
- Mohammadi, M.; Dikic, I.; Sorokin, A.; Burgess, W.H.; Jaye, M.; Schlessinger, J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol. Cell. Biol. 1996, 16, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Honegger, A.M.; Rotin, D.; Fischer, R.; Bellot, F.; Li, W.; Dionne, C.A.; Jaye, M.; Rubinstein, M.; Schlessinger, J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth-factor receptor (FLG) is a binding-site for the sh2 domain of phospholipase C-gamma-1. Mol. Cell. Biol. 1991, 11, 5068–5078. [Google Scholar] [CrossRef]
- Francavilla, C.; Rigbolt, K.T.G.; Emdal, K.B.; Carraro, G.; Vernet, E.; Bekker-Jensen, D.B.; Streicher, W.; Wikstrom, M.; Sundstrom, M.; Bellusci, S.; et al. Functional Proteomics Defines the Molecular Switch Underlying FGF Receptor Trafficking and Cellular Outputs. Mol. Cell 2013, 51, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Salazar, L.; Kashiwada, T.; Krejci, P.; Muchowski, P.; Donoghue, D.; Wilcox, W.R.; Thompson, L.M. A novel interaction between fibroblast growth factor receptor 3 and the p85 subunit of phosphoinositide 3-kinase: Activation-dependent regulation of ERK by p85 in multiple myeloma cells. Hum. Mol. Genet. 2009, 18, 1951–1961. [Google Scholar] [CrossRef]
- Kouhara, H.; Hadari, Y.R.; SpivakKroizman, T.; Schilling, J.; BarSagi, D.; Lax, I.; Schlessinger, J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 1997, 89, 693–702. [Google Scholar] [CrossRef]
- Ong, S.H.; Guy, G.R.; Hadari, Y.R.; Laks, S.; Gotoh, N.; Schlessinger, J.; Lax, I. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol. 2000, 20, 979–989. [Google Scholar] [CrossRef]
- Hadari, Y.R.; Kouhara, H.; Lax, I.; Schlessinger, J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell. Biol. 1998, 18, 3966–3973. [Google Scholar] [CrossRef]
- Larsson, H.; Klint, P.; Landgren, E.; Claesson-Welsh, L. Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J. Biol. Chem. 1999, 274, 25726–25734. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Suenaga, A.; Hatakeyama, M.; Taiji, M.; Imamoto, A. Structural and Functional Basis of a Role for CRKL in a Fibroblast Growth Factor 8-Induced Feed-Forward Loop. Mol. Cell. Biol. 2009, 29, 3076–3087. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.N.; Mao, Y.Y.; Li, H.G.; Bouaziz, M.; Hong, A.; Feng, G.S.; Wang, F.; Quilliam, L.A.; Chen, L.; Park, T.; et al. Crk proteins transduce FGF signaling to promote lens fiber cell elongation. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.M.; Guris, D.L.; Seo, J.H.; Li, L.M.; Hammond, J.; Talbot, A.; Imamoto, A. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev. Cell 2006, 10, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Fafilek, B.; Balek, L.; Bosakova, M.K.; Varecha, M.; Nita, A.; Gregor, T.; Gudernova, I.; Krenova, J.; Ghosh, S.; Piskacek, M.; et al. The inositol phosphatase SHIP2 enables sustained ERK activation downstream of FGF receptors by recruiting Src kinases. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Krejci, P.; Aklian, A.; Kaucka, M.; Sevcikova, E.; Prochazkova, J.; Masek, J.K.; Mikolka, P.; Pospisilova, T.; Spoustova, T.; Weis, M.; et al. Receptor Tyrosine Kinases Activate Canonical WNT/beta-Catenin Signaling via MAP Kinase/LRP6 Pathway and Direct beta-Catenin Phosphorylation. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Elf, S.; Dong, S.; Hitosugi, T.; Guo, A.; Ruan, H.; Lonial, S.; Khoury, H.; Williams, I.; Karsenty, G.; et al. FGFR3 Associates with and Tyrosine-Phosphorylates p90RSK2, Leading to RSK2 Activation That Mediates Hematopoietic Transformation. Blood 2008, 112, 1276. [Google Scholar] [CrossRef]
- Kang, S.; Elf, S.; Dong, S.; Hitosugi, T.; Lythgoe, K.; Guo, A.; Ruan, H.; Lonial, S.; Khoury, H.J.; Williams, I.R.; et al. Fibroblast Growth Factor Receptor 3 Associates with and Tyrosine Phosphorylates p90 RSK2, Leading to RSK2 Activation That Mediates Hematopoietic Transformation. Mol. Cell. Biol. 2009, 29, 2105–2117. [Google Scholar] [CrossRef][Green Version]
- Ong, S.H.; Hadari, Y.R.; Gotoh, N.; Guy, G.R.; Schlessinger, J.; Lax, I. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 6074–6079. [Google Scholar] [CrossRef]
- Weylie, B.; Zhu, J.; Singh, U.; Ambrus, S.; Forough, R. Phosphatidylinositide 3-kinase is important in late-stage fibroblast growth factor-1-mediated angiogenesis in vivo. J. Vasc. Res. 2006, 43, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Forough, R.; Weylie, B.; Patel, C.; Ambrus, S.; Singh, L.S.; Zhu, J. Role of AKT/PKB signaling in fibroblast growth factor-1 (FGF-1)-induced angiogenesis in the chicken chorioallantoic membrane (CAM). J. Cell. Biochem. 2005, 94, 109–116. [Google Scholar] [CrossRef]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.P.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087. [Google Scholar] [CrossRef]
- Yang, X.H.; Qiao, D.H.; Meyer, K.; Friedl, A. Signal Transducers and Activators of Transcription Mediate Fibroblast Growth Factor-Induced Vascular Endothelial Morphogenesis. Cancer Res. 2009, 69, 1668–1677. [Google Scholar] [CrossRef]
- Su, W.C.S.; Kitagawa, M.; Xue, N.R.; Xie, B.; Garofalo, S.; Cho, J.; Deng, C.X.; Horton, W.A.; Fu, X.Y. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature 1997, 386, 288–292. [Google Scholar] [CrossRef]
- Hart, K.C.; Robertson, S.C.; Kanemitsu, M.Y.; Meyer, A.N.; Tynan, J.A.; Donoghue, D.J. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene 2000, 19, 3309–3320. [Google Scholar] [CrossRef] [PubMed]
- Dudka, A.A.; Sweet, S.M.M.; Heath, J.K. Signal Transducers and Activators of Transcription-3 Binding to the Fibroblast Growth Factor Receptor Is Activated by Receptor Amplification. Cancer Res. 2010, 70, 3391–3401. [Google Scholar] [CrossRef] [PubMed]
- Lonic, A.; Barry, E.F.; Quach, C.; Kobe, B.; Saunders, N.; Guthridge, M.A. Fibroblast growth factor receptor 2 phosphorylation on serine 779 couples to 14-3-3 and regulates cell survival and proliferation. Mol. Cell. Biol. 2008, 28, 3372–3385. [Google Scholar] [CrossRef][Green Version]
- Lonic, A.; Powell, J.A.; Kong, Y.; Thomas, D.; Holien, J.K.; Truong, N.; Parker, M.W.; Guthridge, M.A. Phosphorylation of Serine 779 in Fibroblast Growth Factor Receptor 1 and 2 by Protein Kinase C epsilon Regulates Ras/Mitogen-activated Protein Kinase Signaling and Neuronal Differentiation. J. Biol. Chem. 2013, 288, 14874–14885. [Google Scholar] [CrossRef]
- Zakrzewska, M.; Haugsten, E.M.; Nadratowska-Wesolowska, B.; Oppelt, A.; Hausott, B.; Jin, Y.X.; Otlewski, J.; Wesche, J.; Wiedlocha, A. ERK-Mediated Phosphorylation of Fibroblast Growth Factor Receptor 1 on Ser(777) Inhibits Signaling. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef]
- Lax, I.; Wong, A.; Lamothe, B.; Lee, A.; Frost, A.; Hawes, J.; Schlessinger, J. The docking protein FRS2 alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol. Cell 2002, 10, 709–719. [Google Scholar] [CrossRef]
- Zakrzewska, M.; Opalinski, L.; Haugsten, E.M.; Otlewski, J.; Wiedlocha, A. Crosstalk between p38 and Erk 1/2 in Downregulation of FGF1-Induced Signaling. Int. J. Mol. Sci. 2019, 20, 1826. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Z.M.; Yuan, H.; Ji, W.X.; Wang, K.X.; Lu, T.T.; Yu, Y.F.; Zeng, Q.Y.; Li, F.; Xia, W.L.; et al. Reciprocal regulatory mechanism between miR-214-3p and FGFR1 in FGFR1-amplified lung cancer. Oncogenesis 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Schelch, K.; Kirschner, M.B.; Williams, M.; Cheng, Y.Y.; van Zandwijk, N.; Grusch, M.; Reid, G. A link between the fibroblast growth factor axis and the miR-16 family reveals potential new treatment combinations in mesothelioma. Mol. Oncol. 2018, 12, 58–73. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.W.; Wang, X.C.; Zheng, C.; Ma, W.D. Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem. Biophys. Res. Commun. 2013, 439, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Tian, Z.; Zhu, H.Y.; Ouyang, X.Y. miRNA-133b targets FGFR1 and presents multiple tumor suppressor activities in osteosarcoma. Cancer Cell Int. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cheng, Y.; Zhang, Y.; Han, K. MicroRNA-889-3p targets FGFR2 to inhibit cervical cancer cell viability and invasion. Exp. Ther. Med. 2019, 18, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.Y.; Zhao, N.; Xie, N.L.; Ye, M.X.; Zhang, Y.; Zhang, Z.P.; Li, M.Y.; Lai, X.F.; Zhang, J.; Gu, Z.P. miR-24-3p/FGFR3 Signaling as a Novel Axis Is Involved in Epithelial-Mesenchymal Transition and Regulates Lung Adenocarcinoma Progression. J. Immunol. Res. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Lo, T.L.; Yusoff, P.; Fong, C.W.; Guo, K.; McCaw, B.J.; Phillips, W.A.; Yang, H.; Wong, E.S.M.; Leong, H.F.; Zeng, Q.; et al. The Ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res. 2004, 64, 6127–6136. [Google Scholar] [CrossRef]
- Sasaki, A.; Taketomi, T.; Wakioka, T.; Kato, R.; Yoshimura, A. Identification of a dominant negative mutant of sprouty that potentiates fibroblast growth factor-but not epidermal growth factor-induced ERK activation. J. Biol. Chem. 2001, 276, 36804–36808. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.W.; Leong, H.F.; Wong, E.S.M.; Lim, J.; Yusoff, P.; Guy, G.R. Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J. Biol. Chem. 2003, 278, 33456–33464. [Google Scholar] [CrossRef]
- Rubin, C.; Zwang, Y.; Vaisman, N.; Ron, D.; Yarden, Y. Phosphorylation of carboxyl-terminal tyrosines modulates the specificity of sprouty-2 inhibition of different signaling pathways. J. Biol. Chem. 2005, 280, 9735–9744. [Google Scholar] [CrossRef]
- Mason, J.M.; Morrison, D.J.; Bassit, B.; Dimri, M.; Band, H.; Licht, J.D.; Gross, I. Tyrosine phosphorylation of sprouty proteins regulates their ability to inhibit growth factor signaling: A dual feedback loop. Mol. Biol. Cell 2004, 15, 2176–2188. [Google Scholar] [CrossRef]
- Lao, D.H.; Yusoff, P.; Chandramouli, S.; Philp, R.J.; Fong, C.W.; Jackson, R.A.; Saw, T.Y.; Yu, C.Y.; Guy, G.R. Direct binding of PP2A to Sprouty2 and phosphorylation changes are a prerequisite for ERK inhibition downstream of fibroblast growth factor receptor stimulation. J. Biol. Chem. 2007, 282, 9117–9126. [Google Scholar] [CrossRef]
