A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease
Abstract
1. Introduction
2. Structure and Functional Data on the TOM Complex
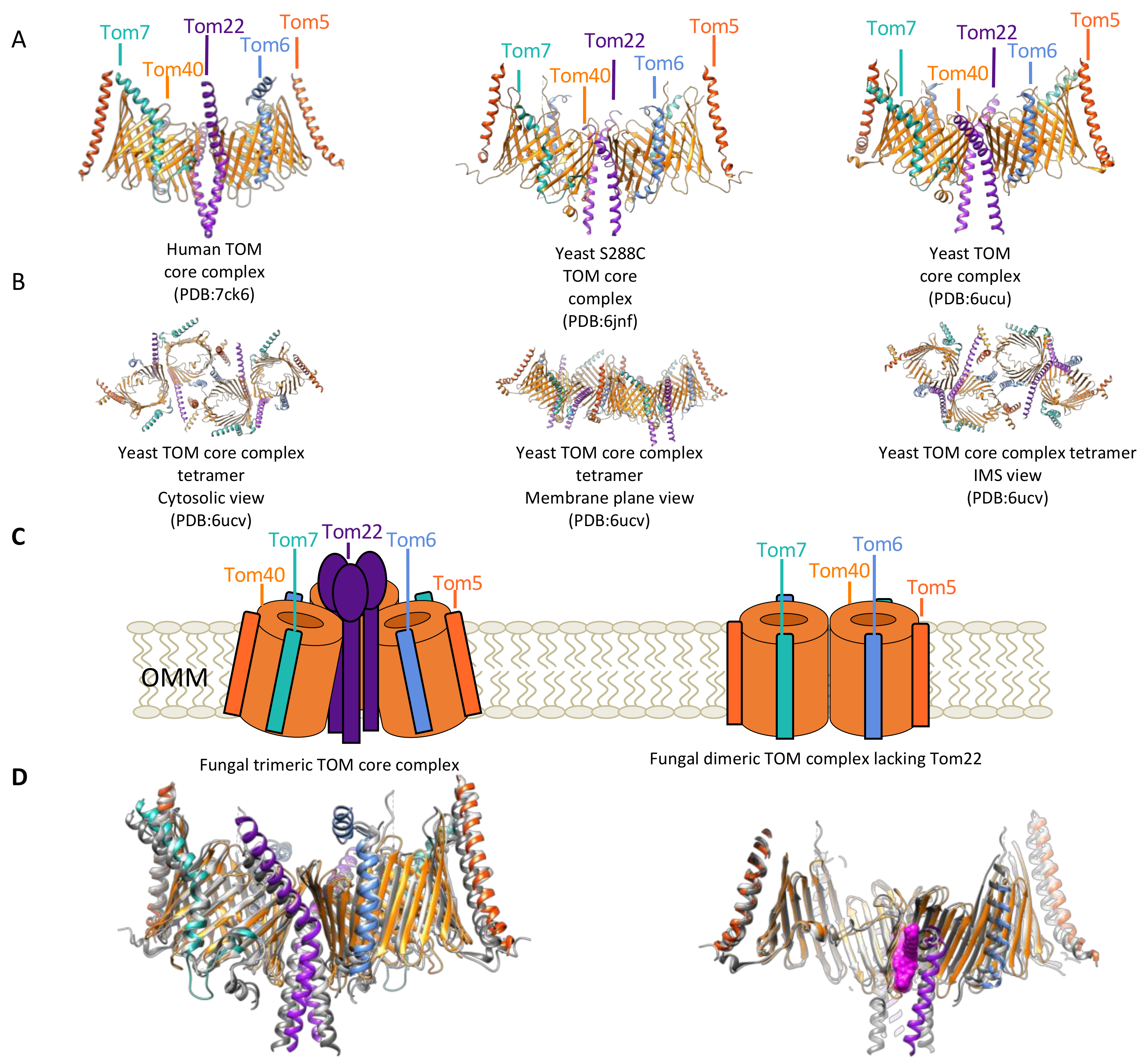
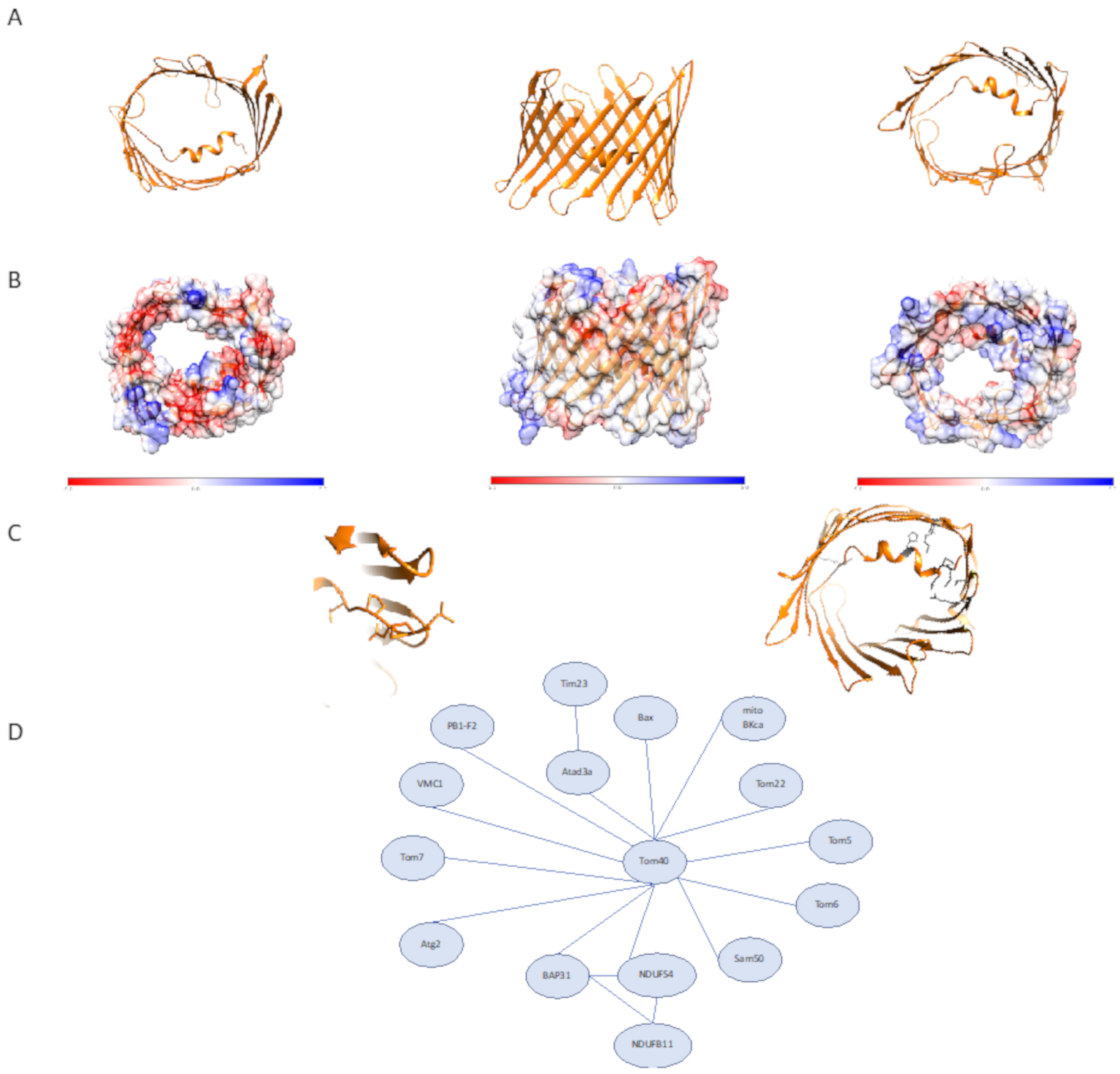
2.1. Biochemical and Structural Characterization of the Components of the TOM Complex
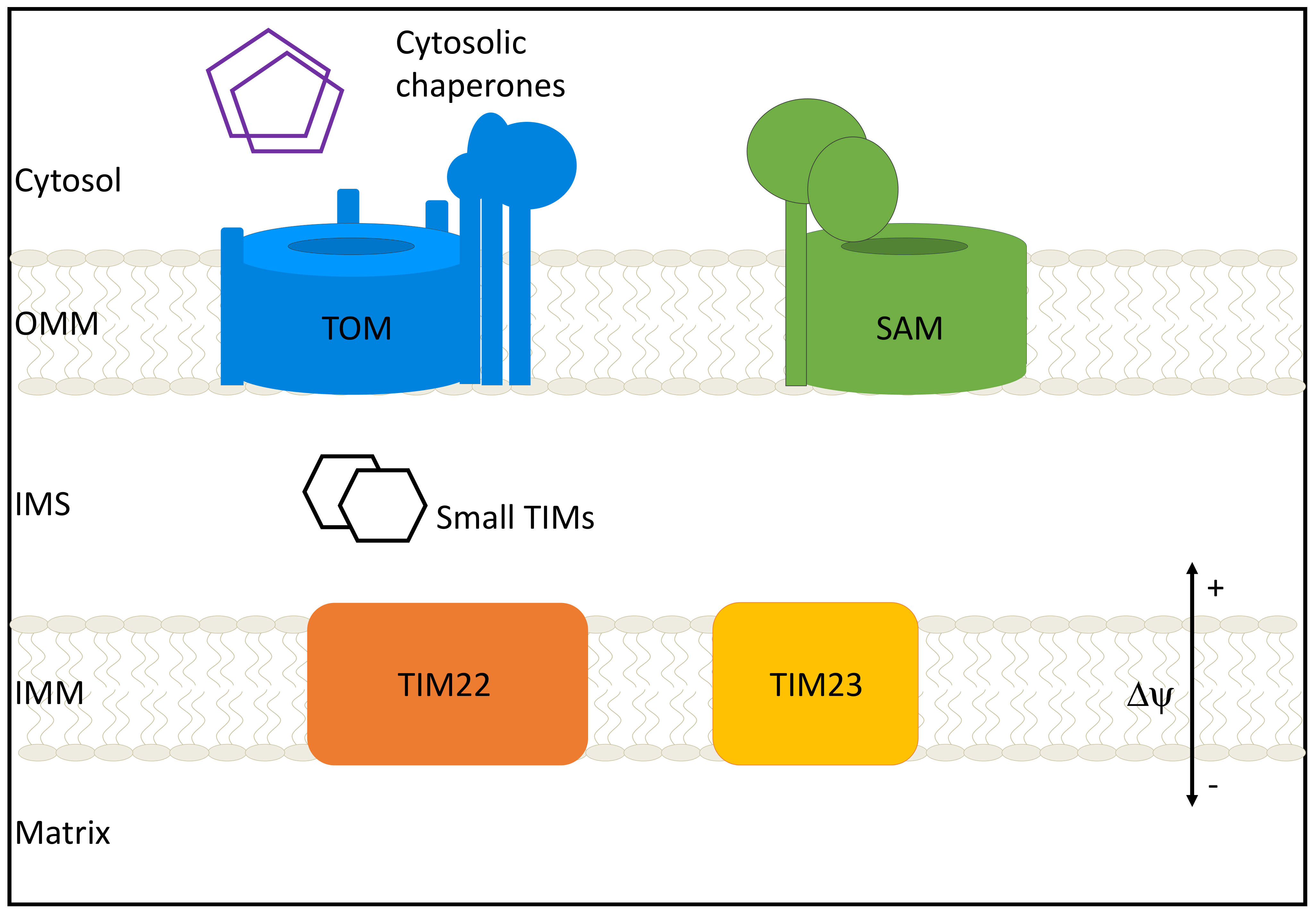
2.2. Observed Structural Arrangements of the TOM Complex and Interplay with Other Mitochondrial Membrane Proteins
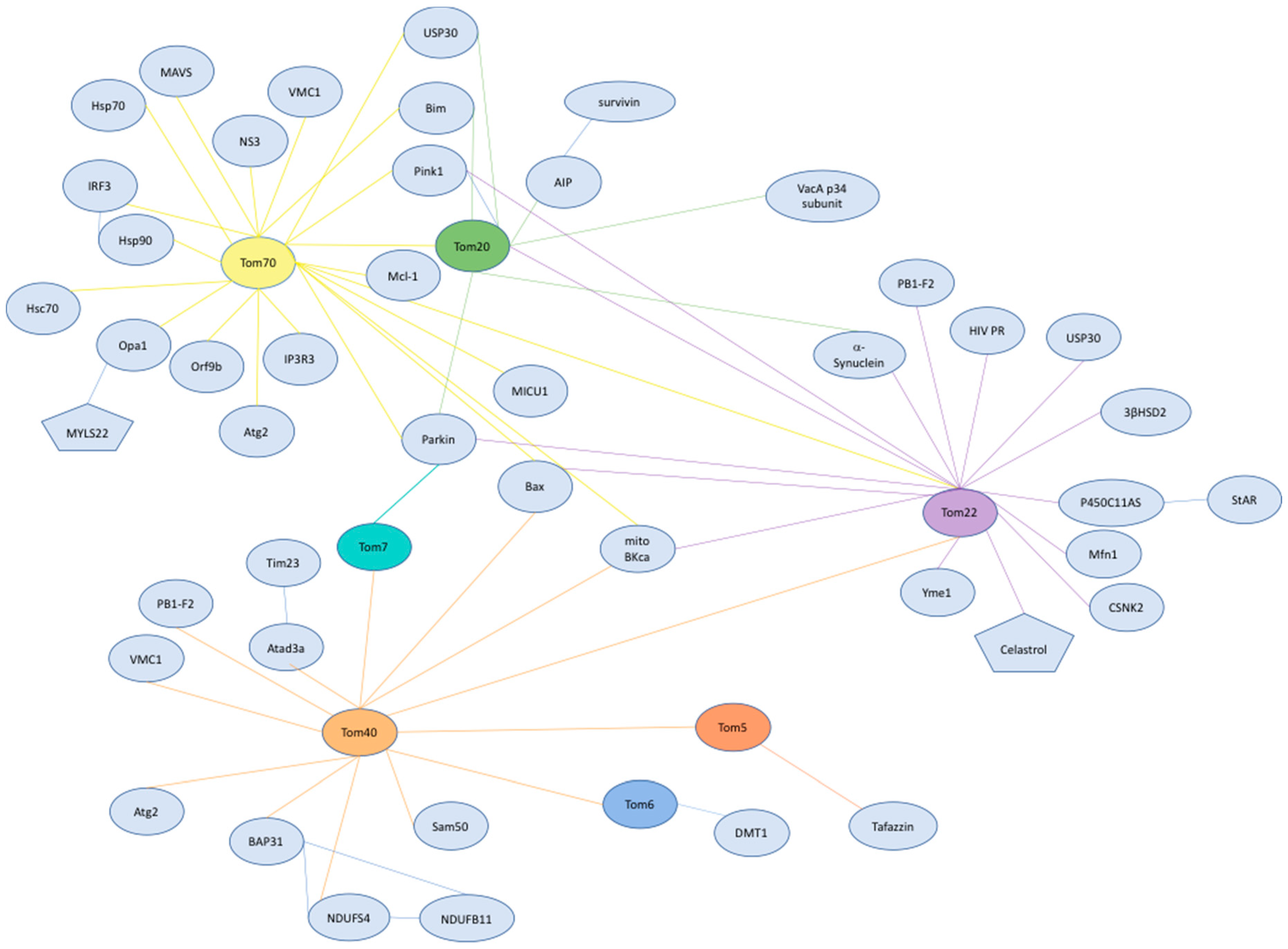
3. The Role of the TOM Complex Subunits in Human Health and Disease
3.1. The Role of Tom40 in Mitochondrial Homeostasis, Human Disease, and Therapeutics
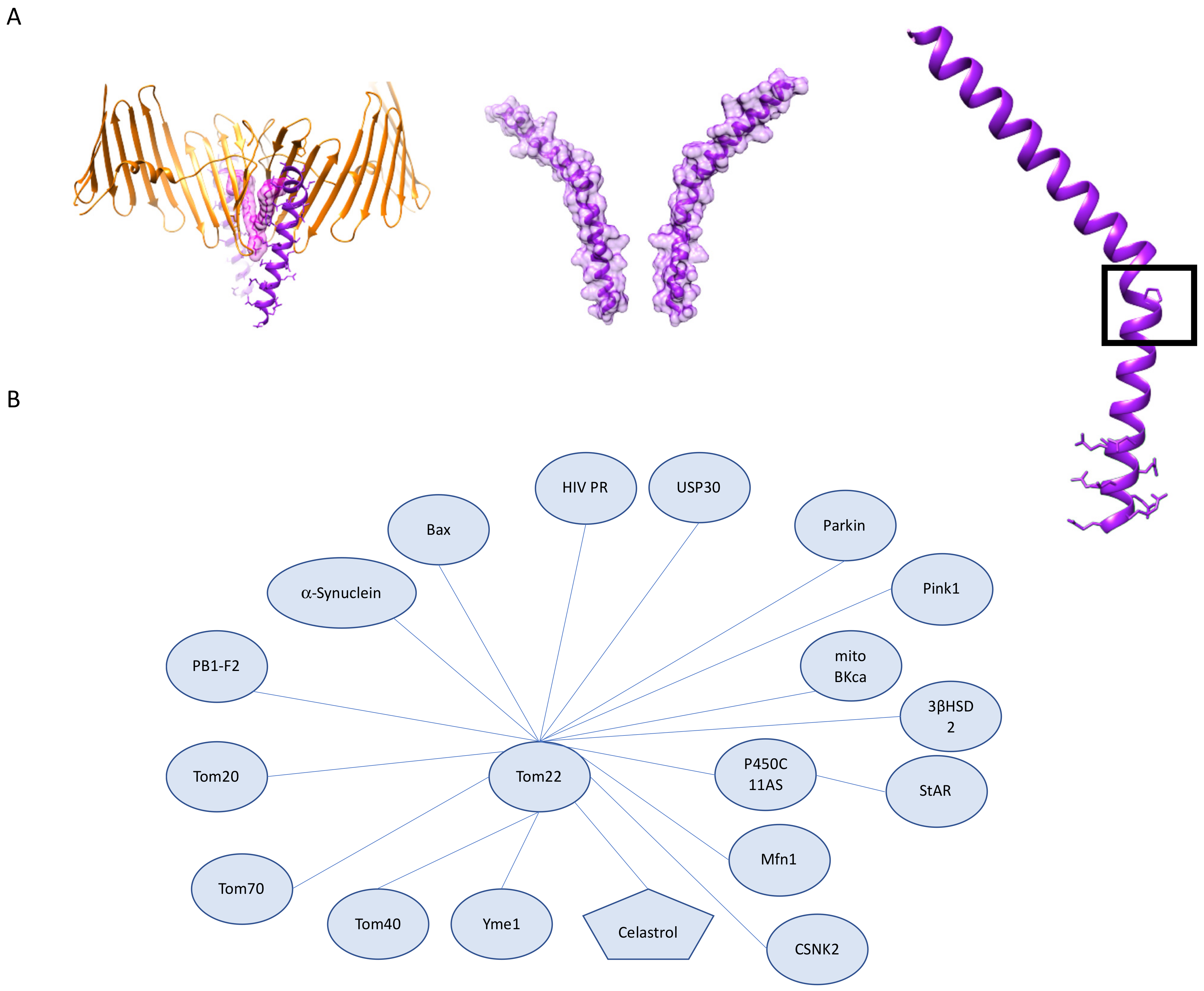