- Li, X.; Brunton, V.G.; Burgar, H.R.; Wheldon, L.M.; Heath, J.K. FRS2-dependent SRC activation is required for fibroblast growth factor receptor-induced phosphorylation of sprouty and suppression of ERK activity. J. Cell Sci. 2004, 117, 6007–6017. [Google Scholar] [CrossRef][Green Version]
- Hanafusa, H.; Torii, S.; Yasunaga, T.; Matsumoto, K.; Nishida, E. Shp2, an SH2-containing protein-tyrosine phosphatase, positively regulates receptor tyrosine kinase signaling by dephosphorylating and inactivating the inhibitor sprouty. J. Biol. Chem. 2004, 279, 22992–22995. [Google Scholar] [CrossRef] [PubMed]
- Kostas, M.; Haugsten, E.M.; Zhen, Y.; Sorensen, V.; Szybowska, P.; Fiorito, E.; Lorenz, S.; Jones, N.; de Souza, G.A.; Wiedlocha, A.; et al. Protein Tyrosine Phosphatase Receptor Type G (PTPRG) Controls Fibroblast Growth Factor Receptor (FGFR) 1 Activity and Influences Sensitivity to FGFR Kinase Inhibitors. Mol. Cell. Proteom. 2018, 17, 850–870. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Mizuno, H.; Sase, H.; Fujii, T.; Sakata, K.; Akiyama, N.; Aoki, Y.; Aoki, M.; Ishii, N. ERK Signal Suppression and Sensitivity to CH5183284/Debio 1347, a Selective FGFR Inhibitor. Mol. Cancer Ther. 2015, 14, 2831–2839. [Google Scholar] [CrossRef] [PubMed]
- Browaeys-Poly, E.; Blanquart, C.; Perdereau, D.; Antoine, A.F.; Goenaga, D.; Luzy, J.P.; Chen, H.X.; Garbay, C.; Issad, T.; Cailliau, K.; et al. Grb14 inhibits FGF receptor signaling through the regulation of PLC gamma recruitment and activation. FEBS Lett. 2010, 584, 4383–4388. [Google Scholar] [CrossRef]
- Cailliau, K.; Perdereau, D.; Lescuyer, A.; Chen, H.X.; Garbay, C.; Vilain, J.P.; Burnol, A.F.; Browaeys-Poly, E. FGF receptor phosphotyrosine 766 is a target for Grb14 to inhibit MDA-MB-231 human breast cancer cell signaling. Anticancer Res. 2005, 25, 3877–3882. [Google Scholar]
- Reilly, J.F.; Mickey, G.; Maher, P.A. Association of fibroblast growth factor receptor 1 with the adaptor protein Grb14—Characterization of a new receptor binding partner. J. Biol. Chem. 2000, 275, 7771–7778. [Google Scholar] [CrossRef]
- Mohapatra, B.; Ahmad, G.; Nadeau, S.; Zutshi, N.; An, W.; Scheffe, S.; Dong, L.; Feng, D.; Goetz, B.; Arya, P.; et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of Cbl-family ubiquitin ligases. Biochim. Et Biophys. Acta Mol. Cell Res. 2013, 1833, 122–139. [Google Scholar] [CrossRef]
- Wong, A.; Lamothe, B.; Li, A.; Schlessinger, J.; Lax, I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc. Natl. Acad. Sci. USA 2002, 99, 6684–6689. [Google Scholar] [CrossRef] [PubMed]
- Dufour, C.; Guenou, H.; Kaabeche, K.; Bouvard, D.; Sanjay, A.; Marie, P.J. FGFR2-Cb1 interaction in lipid rafts triggers attenuation of PI3K/Akt signaling and osteoblast survival. Bone 2008, 42, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Haugsten, E.M.; Malecki, J.; Bjorklund, S.M.S.; Olsnes, S.; Wesche, J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 2008, 19, 3390–3403. [Google Scholar] [CrossRef]
- Haugsten, E.M.; Sorensen, V.; Brech, A.; Olsnes, S.; Wesche, J. Different intracellular trafficking of FGF1 endocytosed by the four homologous FGF receptors. J. Cell Sci. 2005, 118, 3869–3881. [Google Scholar] [CrossRef]
- Belleudi, F.; Leone, L.; Nobili, V.; Raffa, S.; Francescangeli, F.; Maggio, M.; Morrone, S.; Marchese, C.; Torrisi, M.R. Keratinocyte growth factor receptor ligands target the receptor to different intracellular pathways. Traffic 2007, 8, 1854–1872. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef] [PubMed]
- McNiel, E.A.; Tsichlis, P.N. Analyses of publicly available genomics resources define FGF-2-expressing bladder carcinomas as EMT-prone, proliferative tumors with low mutation rates and high expression of CTLA-4, PD-1 and PD-L1. Signal Transduct. Target. Ther. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Santolla, M.F.; Vivacqua, A.; Lappano, R.; Rigiracciolo, D.C.; Cirillo, F.; Galli, G.R.; Talia, M.; Brunetti, G.; Miglietta, A.M.; Belfiore, A.; et al. GPER Mediates a Feedforward FGF2/FGFR1 Paracrine Activation Coupling CAFs to Cancer Cells toward Breast Tumor Progression. Cells 2019, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Birck, A.; Kirkin, A.F.; Zeuthen, J.; Hou-Jensen, K. Expression of basic fibroblast growth factor and vascular endothelial growth factor in primary and metastatic melanoma from the same patients. Melanoma Res. 1999, 9, 375–382. [Google Scholar] [CrossRef]
- Manousakidi, S.; Guillaume, A.; Pirou, C.; Bouleau, S.; Mignotte, B.; Renaud, F.; Le Floch, N. FGF1 induces resistance to chemotherapy in ovarian granulosa tumor cells through regulation of p53 mitochondrial localization. Oncogenesis 2018, 7. [Google Scholar] [CrossRef]
- Awaji, M.; Futakuchi, M.; Heavican, T.; Iqbal, J.; Singh, R.K. Cancer-Associated Fibroblasts Enhance Survival and Progression of the Aggressive Pancreatic Tumor Via FGF-2 and CXCL8. Cancer Microenviron. 2019, 12, 37–46. [Google Scholar] [CrossRef]
- Ruotsalainen, T.; Joensuu, H.; Mattson, K.; Salven, P. High pretreatment serum concentration of basic fibroblast growth factor is a predictor of poor prognosis in small cell lung cancer. Cancer Epidemiol. Biomark. Prev. 2002, 11, 1492–1495. [Google Scholar]
- Han, D.X.; Wang, M.M.; Yu, Z.K.; Yin, L.; Liu, C.L.; Wang, J.M.; Liu, Y.J.; Jiang, S.Y.; Ren, Z.W.; Yin, J. FGF5 promotes osteosarcoma cells proliferation via activating MAPK signaling pathway. Cancer Manag. Res. 2019, 11, 6457–6466. [Google Scholar] [CrossRef] [PubMed]
- Urbini, M.; Indio, V.; Tarantino, G.; Ravegnini, G.; Angelini, S.; Nannini, M.; Saponara, M.; Santini, D.; Ceccarelli, C.; Fiorentino, M.; et al. Gain of FGF4 is a frequent event in KIT/PDGFRA/SDH/RAS-P WT GIST. Genes Chromosomes Cancer 2019, 58, 636–642. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Drier, Y.; Johnstone, S.E.; Hemming, M.L.; Tarjan, D.R.; Hegazi, E.; Shareef, S.J.; Javed, N.M.; Raut, C.P.; Eschle, B.K.; et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 2019, 575, 229. [Google Scholar] [CrossRef] [PubMed]
- Allerstorfer, S.; Sonvilla, G.; Fischer, H.; Spiegl-Kreinecker, S.; Gauglhofer, C.; Setinek, U.; Czech, T.; Marosi, C.; Buchroithner, J.; Pichler, J.; et al. FGF5 as an oncogenic factor in human glioblastoma multiforme: Autocrine and paracrine activities. Oncogene 2008, 27, 4180–4190. [Google Scholar] [CrossRef]
- Huang, T.T.; Wang, L.; Liu, D.; Li, P.; Xiong, H.H.; Zhuang, L.; Sun, L.; Yuan, X.L.; Qiu, H. FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin-1. Int. J. Oncol. 2017, 50, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Mei, Y.B.; Wang, Y.; Feng, Z.C. miR-5582-5p inhibits cell proliferation of non-small cell lung cancer through targeting FGF-10. Int. J. Clin. Exp. Pathol. 2019, 12, 1087–1094. [Google Scholar] [PubMed]
- Liu, H.Y.; Zhao, H.; Li, W.X. Integrated Analysis of Transcriptome and Prognosis Data Identifies FGF22 as a Prognostic Marker of Lung Adenocarcinoma. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef]
- Nomura, S.; Yoshitomi, H.; Takano, S.; Shida, T.; Kobayashi, S.; Ohtsuka, M.; Kimura, F.; Shimizu, H.; Yoshidome, H.; Kato, A.; et al. FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br. J. Cancer 2008, 99, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Harpain, F.; Ahmed, M.A.; Hudec, X.; Timelthaler, G.; Jomrich, G.; Mullauer, L.; Selzer, E.; Dorr, W.; Bergmann, M.; Holzmann, K.; et al. FGF8 induces therapy resistance in neoadjuvantly radiated rectal cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 77–86. [Google Scholar] [CrossRef]
- Gauglhofer, C.; Sagmeister, S.; Schrottmaier, W.; Fischer, C.; Rodgarkia-Dara, C.; Mohr, T.; Stattner, S.; Bichler, C.; Kandioler, D.; Wrba, F.; et al. Up-Regulation of the Fibroblast Growth Factor 8 Subfamily in Human Hepatocellular Carcinoma for Cell Survival and Neoangiogenesis. Hepatology 2011, 53, 854–864. [Google Scholar] [CrossRef]
- Hao, Y.L.; Xiao, Y.X.; Liao, X.Y.; Tang, S.Y.; Xie, X.Y.; Liu, R.; Chen, Q.M. FGF8 induces epithelial-mesenchymal transition and promotes metastasis in oral squamous cell carcinoma. Int. J. Oral Sci. 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Heer, R.; Douglas, D.; Mathers, M.E.; Robson, C.N.; Leung, H.Y. Fibroblast growth factor 17 is over-expressed in human prostate cancer. J. Pathol. 2004, 204, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, Z.Y.; Ye, F.D.; Mou, Z.Z.; Chen, X.N.; Ou, Y.X.; Xu, C.Y.; Wu, S.Q.; Cheng, Z.; Hu, J.M.; et al. FGF18 Inhibits Clear Cell Renal Cell Carcinoma Proliferation and Invasion via Regulating Epithelial-Mesenchymal Transition. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Mizukami, T.; Togashi, Y.; Naruki, S.; Banno, E.; Terashima, M.; de Velasco, M.A.; Sakai, K.; Yoneshige, A.; Hayashi, H.; Fujita, Y.; et al. Significance of FGF9 gene in resistance to anti-EGFR therapies targeting colorectal cancer: A subset of colorectal cancer patients with FGF9 upregulation may be resistant to anti-EGFR therapies. Mol. Carcinog. 2017, 56, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Seitz, T.; Freese, K.; Dietrich, P.; Thasler, W.E.; Bosserhoff, A.; Hellerbrand, C. Fibroblast Growth Factor 9 is expressed by activated hepatic stellate cells and promotes progression of hepatocellular carcinoma. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Teishima, J.; Shoji, K.; Hayashi, T.; Miyamoto, K.; Ohara, S.; Matsubara, A. Relationship between the localization of fibroblast growth factor 9 in prostate cancer cells and postoperative recurrence. Prostate Cancer Prostatic Dis. 2012, 15, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Teishima, J.; Yano, S.; Shoji, K.; Hayashi, T.; Goto, K.; Kitano, H.; Oka, K.; Nagamatsu, H.; Matsubara, A. Accumulation of FGF9 in Prostate Cancer Correlates with Epithelial-to-Mesenchymal Transition and Induction of VEGF-A Expression. Anticancer Res. 2014, 34, 695–700. [Google Scholar]
- Qian, J.; Tikk, K.; Weigl, K.; Balavarca, Y.; Brenner, H. Fibroblast growth factor 21 as a circulating biomarker at various stages of colorectal carcinogenesis. Br. J. Cancer 2018, 119, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Cymbaluk-Ploska, A.; Gargulinska, P.; Chudecka-Glaz, A.; Kwiatkowski, S.; Pius-Sadowska, E.; Machalinski, B. The Suitability of FGF21 and FGF23 as New Biomarkers in Endometrial Cancer Patients. Diagnostics 2020, 10, 414. [Google Scholar] [CrossRef]
- Kanzaki, H.; Chiba, T.; Ao, J.; Koroki, K.; Kanayama, K.; Maruta, S.; Maeda, T.; Kusakabe, Y.; Kobayashi, K.; Kanogawa, N.; et al. The impact of FGF19/FGFR4 signaling inhibition in antitumor activity of multi-kinase inhibitors in hepatocellular carcinoma. Sci. Rep. 2021, 11, 5303. [Google Scholar] [CrossRef]
- Feng, S.; Dakhova, O.; Creighton, C.J.; Ittmann, M. Endocrine Fibroblast Growth Factor FGF19 Promotes Prostate Cancer Progression. Cancer Res. 2013, 73, 2551–2562. [Google Scholar] [CrossRef] [PubMed]
- Su, T.H.; Huang, L.L.; Zhang, N.; Peng, S.; Li, X.X.; Wei, G.Y.; Zhai, E.T.; Zeng, Z.R.; Xu, L.X. FGF14 Functions as a Tumor Suppressor through Inhibiting PI3K/AKT/mTOR Pathway in Colorectal Cancer. J. Cancer 2020, 11, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Li, L.; Yeh, S.Y.; Cui, Y.; Li, X.; Chang, H.C.; Jin, J.; Chang, C.S. Infiltrating T cells promote prostate cancer metastasis via modulation of FGF11 -> miRNA-541 -> androgen receptor (AR) -> MMP9 signaling. Mol. Oncol. 2015, 9, 44–57. [Google Scholar] [CrossRef]
- Yu, L.; Toriseva, M.; Tuomala, M.; Seikkula, H.; Elo, T.; Tuomela, J.; Kallajoki, M.; Mirtti, T.; Taimen, P.; Bostrom, P.J.; et al. Increased expression of fibroblast growth factor 13 in prostate cancer is associated with shortened time to biochemical recurrence after radical prostatectomy. Int. J. Cancer 2016, 139, 140–152. [Google Scholar] [CrossRef]
- Johnstone, C.N.; Pattison, A.D.; Harrison, P.F.; Powell, D.R.; Lock, P.; Ernst, M.; Anderson, R.L.; Beilharz, T.H. FGF13 promotes metastasis of triple-negative breast cancer. Int. J. Cancer 2020, 147, 230–243. [Google Scholar] [CrossRef]
- Mignatti, P.; Morimoto, T.; Rifkin, D.B. Basic fibroblast growth-factor, a protein devoid of secretory signal sequence, is released by cells via a pathway independent of the endoplasmic-reticulum golgi-complex. J. Cell. Physiol. 1992, 151, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Prudovsky, I.; Kumar, T.K.S.; Sterling, S.; Neivandt, D. Protein-Phospholipid Interactions in Nonclassical Protein Secretion: Problem and Methods of Study. Int. J. Mol. Sci. 2013, 14, 3734–3772. [Google Scholar] [CrossRef]
- Zhang, Z.; Coomans, C.; David, G. Membrane heparan sulfate proteoglycan-supported FGF2-FGFR1 signaling—Evidence in support of the “cooperative end structures” model. J. Biol. Chem. 2001, 276, 41921–41929. [Google Scholar] [CrossRef]
- Aviezer, D.; Hecht, D.; Safran, M.; Eisinger, M.; David, G.; Yayon, A. Perlecan, basal lamina proteoglycan, promotes basic fibroblast growth factor-receptor binding, mitogenesis, and angiogenesis. Cell 1994, 79, 1005–1013. [Google Scholar] [CrossRef]
- Mali, M.; Elenius, K.; Miettinen, H.M.; Jalkanen, M. Inhibition of basic fibroblast growth factor-induced growth promotion by overexpression of syndecan-1. J. Biol. Chem. 1993, 268, 24215–24222. [Google Scholar] [CrossRef]
- Chang, Z.; Meyer, K.; Rapraeger, A.C.; Friedl, A. Differential ability of heparan sulfate proteoglycans to assemble the fibroblast growth factor receptor complex in situ. FASEB J. 2000, 14, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, R.; Van DenBerghe, H.; David, G. Stimulation of fibroblast growth factor receptor-1 occupancy and signaling by cell surface-associated syndecans and glypican. J. Cell Biol. 1996, 133, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; John, E.M.; Stern, M.C.; Herrick, J.; Lundgreen, A.; Giuliano, A.R.; Hines, L.; Baumgartner, K.B.; Torres-Mejia, G.; Wolff, R.K. Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: The Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 2013, 140, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Birrer, M.J.; Johnson, M.E.; Hao, K.; Wong, K.K.; Park, D.C.; Bell, A.; Welch, W.R.; Berkowitz, R.S.; Mok, S.C. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor I as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J. Clin. Oncol. 2007, 25, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Ng, M.T.H.; Shepherd, L.; Herrington, C.S.; Gourley, C.; Ferguson, M.J.; Wolf, C.R. Individuality in FGF1 expression significantly influences platinum resistance and progression-free survival in ovarian cancer. Br. J. Cancer 2012, 107, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Z.; Fan, X.L.; Zhang, Q.; Shi, X.Y.; Xu, G.W.; Zou, C.M. Cancer-associated fibroblasts secrete FGF-1 to promote ovarian proliferation, migration, and invasion through the activation of FGF-1/FGFR4 signaling. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Giulianelli, S.; Cerliani, J.P.; Lamb, C.A.; Fabris, V.T.; Bottino, M.C.; Gorostiaga, M.A.; Novaro, V.; Gongora, A.; Baldi, A.; Molinolo, A.; et al. Carcinoma-associated fibroblasts activate progesterone receptors and induce hormone independent mammary tumor growth: A role for the FGF-2/FGFR-2 axis. Int. J. Cancer 2008, 123, 2518–2531. [Google Scholar] [CrossRef] [PubMed]
- Giulianelli, S.; Riggio, M.; Guillardoy, T.; Pinero, C.P.; Gorostiaga, M.A.; Sequeira, G.; Pataccini, G.; Abascal, M.F.; Toledo, M.F.; Jacobsen, B.M.; et al. FGF2 induces breast cancer growth through ligand-independent activation and recruitment of ER alpha and PRB Delta 4 isoform to MYC regulatory sequences. Int. J. Cancer 2019, 145, 1874–1888. [Google Scholar] [CrossRef]
- Shee, K.; Yang, W.; Hinds, J.W.; Hampsch, R.A.; Varn, F.S.; Traphagen, N.A.; Patel, K.; Cheng, C.; Jenkins, N.P.; Kettenbach, A.N.; et al. Therapeutically targeting tumor microenvironment-mediated drug resistance in estrogen receptor-positive breast cancer. J. Exp. Med. 2018, 215, 895–910. [Google Scholar] [CrossRef] [PubMed]
- Kottakis, F.; Polytarchou, C.; Foltopoulou, P.; Sanidas, I.; Kampranis, S.C.; Tsichlis, P.N. FGF-2 Regulates Cell Proliferation, Migration, and Angiogenesis through an NDY1/KDM2B-miR-101-EZH2 Pathway. Mol. Cell 2011, 43, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Zaravinos, A.; Volanis, D.; Lambrou, G.I.; Delakas, D.; Spandidos, D.A. Role of the angiogenic components, VEGFA, FGF2, OPN and RHOC, in urothelial cell carcinoma of the urinary bladder. Oncol. Rep. 2012, 28, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, K.; Miyano, S.W.; Minoshima, Y.; Matsui, J.; Funahashi, Y. Activated FGF2 signaling pathway in tumor vasculature is essential for acquired resistance to anti-VEGF therapy. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Bisping, G.; Leo, R.; Wenning, D.; Dankbar, B.; Padro, T.; Kropff, M.; Scheffold, C.; Kroger, M.; Mesters, R.M.; Berdel, W.E.; et al. Paracrine interactions of basic fibroblast growth factor and interleukin-6 in multiple myeloma. Blood 2003, 101, 2775–2783. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, A.; Sethi, G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef]
- Ronca, R.; Ghedini, G.C.; Maccarinelli, F.; Sacco, A.; Locatelli, S.L.; Foglio, E.; Taranto, S.; Grillo, E.; Matarazzo, S.; Castelli, R.; et al. FGF Trapping Inhibits Multiple Myeloma Growth through c-Myc Degradation-Induced Mitochondrial Oxidative Stress. Cancer Res. 2020, 80, 2340–2354. [Google Scholar] [CrossRef] [PubMed]
- Pintucci, G.; Quarto, N.; Rifkin, D.B. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol. Biol. Cell 1996, 7, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yang, L.J.; Shi, L.; Li, Q.; Zhang, G.S.; Wu, J.L.; Zheng, J.; Jiao, B.H. Nuclear translocation of fibroblast growth factor-2 (FGF2) is regulated by Karyopherin-beta 2 and Ran GTPase in human glioblastoma cells. Oncotarget 2015, 6, 21468–21478. [Google Scholar] [CrossRef][Green Version]
- Joy, A.; Moffett, J.; Neary, K.; Mordechai, E.; Stachowiak, E.K.; Coons, S.; RankinShapiro, J.; Florkiewicz, R.Z.; Stachowiak, M.K. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene 1997, 14, 171–183. [Google Scholar] [CrossRef]
- Coleman, S.J.; Chioni, A.M.; Ghallab, M.; Anderson, R.K.; Lemoine, N.R.; Kocher, H.M.; Grose, R.P. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol. Med. 2014, 6, 467–481. [Google Scholar] [CrossRef]
- Bong, S.M.; Bae, S.H.; Song, B.; Gwak, H.; Yang, S.W.; Kim, S.; Nam, S.; Rajalingam, K.; Oh, S.J.; Kim, T.W.; et al. Regulation of mRNA export through API5 and nuclear FGF2 interaction. Nucleic Acids Res. 2020, 48, 6340–6352. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Xu, J.S.; Colvin, J.S.; McEwen, D.G.; MacArthur, C.A.; Coulier, F.; Gao, G.X.; Goldfarb, M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996, 271, 15292–15297. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.M.; Steringer, J.P.; Wegehingel, S.; Bleicken, S.; Munster, M.; Dimou, E.; Unger, S.; Weidmann, G.; Andreas, H.; Garcia-Saez, A.J.; et al. Formation of Disulfide Bridges Drives Oligomerization, Membrane Pore Formation, and Translocation of Fibroblast Growth Factor 2 to Cell Surfaces. J. Biol. Chem. 2015, 290, 8925–8937. [Google Scholar] [CrossRef] [PubMed]
- Kwek, S.S.; Roy, R.; Zhou, H.; Climent, J.; Martinez-Climent, J.A.; Fridlyand, J.; Albertson, D.G. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene 2009, 28, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Schwaederle, M.; Kurzrock, R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: Biologic and clinical implications. Cancer Metastasis Rev. 2015, 34, 479–496. [Google Scholar] [CrossRef]
- Yasuda, K.; Torigoe, T.; Mariya, T.; Asano, T.; Kuroda, T.; Matsuzaki, J.; Ikeda, K.; Yamauchi, M.; Emori, M.; Asanuma, H.; et al. Fibroblasts induce expression of FGF4 in ovarian cancer stem-like cells/cancer-initiating cells and upregulate their tumor initiation capacity. Lab. Investig. 2014, 94, 1355–1369. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Ibrahimi, O.A.; Olsen, S.K.; Umemori, H.; Mohammadi, M.; Ornitz, D.M. Receptor specificity of the fibroblast growth factor family—The complete mammalian FGF family. J. Biol. Chem. 2006, 281, 15694–15700. [Google Scholar] [CrossRef] [PubMed]