3.2. The Role of Tom22 in Mitochondrial Homeostasis, Human Disease, and Therapeutics
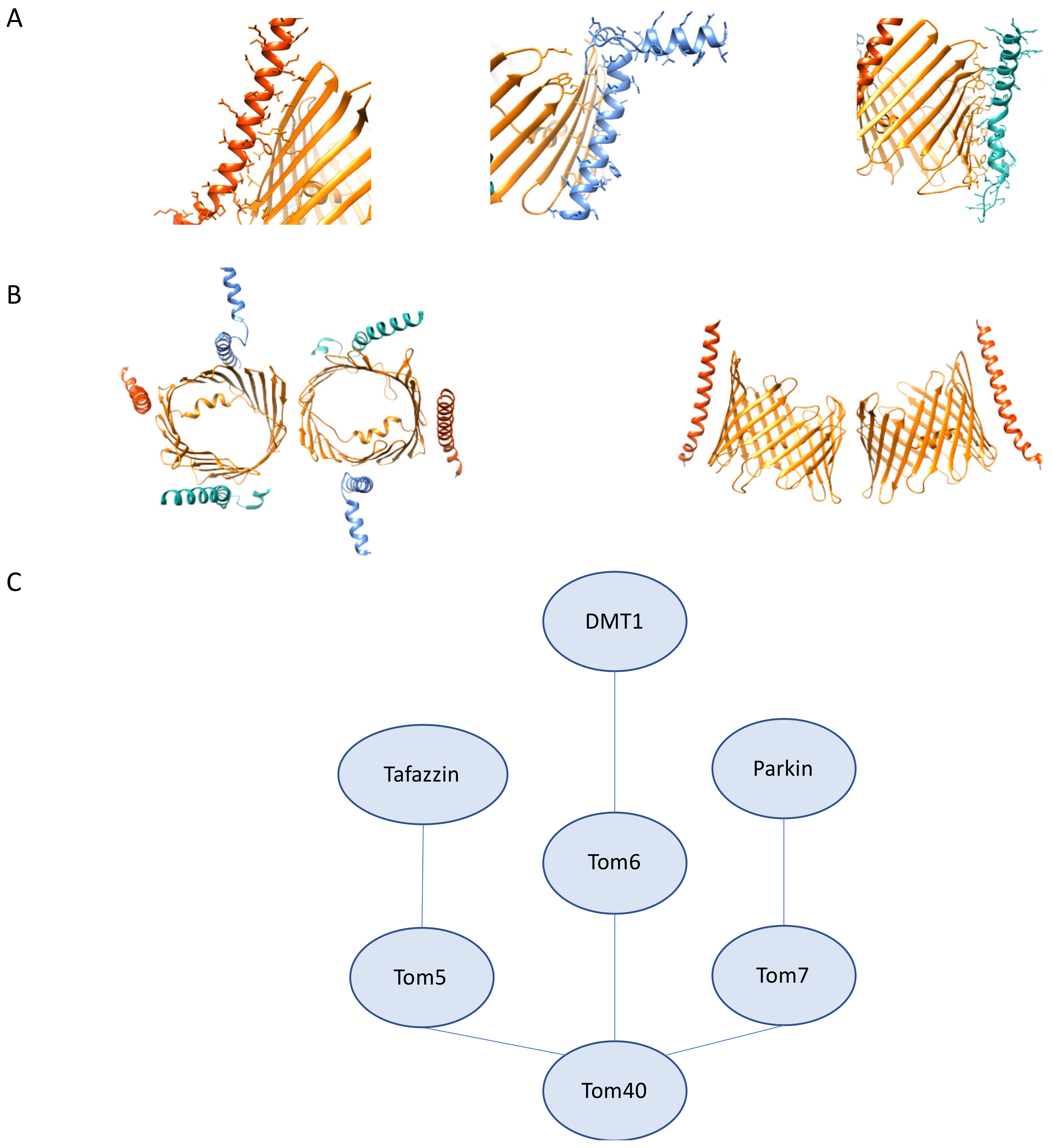
3.3. The Roles Tom5, Tom6, and Tom7 in Mitochondrial Homeostasis, Human Disease, and Therapeutics
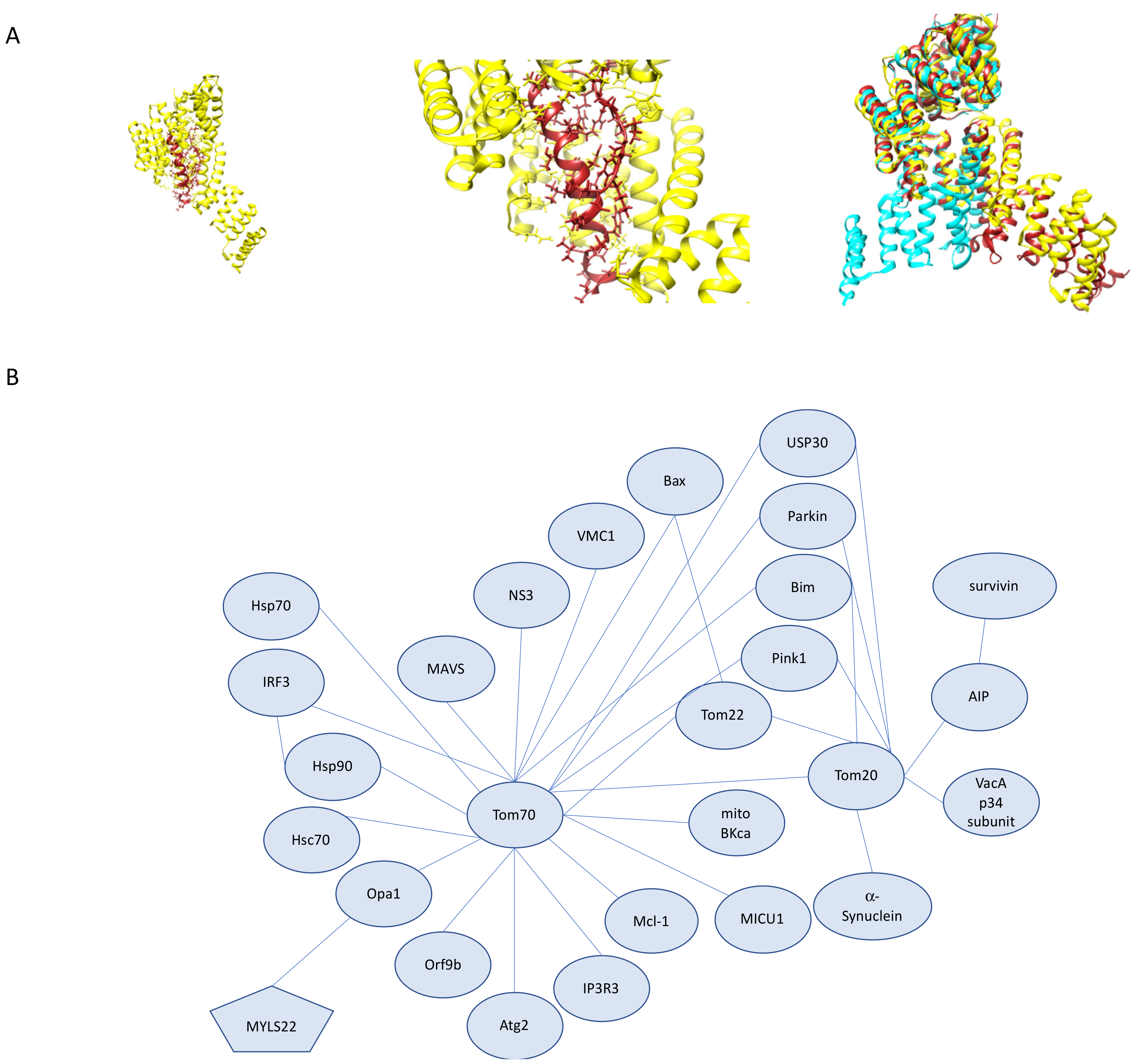
3.4. The Roles Tom20 and Tom70 in Mitochondrial Homeostasis, Human Disease, and Therapeutics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing Mitochondrial Proteins: Machineries and Mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Lotz, C.; Lin, A.J.; Black, C.M.; Zhang, J.; Lau, E.; Deng, N.; Wang, Y.; Zong, N.C.; Choi, J.H.; Xu, T.; et al. Characterization, Design, and Function of the Mitochondrial Proteome: From Organs to Organisms. J. Proteome Res. 2014, 13, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, W.K.; Lutz, M.W.; He, Y.T.; Saunders, A.M.; Burns, D.K.; Roses, A.D.; Chiba-Falek, O. The Broad Impact of TOM40 on Neurodegenerative Diseases in Aging. J. Parkinsons Dis. Alzheimers Dis. 2014, 1, 1. [Google Scholar]
- Cotter, D. MitoProteome: Mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004, 32, 463D–467D. [Google Scholar] [CrossRef]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2011, 40, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Treviño, P.; Velásquez, M.; García, N. Mechanisms of mitochondrial DNA escape and its relationship with different metabolic diseases. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165761. [Google Scholar] [CrossRef]
- Martincová, E.; Voleman, L.; Pyrih, J.; Zarsky, V.; Vondráčková, P.; Kolisko, M.; Tachezy, J.; Doležal, P. Probing the Biology of Giardia intestinalis Mitosomes UsingIn VivoEnzymatic Tagging. Mol. Cell. Biol. 2015, 35, 2864–2874. [Google Scholar] [CrossRef]
- Taylor, R.D.; McHale, B.J.; Nargang, F.E. Characterization of Neurospora crassa Tom40-deficient Mutants and Effect of Specific Mutations on Tom40 Assembly. J. Biol. Chem. 2003, 278, 765–775. [Google Scholar] [CrossRef]
- Wang, W.; Chen, X.; Zhang, L.; Yi, J.; Ma, Q.; Yin, J.; Zhuo, W.; Gu, J.; Yang, M. Atomic structure of human TOM core complex. Cell Discov. 2020, 6, 1–10. [Google Scholar] [CrossRef]