- Yeh, B.K.; Igarashi, M.; Eliseenkova, A.V.; Plotnikov, A.N.; Sher, I.; Ron, D.; Aaronson, S.A.; Mohammadi, M. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc. Natl. Acad. Sci. USA 2003, 100, 2266–2271. [Google Scholar] [CrossRef]
- Zinkle, A.; Mohammadi, M. Structural Biology of the FGF7 Subfamily. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Watson, J.; Francavilla, C. Regulation of FGF10 signaling in development and disease. Front. Genet. 2018, 9, 500. [Google Scholar] [CrossRef]
- Sunmonu, N.A.; Li, K.R.; Li, J.Y.H. Numerous Isoforms of Fgf8 Reflect Its Multiple Roles in the Developing Brain. J. Cell. Physiol. 2011, 226, 1722–1726. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, M.; Naruo, K.; Seko, C.; Matsumoto, S.; Kondo, T.; Kurokawa, T. Molecular-cloning of a novel cytokine cdna-encoding the 9th member of the fibroblast growth-factor family, which has a unique secretion property. Mol. Cell. Biol. 1993, 13, 4251–4259. [Google Scholar] [CrossRef] [PubMed]
- Miyake, A.; Konishi, M.; Martin, F.H.; Hernday, N.A.; Ozaki, K.; Yamamoto, S.; Mikami, T.; Arakawa, T.; Itoh, N. Structure and expression of a novel member, FGF-16, of the fibroblast growth factor family. Biochem. Biophys. Res. Commun. 1998, 243, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, M.; Shimkets, R.; Prayaga, S.; Boldog, F.; Yang, M.J.; Burgess, C.; Fernandes, E.; Rittman, B.; Shimkets, J.; LaRochelle, W.J.; et al. Identification of a novel human fibroblast growth factor and characterization of its role in oncogenesis. Cancer Res. 2001, 61, 3131–3138. [Google Scholar] [PubMed]
- Miyakawa, K.; Hatsuzawa, K.; Kurokawa, T.; Asada, M.; Kuroiwa, T.; Imamura, T. A hydrophobic region locating at the center of fibroblast growth factor-9 is crucial for its secretion. J. Biol. Chem. 1999, 274, 29352–29357. [Google Scholar] [CrossRef]
- Miyakawa, K.; Imamura, T. Secretion of FGF-16 requires an uncleaved bipartite signal sequence. J. Biol. Chem. 2003, 278, 35718–35724. [Google Scholar] [CrossRef]
- Shi, S.Y.; Lu, Y.W.; Richardson, J.; Min, X.S.; Weiszmann, J.; Richards, W.G.; Wang, Z.L.; Zhang, Z.Q.; Zhang, J.; Li, Y. A systematic dissection of sequence elements determining beta-Klotho and FGF interaction and signaling. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Suzuki, M.; Uehara, Y.; Motomura-Matsuzaka, K.; Oki, J.; Koyama, Y.; Kimura, M.; Asada, M.; Komi-Kuramochi, A.; Oka, S.; Imamura, T. Beta Klotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol. Endocrinol. 2008, 22, 1006–1014. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, K.H.; Lee, J.; Oh, J.J.; Cheong, H.S.; Wong, E.L.; Yang, B.S.; Byun, S.S.; Myung, S.C. Single nucleotide polymorphisms in fibroblast growth factor 23 gene, FGF23, are associated with prostate cancer risk. BJU Int. 2014, 114, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Mansinho, A.; Ferreira, A.R.; Casimiro, S.; Alho, I.; Vendrell, I.; Costa, A.L.; Sousa, R.; Abreu, C.; Pulido, C.; Macedo, D.; et al. Levels of Circulating Fibroblast Growth Factor 23 (FGF23) and Prognosis in Cancer Patients with Bone Metastases. Int. J. Mol. Sci. 2019, 20, 695. [Google Scholar] [CrossRef]
- Smallwood, P.M.; MunozSanjuan, I.; Tong, P.; Macke, J.P.; Hendry, S.H.C.; Gilbert, D.J.; Copeland, N.G.; Jenkins, N.A.; Nathans, J. Fibroblast growth factor (FGF) homologous factors: New members of the FGF family implicated in nervous system development. Proc. Natl. Acad. Sci. USA 1996, 93, 9850–9857. [Google Scholar] [CrossRef]
- Liu, C.J.; Dib-Hajj, S.D.; Waxman, S.G. Fibroblast growth factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNa(v)1.9a (NaN). J. Biol. Chem. 2001, 276, 18925–18933. [Google Scholar] [CrossRef]
- Olsen, S.K.; Garbi, M.; Zampieri, N.; Eliseenkova, A.V.; Ornitz, D.M.; Goldfarb, M.; Mohammadi, M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J. Biol. Chem. 2003, 278, 34226–34236. [Google Scholar] [CrossRef] [PubMed]
- Sochacka, M.; Opalinski, L.; Szymczyk, J.; Zimoch, M.B.; Czyrek, A.; Krowarsch, D.; Otlewski, J.; Zakrzewska, M. FHF1 is a bona fide fibroblast growth factor that activates cellular signaling in FGFR-dependent manner. Cell Commun. Signal. 2020, 18. [Google Scholar] [CrossRef]
- Lin, H.P.; Lu, P.Y.; Zhou, M.; Wu, F.F.; Weng, L.; Meng, K.K.; Yang, D.; Li, S.J.; Jiang, C.; Tian, H.S. Purification of recombinant human fibroblast growth factor 13 in E. coli and its molecular mechanism of mitogenesis. Appl. Microbiol. Biotechnol. 2019, 103, 7017–7027. [Google Scholar] [CrossRef]
- Courjal, F.; Cuny, M.; SimonyLafontaine, J.; Louason, G.; Speiser, P.; Zeillinger, R.; Rodriguez, C.; Theillet, C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: Definition of phenotypic groups. Cancer Res. 1997, 57, 4360–4367. [Google Scholar] [PubMed]
- Cuny, M.; Kramar, A.; Courjal, F.; Johannsdottir, V.; Iacopetta, B.; Fontaine, H.; Grenier, J.; Culine, S.; Theillet, C. Relating genotype and phenotype in breast cancer: An analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000, 60, 1077–1083. [Google Scholar] [PubMed]
- Mo, J.; Wen, X.; Kim, T.S.; Kwak, Y.; Cho, N.Y.; Lee, H.S.; Kang, G.H. Fibroblast Growth Factor Receptor 1 (FGFR1) Amplification Detected by Droplet Digital Polymerase Chain Reaction (ddPCR) Is a Prognostic Factor in Colorectal Cancers. Cancer Res. Treat. 2020, 52, 74–84. [Google Scholar] [CrossRef]
- Goke, F.; Goke, A.; von Massenhausen, A.; Franzen, A.; Sharma, R.; Kirsten, R.; Bohm, D.; Kristiansen, G.; Stenzinger, A.; Wynes, M.; et al. Fibroblast Growth Factor Receptor 1 as a Putative Therapy Target in Colorectal Cancer. Digestion 2013, 88, 172–181. [Google Scholar] [CrossRef]
- Astolfi, A.; Pantaleo, M.A.; Indio, V.; Urbini, M.; Nannini, M. The Emerging Role of the FGF/FGFR Pathway in Gastrointestinal Stromal Tumor. Int. J. Mol. Sci. 2020, 21, 3313. [Google Scholar] [CrossRef]
- Singh, D.; Chan, J.M.; Zoppoli, P.; Niola, F.; Sullivan, R.; Castano, A.; Liu, E.M.; Reichel, J.; Porrati, P.; Pellegatta, S.; et al. Transforming Fusions of FGFR and TACC Genes in Human Glioblastoma. Science 2012, 337, 1231–1235. [Google Scholar] [CrossRef]
- Trisolini, E.; El Wardighi, D.; Giry, M.; Bernardi, P.; Boldorini, R.L.; Mokhtari, K.; Sanson, M. Actionable FGFR1 and BRAF mutations in adult circumscribed gliomas. J. Neuro Oncol. 2019, 145, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Freier, K.; Schwaenen, C.; Sticht, C.; Flechtenmacher, C.; Muhling, J.; Hofele, C.; Radlwimmer, B.; Lichter, P.; Joos, S. Recurrent FGFR 1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral. Oncol. 2007, 43, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Sos, M.L.; Seidel, D.; Peifer, M.; Zander, T.; Heuckmann, J.M.; Ullrich, R.T.; Menon, R.; Maier, S.; Soltermann, A.; et al. Frequent and Focal FGFR1 Amplification Associates with Therapeutically Tractable FGFR1 Dependency in Squamous Cell Lung Cancer. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Cihoric, N.; Savic, S.; Schneider, S.; Ackermann, I.; Bichsel-Naef, M.; Schmid, R.A.; Lardinois, D.; Gugger, M.; Bubendorf, L.; Zlobec, I.; et al. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br. J. Cancer 2014, 110, 2914–2922. [Google Scholar] [CrossRef]
- Peifer, M.; Fernandez-Cuesta, L.; Sos, M.L.; George, J.; Seidel, D.; Kasper, L.H.; Plenker, D.; Leenders, F.; Sun, R.P.; Zander, T.; et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012, 44, 1104. [Google Scholar] [CrossRef]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B.; et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008, 455, 1069–1075. [Google Scholar] [CrossRef]
- Miao, J.L.; Zhou, J.H.; Cai, J.J.; Liu, R.J. The association between fibroblast growth factor receptor 1 gene amplification and lung cancer: A meta-analysis. Arch. Med. Sci. 2020, 16, 16–26. [Google Scholar] [CrossRef]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef]
- Lehnen, N.C.; von Massenhausen, A.; Kalthoff, H.; Zhou, H.; Glowka, T.; Schutte, U.; Holler, T.; Riesner, K.; Boehm, D.; Merkelbach-Bruse, S.; et al. Fibroblast growth factor receptor 1 gene amplification in pancreatic ductal adenocarcinoma. Histopathology 2013, 63, 157–166. [Google Scholar] [CrossRef]
- Yan, G.C.; Fukabori, Y.; McBride, G.; Nikolaropolous, S.; McKeehan, W.L. Exon switching and activation of stromal and embryonic fibroblast growth-factor (FGF)-FGF receptor genes in prostate epithelial-cells accompany stromal independence and malignancy. Mol. Cell. Biol. 1993, 13, 4513–4522. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.L.; McKeehan, K.; Guo, W.; Jamna, S.; Ittmann, M.M.; Foster, B.; Greenberg, N.M.; McKeehan, W.L.; Wang, F. Cooperation between ectopic FGFR1 and depression of FGFR2 in induction of prostatic intraepithelial neoplasia in the mouse prostate. Cancer Res. 2003, 63, 8784–8790. [Google Scholar]
- Wang, F.; McKeehan, K.; Yu, C.D.; Ittmann, M.; McKeehan, W.L. Chronic activity of ectopic type I fibroblast growth factor receptor tyrosine kinase in prostate epithelium results in hyperplasia accompanied by intraepithelial neoplasia. Prostate 2004, 58, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ke, Y.P.; Liu, S.Y.; Pan, S.R.; Liu, Z.Y.; Zhang, H.; Fan, Z.C.; Zhou, C.Y.; Liu, J.C.; Wang, F. Ectopic fibroblast growth factor receptor 1 promotes inflammation by promoting nuclear factor-kappa B signaling in prostate cancer cells. J. Biol. Chem. 2018, 293, 14839–14849. [Google Scholar] [CrossRef] [PubMed]
- Schultheis, A.M.; Bos, M.; Schmitz, K.; Wilsberg, L.; Binot, E.; Wolf, J.; Buttner, R.; Schildhaus, H.U. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod. Pathol. 2014, 27, 214–221. [Google Scholar] [CrossRef][Green Version]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.P.; Ozretic, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef]
- Kim, K.B.; Kim, Y.; Rivard, C.J.; Kim, D.W.; Park, K.S. FGFR1 Is Critical for RBL2 Loss-Driven Tumor Development and Requires PLCG1 Activation for Continued Growth of Small Cell Lung Cancer. Cancer Res. 2020, 80, 5051–5062. [Google Scholar] [CrossRef]
- Lei, H.P.; Deng, C.X. Fibroblast Growth Factor Receptor 2 Signaling in Breast Cancer. Int. J. Biol. Sci. 2017, 13, 1163–1171. [Google Scholar] [CrossRef]
- Jain, A.; Borad, M.J.; Kelley, R.K.; Wang, Y.; Abdel-Wahab, R.; Meric-Bernstam, F.; Baggerly, K.A.; Kaseb, A.O.; Al-Shamsi, H.O.; Ahn, D.H.; et al. Cholangiocarcinoma With FGFR Genetic Aberrations: A Unique Clinical Phenotype. JCO Precis. Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Lowery, M.A.; Ptashkin, R.; Jordan, E.; Berger, M.F.; Zehir, A.; Capanu, M.; Kemeny, N.E.; O’Reilly, E.M.; El-Dika, I.; Jarnagin, W.R.; et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin. Cancer Res. 2018, 24, 4154–4161. [Google Scholar] [CrossRef]
- Ross, J.S.; Wang, K.; Gay, L.; Al-Rohil, R.; Rand, J.V.; Jones, D.M.; Lee, H.J.; Sheehan, C.E.; Otto, G.A.; Palmer, G.; et al. New Routes to Targeted Therapy of Intrahepatic Cholangiocarcinomas Revealed by Next-Generation Sequencing. Oncologist 2014, 19, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Jeske, Y.W.; Ali, S.; Byron, S.A.; Gao, F.; Mannel, R.S.; Ghebre, R.G.; DiSilvestro, P.A.; Lele, S.B.; Pearl, M.L.; Schmidt, A.P.; et al. FGFR2 mutations are associated with poor outcomes in endometrioid endometrial cancer: An NRG Oncology/Gynecologic Oncology Group study. Gynecol. Oncol. 2017, 145, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhan, P.; Gavine, P.R.; Morgan, S.; Womack, C.; Ni, X.; Shen, D.; Bang, Y.J.; Im, S.A.; Kim, W.H.; et al. FGFR2 amplification has prognostic significance in gastric cancer: Results from a large international multicentre study. Br. J. Cancer 2014, 110, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; Shin, K.H.; Park, J.G. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001, 61, 3541–3543. [Google Scholar] [PubMed]
- Liao, R.G.; Jung, J.; Tchaicha, J.; Wilkerson, M.D.; Sivachenko, A.; Beauchamp, E.M.; Liu, Q.S.; Pugh, T.J.; Pedamallu, C.S.; Hayes, D.N.; et al. Inhibitor-Sensitive FGFR2 and FGFR3 Mutations in Lung Squamous Cell Carcinoma. Cancer Res. 2013, 73, 5195–5205. [Google Scholar] [CrossRef]
- Fischbach, A.; Rogler, A.; Erber, R.; Stoehr, R.; Poulsom, R.; Heidenreich, A.; Schneevoigt, B.S.; Hauke, S.; Hartmann, A.; Knuechel, R.; et al. Fibroblast growth factor receptor (FGFR) gene amplifications are rare events in bladder cancer. Histopathology 2015, 66, 639–649. [Google Scholar] [CrossRef]
- Guancial, E.A.; Werner, L.; Bellmunt, J.; Bamias, A.; Choueiri, T.K.; Ross, R.; Schutz, F.A.; Park, R.S.; O’Brien, R.J.; Hirsch, M.S.; et al. FGFR3 expression in primary and metastatic urothelial carcinoma of the bladder. Cancer Med. 2014, 3, 835–844. [Google Scholar] [CrossRef]
- Hammam, O.; Aboushousha, T.; El-Hindawi, A.; Khairy, H.; Khalil, H.; Kamel, A.; Akl, M.; Abdel-Hady, A.; Magdy, M.; Badawy, M.; et al. Expression of FGFR3 Protein and Gene Amplification in Urinary Bladder Lesions in Relation to Schistosomiasis. Open Access Maced. J. Med. Sci. 2017, 5, 160–166. [Google Scholar] [CrossRef]
- Cappellen, D.; De Oliveira, C.; Ricol, D.; de Medina, S.G.D.; Bourdin, J.; Sastre-Garau, X.; Chopin, D.; Thiery, J.P.; Radvanyi, F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 1999, 23, 18–20. [Google Scholar] [CrossRef]
- Sfakianos, J.P.; Cha, E.K.; Iyer, G.; Scott, S.N.; Zabor, E.C.; Shah, R.H.; Ren, Q.; Bagrodia, A.; Kim, P.H.; Hakimi, A.A.; et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2015, 68, 970–977. [Google Scholar] [CrossRef]
- Robinson, B.D.; Vlachostergios, P.J.; Bhinder, B.; Liu, W.S.; Li, K.; Moss, T.J.; Bareja, R.; Park, K.; Tavassoli, P.; Cyrta, J.; et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Teo, M.Y.; Mota, J.M.; Whiting, K.A.; Li, H.A.; Funt, S.A.; Lee, C.H.; Solit, D.B.; Al-Ahmadie, H.; Milowsky, M.I.; Balar, A.V.; et al. Fibroblast Growth Factor Receptor 3 Alteration Status is Associated with Differential Sensitivity to Platinum-based Chemotherapy in Locally Advanced and Metastatic Urothelial Carcinoma. Eur. Urol. 2020, 78, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Pal, S.K.; Hahn, A.W.; Nussenzveig, R.H.; Pond, G.R.; Gupta, S.V.; Wang, J.; Bilen, M.A.; Naik, G.; Ghatalia, P.; et al. Characterization of Metastatic Urothelial Carcinoma via Comprehensive Genomic Profiling of Circulating Tumor DNA. Cancer 2018, 124, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.W.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540. [Google Scholar] [CrossRef]
- Carneiro, B.A.; Elvin, J.A.; Kamath, S.D.; Ali, S.M.; Paintal, A.S.; Restrepoe, A.; Berry, E.; Giles, F.J.; Johnson, M.L. FGFR3-TACC3: A novel gene fusion in cervical cancer. Gynecol. Oncol. Rep. 2015, 13, 53–56. [Google Scholar] [CrossRef]
- Tamura, R.; Yoshihara, K.; Saito, T.; Ishimura, R.; Martinez-Ledesma, J.E.; Xin, H.; Ishiguro, T.; Mori, Y.; Yamawaki, K.; Suda, K.; et al. Novel therapeutic strategy for cervical cancer harboring FGFR3-TACC3 fusions. Oncogenesis 2018, 7. [Google Scholar] [CrossRef]
- Qin, A.; Johnson, A.; Ross, J.S.; Miller, V.A.; Ali, S.M.; Schrock, A.B.; Gadgeel, S.M. Detection of Known and Novel FGFR Fusions in Non-Small Cell Lung Cancer by Comprehensive Genomic Profiling. J. Thorac. Oncol. 2019, 14, 54–62. [Google Scholar] [CrossRef]
- Wang, R.; Wang, L.; Li, Y.; Hu, H.C.; Shen, L.; Shen, X.X.; Pan, Y.J.; Ye, T.; Zhang, Y.; Luo, X.Y.; et al. FGFR1/3 Tyrosine Kinase Fusions Define a Unique Molecular Subtype of Non-Small Cell Lung Cancer. Clin. Cancer Res. 2014, 20, 4107–4114. [Google Scholar] [CrossRef]
- Brito, L.P.; Ribeiro, T.C.; Almeida, M.Q.; Jorge, A.A.D.; Soares, I.C.; Latronico, A.C.; Mendonca, B.B.; Fragoso, M.; Lerario, A.M. The role of fibroblast growth factor receptor 4 overexpression and gene amplification as prognostic markers in pediatric and adult adrenocortical tumors. Endocr. Relat. Cancer 2012, 19, L11–L13. [Google Scholar] [CrossRef]
- Yang, W.; Yao, Y.W.; Zeng, J.L.; Liang, W.J.; Wang, L.; Bai, C.Q.; Liu, C.H.; Song, Y. Prognostic value of FGFR1 gene copy number in patients with non-small cell lung cancer: A meta-analysis. J. Thorac. Dis. 2014, 6, 803–809. [Google Scholar] [CrossRef]
- Heist, R.S.; Mino-Kenudson, M.; Sequist, L.V.; Tammireddy, S.; Morrissey, L.; Christiani, D.C.; Engelman, J.A.; Iafrate, A.J. FGFR1 Amplification in Squamous Cell Carcinoma of The Lung. J. Thorac. Oncol. 2012, 7, 1775–1780. [Google Scholar] [CrossRef]
- Giacomini, A.; Taranto, S.; Rezzola, S.; Matarazzo, S.; Grillo, E.; Bugatti, M.; Scotuzzi, A.; Guerra, J.; Di Trani, M.; Presta, M.; et al. Inhibition of the FGF/FGFR System Induces Apoptosis in Lung Cancer Cells via c-Myc Downregulation and Oxidative Stress. Int. J. Mol. Sci. 2020, 21, 9376. [Google Scholar] [CrossRef]
- Turner, N.; Pearson, A.; Sharpe, R.; Lambros, M.; Geyer, F.; Lopez-Garcia, M.A.; Natrajan, R.; Marchio, C.; Iorns, E.; Mackay, A.; et al. FGFR1 Amplification Drives Endocrine Therapy Resistance and Is a Therapeutic Target in Breast Cancer. Cancer Res. 2010, 70, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.S.; Simpson, P.T.; Turner, N.C.; Lambros, M.B.; Jones, C.; Mackay, A.; Grigoriadis, A.; Sarrio, D.; Savage, K.; Dexter, T.; et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin. Cancer Res. 2006, 12, 6652–6662. [Google Scholar] [CrossRef]
- Gelsi-Boyer, W.; Orsetti, B.; Cervera, N.; Finetti, P.; Sircoulomb, F.; Rouge, C.; Lasorsa, L.; Letessier, A.; Ginestier, C.; Monville, F.; et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol. Cancer Res. 2005, 3, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Krishna, S.; Witton, C.J.; Bartlett, J.M.S. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin. Cancer Res. 2003, 9, 5271–5281. [Google Scholar]
- Ishiwata, T.; Matsuda, Y.; Yamamoto, T.; Uchida, E.; Korc, M.; Naito, Z. Enhanced Expression of Fibroblast Growth Factor Receptor 2 IIIc Promotes Human Pancreatic Cancer Cell Proliferation. Am. J. Pathol. 2012, 180, 1928–1941. [Google Scholar] [CrossRef]
- Kornmann, M.; Ishiwata, T.; Matsuda, K.; Lopez, M.E.; Fukahi, K.; Asano, G.; Beger, H.G.; Korc, M. IIIc isoform of fibroblast growth factor receptor 1 is overexpressed in human pancreatic cancer and enhances tumorigenicity of hamster ductal cells. Gastroenterology 2002, 123, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y.; Chao, J.; Kim, K.; Kim, S.T.; Kim, K.M.; Klempner, S.J.; Lee, J. High-level FGFR2 amplification is associated with poor prognosis and Lower response to chemotherapy in gastric cancers. Pathol. Res. Pract. 2020, 216. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Abate, R.E.; Rachiglio, A.M.; Maiello, M.R.; Esposito, C.; Schettino, C.; Izzo, F.; Nasti, G.; Normanno, N. FGFR Fusions in Cancer: From Diagnostic Approaches to Therapeutic Intervention. Int. J. Mol. Sci. 2020, 21, 6856. [Google Scholar] [CrossRef]
- Arai, Y.; Totoki, Y.; Hosoda, F.; Shirota, T.; Hama, N.; Nakamura, H.; Ojima, H.; Furuta, K.; Shimada, K.; Okusaka, T.; et al. Fibroblast Growth Factor Receptor 2 Tyrosine Kinase Fusions Define a Unique Molecular Subtype of Cholangiocarcinoma. Hepatology 2014, 59, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Patani, H.; Bunney, T.D.; Thiyagarajan, N.; Norman, R.A.; Ogg, D.; Breed, J.; Ashford, P.; Potterton, A.; Edwards, M.; Williams, S.V.; et al. Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use. Oncotarget 2016, 7, 24252–24268. [Google Scholar] [CrossRef]
- Gergely, F.; Karlsson, C.; Still, I.; Cowell, J.; Kilmartin, J.; Raff, J.W. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA 2000, 97, 14352–14357. [Google Scholar] [CrossRef]
- Jaakkola, S.; Salmikangas, P.; Nylund, S.; Partanen, J.; Armstrong, E.; Pyrhonen, S.; Lehtovirta, P.; Nevanlinna, H. Amplification of FGFR4 gene in human breast and gynecological cancers. Int. J. Cancer 1993, 54, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Bange, J.; Prechtl, D.; Cheburkin, Y.; Specht, K.; Harbeck, N.; Schmitt, M.; Knyazeva, T.; Muller, S.; Gartner, S.; Sures, I.; et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002, 62, 840–847. [Google Scholar] [PubMed]