- Humphries, A.D.; Streimann, I.C.; Stojanovski, D.; Johnston, A.J.; Yano, M.; Hoogenraad, N.J.; Ryan, M.T. Dissection of the Mitochondrial Import and Assembly Pathway for Human Tom. J. Biol. Chem. 2005, 280, 11535–11543. [Google Scholar] [CrossRef]
- Chan, N.C.; Lithgow, T. The Peripheral Membrane Subunits of the SAM Complex Function Codependently in Mitochondrial Outer Membrane Biogenesis. Mol. Biol. Cell 2008, 19, 126–136. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Chakrabarty, S.; Shukla, V.; Varghese, V.K.; Singh, K.K.; Thangaraj, K.; Satyamoorthy, K. Mitochondrial biology: From molecules to diseases. Mitochondrion 2015, 24, 93–98. [Google Scholar] [CrossRef]
- Bolliger, L.; Junne, T.; Schatz, G.; Lithgow, T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995, 14, 6318–6326. [Google Scholar] [CrossRef]
- Dietmeier, K.; Hönlinger, A.; Bömer, U.; Dekker, P.J.T.; Eckerskorn, C.; Lottspeich, F.; Kübrich, M.; Pfanner, N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nat. Cell Biol. 1997, 388, 195–200. [Google Scholar] [CrossRef]
- Schatz, G. Just follow the acid chain. Nat. Cell Biol. 1997, 388, 121–122. [Google Scholar] [CrossRef]
- Komiya, T.; Rospert, S.; Koehler, C.; Looser, R.; Schatz, G.; Mihara, K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: Evidence for the ’acid chain’ hypothesis. EMBO J. 1998, 17, 3886–3898. [Google Scholar] [CrossRef]
- Moczko, M.; Bömer, U.; Kübrich, M.; Zufall, N.; Hönlinger, A.; Pfanner, N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 1997, 17, 6574–6584. [Google Scholar] [CrossRef] [PubMed]