- Frullanti, E.; Berking, C.; Harbeck, N.; Jezequel, P.; Haugen, A.; Mawrin, C.; Parise, O.; Sasaki, H.; Tsuchiya, N.; Dragani, T.A.; et al. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur. J. Cancer Prev. 2011, 20, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Sun, Y.Y.; Lv, Z.W.; Zhang, Z.; Su, Q.X.; Wu, H.; Zhang, W.; Yuan, W.; Zuo, L.; Shi, L.; et al. Effects of FGFR4 G388R, V10I polymorphisms on the likelihood of cancer. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Yang, X.C.; Steinberg, F.; Zhuang, L.; Bessey, R.; Trueb, B. Receptor FGFRL1 does not promote cell proliferation but induces cell adhesion. Int. J. Mol. Med. 2016, 38, 30–38. [Google Scholar] [CrossRef]
- Rieckmann, T.; Kotevic, I.; Trueb, B. The cell surface receptor FGFRL1 forms constitutive dimers that promote cell adhesion. Exp. Cell Res. 2008, 314, 1071–1081. [Google Scholar] [CrossRef]
- Nurcombe, V.; Smart, C.E.; Chipperfield, H.; Cool, S.M.; Boilly, B.; Hondermarck, H. The proliferative and migratory activities of breast cancer cells can be differentially regulated by heparan sulfates. J. Biol. Chem. 2000, 275, 30009–30018. [Google Scholar] [CrossRef]
- Gotte, M.; Kersting, C.; Radke, I.; Kiesel, L.; Wulfing, P. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast Cancer Res. 2007, 9. [Google Scholar] [CrossRef] [PubMed]
- Mundhenke, C.; Meyer, K.; Drew, S.; Friedl, A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 receptor binding in breast carcinomas. Am. J. Pathol. 2002, 160, 185–194. [Google Scholar] [CrossRef]
- Wu, X.C.; Kan, M.K.; Wang, F.; Jin, C.L.; Yu, C.D.; McKeehan, W.L. A rare premalignant prostate tumor epithelial cell syndecan-1 forms a fibroblast growth factor-binding complex with progression-promoting ectopic fibroblast growth factor receptor 1. Cancer Res. 2001, 61, 5295–5302. [Google Scholar]
- Filla, M.S.; Dam, P.; Rapraeger, A.C. The cell surface proteoglycan syndecan-1 mediates fibroblast growth factor-2 binding and activity. J. Cell. Physiol. 1998, 174, 310–321. [Google Scholar] [CrossRef]
- Zong, F.; Fthenou, E.; Wolmer, N.; Hollosi, P.; Kovalszky, I.; Szilak, L.; Mogler, C.; Nilsonne, G.; Tzanakakis, G.; Dobra, K. Syndecan-1 and FGF-2, but Not FGF Receptor-1, Share a Common Transport Route and Co-Localize with Heparanase in the Nuclei of Mesenchymal Tumor Cells. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.H.; Meyer, K.; Mundhenke, C.; Drew, S.A.; Friedl, A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 signaling in brain endothelial cells. J. Biol. Chem. 2003, 278, 16045–16053. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, A.; Gouge, J.; Kontovounisios, C.; Nikolaou, S.; Ashworth, A.; Lim, K.; Chong, I.E. Klotho and the Treatment of Human Malignancies. Cancers 2020, 12, 1665. [Google Scholar] [CrossRef] [PubMed]
- Wolf, I.; Levanon-Cohen, S.; Bose, S.; Ligumsky, H.; Sredni, B.; Kanety, H.; Kuro-o, M.; Karlan, B.; Kaufman, B.; Koeffler, H.P.; et al. Klotho: A tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene 2008, 27, 7094–7105. [Google Scholar] [CrossRef]
- Lee, S.; Choi, J.; Mohanty, J.; Sousa, L.P.; Tome, F.; Pardon, E.; Steyaert, J.; Lemmon, M.A.; Lax, I.; Schlessinger, J. Structures of beta-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature 2018, 553, 501. [Google Scholar] [CrossRef]
- Li, F.; Li, Z.M.; Han, Q.; Cheng, Y.R.; Ji, W.X.; Yang, Y.; Lu, S.; Xia, W.L. Enhanced autocrine FGF19/FGFR4 signaling drives the progression of lung squamous cell carcinoma, which responds to mTOR inhibitor AZD2104. Oncogene 2020, 39, 3507–3521. [Google Scholar] [CrossRef]
- Poh, W.J.; Wong, W.N.; Ong, H.M.; Aung, M.O.; Lim, S.G.; Chua, B.T.; Ho, H.K. Klotho-beta overexpression as a novel target for suppressing proliferation and fibroblast growth factor receptor-4 signaling in hepatocellular carcinoma. Mol. Cancer 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.Z.; Martin, R.C.; Jin, H.; Liu, X.K.; Pandit, H.; Zhao, H.J.; Cai, L.; Zhang, P.; Li, W.; Li, Y. Up-regulation of FGF15/19 signaling promotes hepatocellular carcinoma in the background of fatty liver. J. Exp. Clin. Cancer Res. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Z.; Hsu, C.; Jeng, Y.M.; Hu, F.C.; Pan, H.W.; Wu, Y.M.; Hsu, H.C.; Cheng, A.L. Klotho-beta and fibroblast growth factor 19 expression correlates with early recurrence of resectable hepatocellular carcinoma. Liver Int. 2019, 39, 1682–1691. [Google Scholar] [CrossRef]
- Lee, C.H.; Su, S.Y.; Sittampalam, K.; Chen, P.C.H.; Petersson, F.; Kao, Y.C.; Carpenter, T.O.; Hsieh, T.H.; Konishi, E.; Tsai, J.W.; et al. Frequent overexpression of klotho in fusion-negative phosphaturic mesenchymal tumors with tumorigenic implications. Mod. Pathol. 2020, 33, 858–870. [Google Scholar] [CrossRef]
- Ronca, R.; Di Salle, E.; Giacomini, A.; Leali, D.; Alessi, P.; Coltrini, D.; Ravelli, C.; Matarazzo, S.; Ribatti, D.; Vermi, W.; et al. Long Pentraxin-3 Inhibits Epithelial-Mesenchymal Transition in Melanoma Cells. Mol. Cancer Ther. 2013, 12, 2760–2771. [Google Scholar] [CrossRef]
- Suvannasankha, A.; Tompkins, D.R.; Edwards, D.F.; Petyaykina, K.V.; Crean, C.D.; Fournier, P.G.; Parker, J.M.; Sandusky, G.E.; Ichikawa, S.; Imel, E.A.; et al. FGF23 is elevated in multiple myeloma and increases heparanase expression by tumor cells. Oncotarget 2015, 6, 19647–19660. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, H.; Ding, S.T.; Qi, S.S.; Liu, S.; Sun, D.Q.; Dong, W.; Yin, L.; Li, M.J.; Zhao, X.B.; et al. Klotho inhibits androgen/androgen receptor-associated epithelial-mesenchymal transition in prostate cancer through inactivation of ERK1/2 signaling. Oncol. Rep. 2018, 40, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Trost, N.; Pena-Llopis, S.; Koirala, S.; Stojan, J.; Potts, P.R.; Tacer, K.F.; Martinez, E.D. Gamma Klotho is a novel marker and cell survival factor in a subset of triple negative breast cancers. Oncotarget 2016, 7, 2611–2628. [Google Scholar] [CrossRef] [PubMed]
- Choy, C.T.; Kim, H.; Lee, J.Y.; Williams, D.M.; Palethorpe, D.; Fellows, G.; Wright, A.J.; Laing, K.; Bridges, L.R.; Howe, F.A.; et al. Anosmin-1 contributes to brain tumor malignancy through integrin signal pathways. Endocr. Relat. Cancer 2014, 21, 85–99. [Google Scholar] [CrossRef]
- Kilkenny, D.M.; Rocheleau, J.V. Fibroblast growth factor receptor-1 signaling in pancreatic islet beta-cells is modulated by the extracellular matrix. Mol. Endocrinol. 2008, 22, 196–205. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, A.; Ojeda-Marquez, L.; Marrugal, A.; Yague, P.; Ponce-Aix, S.; Salinas, A.; Carnero, A.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. The FGFR4-388arg Variant Promotes Lung Cancer Progression by N-Cadherin Induction. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sawada, T.; Jing, X.F.; Yokote, H.; Yan, X.M.; Sakaguchi, K. Regulation of ephexin1, a guanine nucleotide exchange factor of Rho family GTPases, by fibroblast growth factor receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 2007, 282, 31103–31112. [Google Scholar] [CrossRef] [PubMed]
- Kucinska, M.; Porebska, N.; Lampart, A.; Latko, M.; Knapik, A.; Zakrzewska, M.; Otlewski, J.; Opalinski, L. Differential regulation of fibroblast growth factor receptor 1 trafficking and function by extracellular galectins. Cell Commun. Signal. 2019, 17. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Park, M.S.; Nandu, T.S.; Gadad, S.; Kim, S.C.; Kim, M.Y. GALNT14 promotes lung-specific breast cancer metastasis by modulating self-renewal and interaction with the lung microenvironment. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Mori, S.; Tran, V.; Nishikawa, K.; Kaneda, T.; Hamada, Y.; Kawaguchi, N.; Fujita, M.; Takada, Y.K.; Matsuura, N.; Zhao, M.; et al. A Dominant-Negative FGF1 Mutant (the R50E Mutant) Suppresses Tumorigenesis and Angiogenesis. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Mori, S.; Kodaira, M.; Ito, A.; Okazaki, M.; Kawaguchi, N.; Hamada, Y.; Takada, Y.; Matsuura, N. Enhanced Expression of Integrin alpha v beta 3 Induced by TGF-beta Is Required for the Enhancing Effect of Fibroblast Growth Factor 1 (FGF1) in TGF-beta-Induced Epithelia-lMesenchymal Transition (EMT) in Mammary Epithelial Cells. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Brown, W.S.; Tan, L.; Smith, A.; Gray, N.S.; Wendt, M.K. Covalent Targeting of Fibroblast Growth Factor Receptor Inhibits Metastatic Breast Cancer. Mol. Cancer Ther. 2016, 15, 2096–2106. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, V.; Temburni, M.K.; Kappes, J.C.; Galileo, D.S. L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor. Clin. Exp. Metastasis 2013, 30, 507–520. [Google Scholar] [CrossRef]
- Anderson, H.J.; Galileo, D.S. Small-molecule inhibitors of FGFR, integrins and FAK selectively decrease L1CAM-stimulated glioblastoma cell motility and proliferation. Cell. Oncol. 2016, 39, 229–242. [Google Scholar] [CrossRef]
- Suyama, K.; Shapiro, I.; Guttman, M.; Hazan, R.B. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2002, 2, 301–314. [Google Scholar] [CrossRef]
- Hulit, J.; Suyama, K.; Chung, S.; Keren, R.; Agiostratidou, G.; Shan, W.S.; Dong, X.Y.; Williams, T.M.; Lisanti, M.P.; Knudsen, K.; et al. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007, 67, 3106–3116. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.; Akhand, S.S.; Paez, J.S.P.; Brown, W.; Pan, L.; Libring, S.; Badamy, M.; Dykuizen, E.; Solorio, L.; Tao, W.A.; et al. Epigenetic targeting of neuropilin-1 prevents bypass signaling in drug-resistant breast cancer. Oncogene 2021, 40, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Das, T.K.; Cagan, R.L. KIF5B-RET Oncoprotein Signals through a Multi-kinase Signaling Hub. Cell Rep. 2017, 20, 2368–2383. [Google Scholar] [CrossRef]
- Knelson, E.H.; Gaviglio, A.L.; Tewari, A.K.; Armstrong, M.B.; Mythreye, K.; Blobe, G.C. Type III TGF-beta receptor promotes FGF2-mediated neuronal differentiation in neuroblastoma. J. Clin. Investig. 2013, 123, 4786–4798. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.P.; Ferguson, H.R.; Ferguson, J.; Zindy, E.; Kowalczyk, K.M.; Kedward, T.; Bates, C.; Parsons, J.; Courjal, J.; Chandler, S.; et al. Reciprocal Priming between RTKs within Recycling Endosomes Orchestrates Cellular Signaling Outputs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Meyer, A.N.; Gastwirt, R.F.; Schlaepfer, D.D.; Donoghue, D.J. The cytoplasmic tyrosine kinase Pyk2 as a novel effector of fibroblast growth factor receptor 3 activation. J. Biol. Chem. 2004, 279, 28450–28457. [Google Scholar] [CrossRef]
- Salazar, L.; Kashiwada, T.; Krejci, P.; Meyer, A.N.; Casale, M.; Hallowell, M.; Wilcox, W.R.; Donoghue, D.J.; Thompson, L.M. Fibroblast Growth Factor Receptor 3 Interacts with and Activates TGF beta-Activated Kinase 1 Tyrosine Phosphorylation and NFkB Signaling in Multiple Myeloma and Bladder Cancer. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Cavallaro, U.; Niedermeyer, J.; Fuxa, M.; Christofori, G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat. Cell Biol. 2001, 3, 650–657. [Google Scholar] [CrossRef]
- Ezzat, S.; Zheng, L.; Asa, S.L. Pituitary tumor-derived fibroblast growth factor receptor 4 isoform disrupts neural cell-adhesion molecule/N-cadherin signaling to diminish cell adhesiveness: A mechanism underlying pituitary neoplasia. Mol. Endocrinol. 2004, 18, 2543–2552. [Google Scholar] [CrossRef]
- Xu, S.S.; Xu, H.X.; Wang, W.Q.; Li, S.; Li, H.; Li, T.J.; Zhang, W.H.; Yu, X.J.; Liu, L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019, 17. [Google Scholar] [CrossRef]
- Efthymiou, G.; Saint, A.; Ruff, M.; Rekad, Z.; Ciais, D.; Van Obberghen-Schilling, E. Shaping Up the Tumor Microenvironment With Cellular Fibronectin. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Cao, S.; Kang, N.L.; Huebert, R.C.; Shah, V.H. Fibronectin Induces Endothelial Cell Migration through beta 1 Integrin and Src-dependent Phosphorylation of Fibroblast Growth Factor Receptor-1 at Tyrosines 653/654 and 766. J. Biol. Chem. 2012, 287, 7190–7202. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Huergo, S.P.; Blidner, A.G.; Rabinovich, G.A. Galectins: Emerging regulatory checkpoints linking tumor immunity and angiogenesis. Curr. Opin. Immunol. 2017, 45, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Mori, S.; Wu, C.Y.; Yamaji, S.; Saegusa, J.; Shi, B.; Ma, Z.; Kuwabara, Y.; Lam, K.S.; Isseroff, R.R.; Akada, Y.K.; et al. Direct binding of integrin alpha v beta 3 to FGF1 plays a role in FGF1 signaling. J. Biol. Chem. 2008, 283, 18066–18075. [Google Scholar] [CrossRef] [PubMed]
- Geissinger, E.; Weisser, C.; Fischer, P.; Schartl, M.; Wellbrock, C. Autocrine stimulation by osteopontin contributes to antiapoptotic signalling of melanocytes in dermal collagen. Cancer Res. 2002, 62, 4820–4828. [Google Scholar]
- Ruan, S.S.; Lin, M.; Zhu, Y.; Lum, L.; Thakur, A.; Jin, R.M.; Shao, W.L.; Zhang, Y.L.; Hu, Y.Y.; Huang, S.; et al. Integrin beta 4-Targeted Cancer Immunotherapies Inhibit Tumor Growth and Decrease Metastasis. Cancer Res. 2020, 80, 771–783. [Google Scholar] [CrossRef]
- Ge, D.; Kong, X.Q.; Liu, W.Y.; Zhao, J.; Su, L.; Zhang, S.L.; Zhang, Y.; Zhao, B.X.; Miao, J.Y. Phosphorylation and nuclear translocation of integrin beta 4 induced by a chemical small molecule contribute to apoptosis in vascular endothelial cells. Apoptosis 2013, 18, 1120–1131. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, D.; Kim, S.H.; Hu, Y.L.; Guimond, S.; Schofield, J.; Winyard, P.; Vannelli, G.B.; Turnbull, J.; Bouloux, P.M. Anosmin-1 modulates fibroblast growth factor receptor 1 signaling in human gonadotropin-releasing hormone olfactory neuroblasts through a heparan sulfate-dependent mechanism. J. Neurosci. 2004, 24, 10384–10392. [Google Scholar] [CrossRef]
- Korsensky, L.; Ron, D. Regulation of FGF signaling: Recent insights from studying positive and negative modulators. Semin. Cell Dev. Biol. 2016, 53, 101–114. [Google Scholar] [CrossRef]
- Wai Wong, C.; Dye, D.E.; Coombe, D.R. The Role of Immunoglobulin Superfamily Cell Adhesion Molecules in Cancer Metastasis. Int. J. Cell Biol. 2012, 2012, 340296. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, C.; Cattaneo, P.; Berezin, V.; Bock, E.; Ami, D.; de Marco, A.; Christofori, G.; Cavallaro, U. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J. Cell Biol. 2009, 187, 1101–1116. [Google Scholar] [CrossRef] [PubMed]
- Zivotic, M.; Tampe, B.; Muller, G.; Muller, C.; Lipkovski, A.; Xu, X.B.; Nyamsuren, G.; Zeisberg, M.; Markovic-Lipkovski, J. Modulation of NCAM/FGFR1 signaling suppresses EMT program in human proximal tubular epithelial cells. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.N.; Yang, L.; Li, T.; Zhang, Y. Cadherin Signaling in Cancer: Its Functions and Role as a Therapeutic Target. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Sanchez-Heras, E.; Howell, F.V.; Williams, G.; Doherty, P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J. Biol. Chem. 2006, 281, 35208–35216. [Google Scholar] [CrossRef] [PubMed]
- Agiostratidou, G.; Hulit, J.; Phillips, G.R.; Hazan, R.B. Differential cadherin expression: Potential markers for epithelial to mesenchymal transformation during tumor progression. J. Mammary Gland Biol. Neoplasia 2007, 12, 127–133. [Google Scholar] [CrossRef]
- Nguyen, T.; Mege, R.M. N-Cadherin and Fibroblast Growth Factor Receptors crosstalk in the control of developmental and cancer cell migrations. Eur. J. Cell Biol. 2016, 95, 415–426. [Google Scholar] [CrossRef]
- Qian, X.; Anzovino, A.; Kim, S.; Suyama, K.; Yao, J.; Hulit, J.; Agiostratidou, G.; Chandiramani, N.; McDaid, H.M.; Nagi, C.; et al. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene 2014, 33, 3411–3421. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, A.; Ferrer, I.; Guruceaga, E.; Cirauqui, C.; Marrugal, A.; Ojeda, L.; Garcia, S.; Zugazagoitia, J.; Munoz-Galvan, S.; Lopez-Rios, F.; et al. FGFR1 and FGFR4 oncogenicity depends on n-cadherin and their co-expression may predict FGFR-targeted therapy efficacy. Ebiomedicine 2020, 53. [Google Scholar] [CrossRef]
- Boscher, C.; Mege, R.M. Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell. Signal. 2008, 20, 1061–1072. [Google Scholar] [CrossRef]
- Nguyen, T.; Duchesne, L.; Narayana, G.; Boggetto, N.; Fernig, D.D.; Murade, C.U.; Ladoux, B.; Mege, R.M. Enhanced cell-cell contact stability and decreased N-cadherin-mediated migration upon fibroblast growth factor receptor-N-cadherin cross talk. Oncogene 2019, 38, 6283–6300. [Google Scholar] [CrossRef] [PubMed]
- El-Hariry, I.; Pignatelli, M.; Lemoine, N.R. FGF-1 and FGF-2 regulate the expression of E-cadherin and catenins in pancreatic adenocarcinoma. Int. J. Cancer 2001, 94, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Wylie, F.G.; Stow, J.L. Regulation of endocytosis, nuclear translocation, and signaling of fibroblast growth factor receptor 1 by E-cadherin. Mol. Biol. Cell 2005, 16, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Bojesen, K.B.; Clausen, O.; Rohde, K.; Christensen, C.; Zhang, L.J.; Li, S.Z.; Kohler, L.; Nielbo, S.; Nielsen, J.; Gjorlund, M.D.; et al. Nectin-1 Binds and Signals through the Fibroblast Growth Factor Receptor. J. Biol. Chem. 2012, 287, 37420–37433. [Google Scholar] [CrossRef] [PubMed]
- Kedashiro, S.; Sugiura, A.; Mizutani, K.; Takai, Y. Nectin-4 cis-interacts with ErbB2 and its trastuzumab-resistant splice variants, enhancing their activation and DNA synthesis. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Friesel, R.; Kudoh, T.; Dawid, I.B. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 2002, 4, 165–169. [Google Scholar] [CrossRef]
- Ren, Y.M.; Li, Z.Y.; Rong, Z.L.; Cheng, L.; Li, Y.H.; Wang, Z.; Chang, Z.J. Tyrosine 330 in hSef is critical for the localization and the inhibitory effect on FGF signaling. Biochem. Biophys. Res. Commun. 2007, 354, 741–746. [Google Scholar] [CrossRef]
- Xiong, S.Q.; Zhao, Q.H.; Rong, Z.L.; Huang, G.R.; Huang, Y.L.; Chen, P.L.; Zhang, S.P.; Liu, L.; Chang, Z.J. hSef inhibits PC-12 cell differentiation by interfering with Ras-mitogen-activated protein kinase MAPK signaling. J. Biol. Chem. 2003, 278, 50273–50282. [Google Scholar] [CrossRef]
- Ren, Y.M.; Cheng, L.; Rong, Z.L.; Li, Z.Y.; Li, Y.H.; Li, H.G.; Wang, Z.; Chang, Z.J. hSef co-localizes and interacts with Ras in the inhibition of Ras/MAPK signaling pathway. Biochem. Biophys. Res. Commun. 2006, 347, 988–993. [Google Scholar] [CrossRef]
- Ren, Y.M.; Cheng, L.; Rong, Z.L.; Li, Z.Y.; Li, Y.H.; Zhang, X.J.; Xiong, S.Q.; Hu, J.; Fu, X.Y.; Chang, Z.J. HSef potentiates EGF-mediated MATK signaling through affecting EGFR trafficking and degradation. Cell. Signal. 2008, 20, 518–533. [Google Scholar] [CrossRef]
- Torii, S.; Kusakabe, M.; Yamamoto, T.; Maekawa, M.; Nishida, E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell 2004, 7, 33–44. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Gong, Y.; Gower, L.; Yang, X.H.; Friesel, R.E. Sef Regulates Epithelial-Mesenchymal Transition in Breast Cancer Cells. J. Cell. Biochem. 2016, 117, 2346–2356. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Wadhwa, K.; Pisupati, V.; Zecchini, V.; Ramos-Montoya, A.; Warren, A.Y.; Neal, D.E.; Gnanapragasam, V.J. Loss of hSef promotes metastasis through upregulation of EMT in prostate cancer. Int. J. Cancer 2017, 140, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.; Darby, S.; Mathers, M.E.; Gnanapragasam, V.J. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J. Pathol. 2010, 220, 452–460. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.B.; Yan, L.; Li, M.J. Similar expression to FGF (Sef) reduces endometrial adenocarcinoma cells proliferation via inhibiting fibroblast growth factor 2-mediated MAPK/ERK signaling pathway. Gynecol. Oncol. 2011, 122, 669–674. [Google Scholar] [CrossRef]
- Chi, L.H.; Burrows, A.D.; Anderson, R.L. Bone morphogenetic protein signaling in breast cancer progression. Growth Factors 2019, 37, 12–28. [Google Scholar] [CrossRef]
- Montesano, R.; Sarkozi, R.; Schramek, H. Bone morphogenetic protein-4 strongly potentiates growth factor-induced proliferation of mammary epithelial cells. Biochem. Biophys. Res. Commun. 2008, 374, 164–168. [Google Scholar] [CrossRef]
- Jubb, A.M.; Strickland, L.A.; Liu, S.D.; Mak, J.; Schmidt, M.; Koeppen, H. Neuropilin-1 expression in cancer and development. J. Pathol. 2012, 226, 50–60. [Google Scholar] [CrossRef]
- Del Piccolo, N.; Sarabipour, S.; Hristova, K. A New Method to Study Heterodimerization of Membrane Proteins and Its Application to Fibroblast Growth Factor Receptors. J. Biol. Chem. 2017, 292, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Bellot, F.; Crumley, G.; Kaplow, J.M.; Schlessinger, J.; Jaye, M.; Dionne, C.A. Ligand-induced transphosphorylation between different FGF receptors. EMBO J. 1991, 10, 2849–2854. [Google Scholar] [CrossRef]
- Kennedy, S.P.; Hastings, J.F.; Han, J.Z.R.; Croucher, D.R. The Under-Appreciated Promiscuity of the Epidermal Growth Factor Receptor Family. Front. Cell Dev. Biol. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Yokote, H.; Fujita, K.; Jing, X.F.; Sawada, T.; Liang, S.T.; Yao, L.; Yan, X.M.; Zhang, Y.Q.; Schlessinger, J.; Sakaguchi, K. Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proc. Natl. Acad. Sci. USA 2005, 102, 18866–18871. [Google Scholar] [CrossRef] [PubMed]
- Sawada, T.; Jing, X.F.; Zhang, Y.Q.; Shimada, E.; Yokote, H.; Miyajima, M.; Sakaguchi, K. Ternary complex formation of EphA4, FGFR and FRS2 alpha plays an important role in the proliferation of embryonic neural stem/progenitor cells. Genes Cells 2010, 15, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.D.; Grubb, H.N.; Hristova, K. Quantifying the strength of heterointeractions among receptor tyrosine kinases from different subfamilies: Implications for cell signaling. J. Biol. Chem. 2020, 295, 9917–9933. [Google Scholar] [CrossRef]
- Fukai, J.; Yokote, H.; Yamanaka, R.; Arao, T.; Nishio, K.; Itakura, T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mol. Cancer Ther. 2008, 7, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Bong, Y.S.; Park, Y.H.; Lee, H.S.; Mood, K.; Ishimura, A.; Daar, I.O. Tyr-298 in ephrinB1 is critical for an interaction with the Grb4 adaptor protein. Biochem. J. 2004, 377, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, I.M.; Goke, M.; Kanai, M.; Reinecker, H.C.; Podolsky, S.K. Epithelial cell kinase-B61: An autocrine loop modulating intestinal epithelial migration and barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 1997, 273, G824–G832. [Google Scholar] [CrossRef] [PubMed]
- Faraone, D.; Aguzzi, M.S.; Ragone, G.; Russo, K.; Capogrossi, M.C.; Facchiano, A. Heterodimerization of FGF-receptor 1 and PDGF-receptor-alpha: A novel mechanism underlying the inhibitory effect of PDGF-BB on FGF-2 in human cells. Blood 2006, 107, 1896–1902. [Google Scholar] [CrossRef][Green Version]
- Chen, P.Y.; Simons, M.; Friesel, R. FRS2 via Fibroblast Growth Factor Receptor 1 Is Required for Platelet-derived Growth Factor Receptor beta-mediated Regulation of Vascular Smooth Muscle Marker Gene Expression. J. Biol. Chem. 2009, 284, 15980–15992. [Google Scholar] [CrossRef] [PubMed]
- De Marchis, F.; Ribatti, D.; Giampietri, C.; Lentini, A.; Faraone, D.; Scoccianti, M.; Capogrossi, M.C.; Facchiano, A. Platelet-derived growth factor inhibits basic fibroblast growth factor angiogenic properties in vitro and in vivo through its alpha receptor. Blood 2002, 99, 2045–2053. [Google Scholar] [CrossRef]
- Facchiano, A.; De Marchis, F.; Turchetti, E.; Facchiano, F.; Guglielmi, M.; Denaro, A.; Palumbo, R.; Scoccianti, M.; Capogrossi, M.C. The chemotactic and mitogenic effects of platelet-derived growth factor-BB on rat aorta smooth muscle cells are inhibited by basic fibroblast growth factor. J. Cell Sci. 2000, 113, 2855–2863. [Google Scholar] [CrossRef]
- Ark, A.V.; Cao, J.C.; Li, X.H. TGF-beta receptors: In and beyond TGF-beta signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef]
- Zhang, H.R.; Wang, X.D.; Yang, X.; Chen, D.; Hao, J.; Cao, R.; Wu, X.Z. An FGFR inhibitor converts the tumor promoting effect of TGF-beta by the induction of fibroblast-associated genes of hepatoma cells. Oncogene 2017, 36, 3831–3841. [Google Scholar] [CrossRef] [PubMed]
- Akatsu, Y.; Takahashi, N.; Yoshimatsu, Y.; Kimuro, S.; Muramatsu, T.; Katsura, A.; Maishi, N.; Suzuki, H.I.; Inazawa, J.; Hida, K.; et al. Fibroblast growth factor signals regulate transforming growth factor-beta-induced endothelial-to-myofibroblast transition of tumor endothelial cells via Elk1. Mol. Oncol. 2019, 13, 1706–1724. [Google Scholar] [CrossRef] [PubMed]
- Bordignon, P.; Bottoni, G.; Xu, X.Y.; Popescu, A.S.; Truan, Z.; Guenova, E.; Kofler, L.; Jafari, P.; Ostano, P.; Roecken, M.; et al. Dualism of FGF and TGF-beta Signaling in Heterogeneous Cancer-Associated Fibroblast Activation with ETV1 as a Critical Determinant. Cell Rep. 2019, 28, 2358. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, H.; Lee, N.Y. Multifaceted roles of TAK1 signaling in cancer. Oncogene 2020, 39, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Butti, R.; Das, S.; Gunasekaran, V.P.; Yadav, A.S.; Kumar, D.; Kundu, G.C. Receptor tyrosine kinases (RTKs) in breast cancer: Signaling, therapeutic implications and challenges. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.L.; Li, S.Q.; Mertins, P.; Cao, S.; Gunawardena, H.P.; Ruggles, K.V.; Mani, D.R.; Clauser, K.R.; Tanioka, M.; Usary, J.; et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.L.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55. [Google Scholar] [CrossRef]
- Arakaki, A.K.S.; Pan, W.A.; Trejo, J. GPCRs in Cancer: Protease-Activated Receptors, Endocytic Adaptors and Signaling. Int. J. Mol. Sci. 2018, 19, 1886. [Google Scholar] [CrossRef]
- Haxho, F.; Neufeld, R.J.; Szewczuk, M.R. Neuraminidase-1: A novel therapeutic target in multistage tumorigenesis. Oncotarget 2016, 7, 40860–40903. [Google Scholar] [CrossRef] [PubMed]
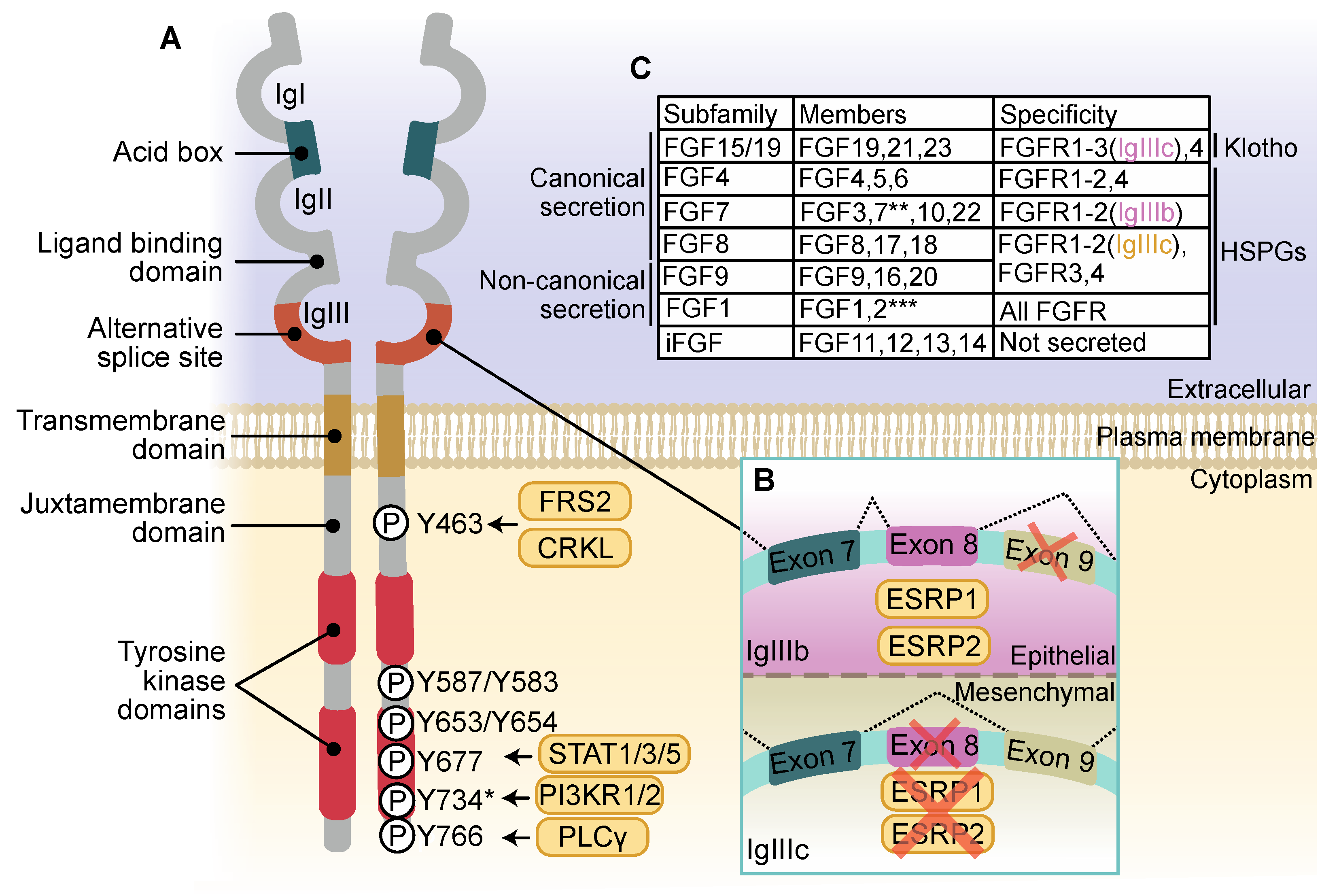
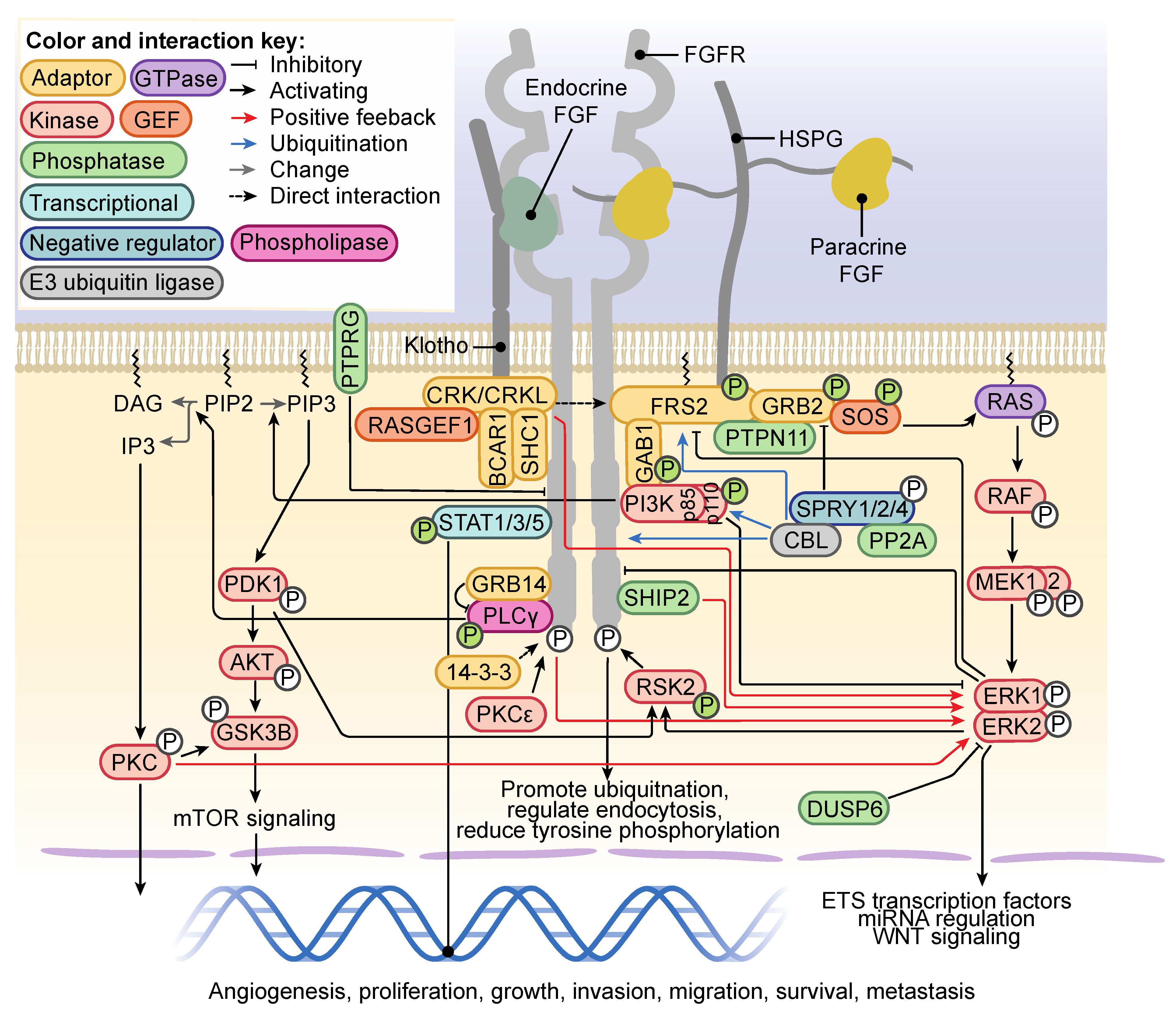
| FGF Subfamily | Cancer Type | Associated References |
|---|---|---|
| FGF1 | Bladder | [95] |
| Breast | [96] | |
| Melanoma | [97] | |
| Ovarian | [98] | |
| Pancreatic ductal adenocarcinoma | [99] | |
| Small cell lung cancer | [100] | |
| FGF4 | Bone | [101] |
| Gastrointestinal | [102,103] | |
| Glioma | [104] | |
| FGF7 | Breast | [42] |
| Gastric | [105] | |
| Non-small cell lung cancer | [106,107] | |
| Pancreatic ductal adenocarcinoma | [108] | |
| FGF8 | Colorectal | [109] |
| Hepatocellular carcinoma | [110] | |
| Head and neck | [111] | |
| Prostate | [112] | |
| Renal cell carcinoma | [113] | |
| FGF9 | Colorectal | [114] |
| Hepatocellular carcinoma | [115] | |
| Prostate | [116,117] | |
| FGF15/19 | Colorectal | [118] |
| Endometrial | [119] | |
| Hepatocellular carcinoma | [120] | |
| Prostate | [121] | |
| iFGFs | Colorectal | [122] |
| Prostate | [123,124] | |
| Triple negative breast cancer | [125] |
| FGFR | Partner | Cancer Subtype | Consequence of Interaction | Associated References |
|---|---|---|---|---|
| FGFR1 | Anosmin-1 | Brain; Glioblastoma | Promotes motility and invasion in FGFR-dependent manner | [259] |
| Collagen type IV | Pancreatic | Sustained ERK1/2 activation | [10,176,177,260] | |
| EGFR | Lung | Increases AKT and STAT3 signaling | [261] | |
| EPHA4 | Glioma | Potentiates FGFR1-signaling | [262] | |
| Galectin-1 | Osteosarcoma | Activated FGFR1, increased proliferation and survival | [263] | |
| Galectin-3 | Osteosarcoma | FGFR1 plasma membrane clustering | [263] | |
| GALNT14 | Breast | Promotes FGFR activation | [264] | |
| Integrin-αVβ3 | Breast | Increased tumorigensis, angiogenesis and EMT | [265,266] | |
| Integrin-β3 | Breast | Disrupts colocalization with E-cadherin and promotes EMT | [267] | |
| L1CAM | Glioma | Promote motility and proliferation | [268,269] | |
| N-cadherin | Breast | Stabilizes FGFR1 at plasma membrane, promoting sustained ERK1/2 signal, increased invasion, motility and metastasis | [270,271] | |
| NRP1 | Breast | FGFR1-NRP1 complex increases during EMT | [272] | |
| RET-KIF5B, EGFR | RET-KIF5B fusion positive cancers | Increases FGFR activation and promotes inavdopodia formation | [273] | |
| TGFBR3 | Neuroblastoma | Activates ERK1/2 signaling, promoting differentiation, suppressing tumor growth and metastasis | [274] | |
| FGFR2 | EGFR | Breast | Induces EGFR T693 phosphorylation, recycling and stabilization and increases cell cycle outputs | [275] |
| EPHA4 | Glioma | Increases proliferation and migration | [262] | |
| RET-KIF5B, EGFR | RET-KIF5B fusion positive cancers | Increases FGFR activation and promotes inavdopodia formation | [273] | |
| FGFR3 | PYK2 | Multiple myeloma | Increases STAT5 activation | [276] |
| TAK1 | Multiple myelomaBladder | Cell adhesion and NFκB-dependent transcription | [277] | |
| FGFR4 | NCAM | Pancreatic | Promote neurite outgrowth | [278] |
| NCAM | Pituitary neoplasia | When interaction is inhibited increases invasive phenotype | [279] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferguson, H.R.; Smith, M.P.; Francavilla, C. Fibroblast Growth Factor Receptors (FGFRs) and Noncanonical Partners in Cancer Signaling. Cells 2021, 10, 1201. https://doi.org/10.3390/cells10051201
Ferguson HR, Smith MP, Francavilla C. Fibroblast Growth Factor Receptors (FGFRs) and Noncanonical Partners in Cancer Signaling. Cells. 2021; 10(5):1201. https://doi.org/10.3390/cells10051201
Chicago/Turabian StyleFerguson, Harriet R., Michael P. Smith, and Chiara Francavilla. 2021. "Fibroblast Growth Factor Receptors (FGFRs) and Noncanonical Partners in Cancer Signaling" Cells 10, no. 5: 1201. https://doi.org/10.3390/cells10051201
APA StyleFerguson, H. R., Smith, M. P., & Francavilla, C. (2021). Fibroblast Growth Factor Receptors (FGFRs) and Noncanonical Partners in Cancer Signaling. Cells, 10(5), 1201. https://doi.org/10.3390/cells10051201