- Saeki, K.; Suzuki, H.; Tsuneoka, M.; Maeda, M.; Iwamoto, R.; Hasuwa, H.; Shida, S.; Takahashi, T.; Sakaguchi, M.; Endo, T.; et al. Identification of Mammalian TOM22 as a Subunit of the Preprotein Translocase of the Mitochondrial Outer Membrane. J. Biol. Chem. 2000, 275, 31996–32002. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Hoogenraad, N.; Terada, K.; Mori, M. Identification and Functional Analysis of Human Tom22 for Protein Import into Mitochondria. Mol. Cell. Biol. 2000, 20, 7205–7213. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Mihara, K. Identification of Tom5 and Tom6 in the preprotein translocase complex of human mitochondrial outer membrane. Biochem. Biophys. Res. Commun. 2008, 369, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Honlinger, A.; Bömer, U.; Alconada, A.; Eckerskorn, C.; Lottspeich, F.; Dietmeier, K.; Pfanner, N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996, 15, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Model, K.; Meisinger, C.; Prinz, T.; Wiedemann, N.; Truscott, K.; Pfanner, N.; Ryan, M.T. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Genet. 2001, 8, 361–370. [Google Scholar] [CrossRef]
- Wu, Y.; Sha, B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006, 13, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Shodai, T.; Muto, T.; Mihara, K.; Torii, H.; Nishikawa, S.-I.; Endo, T.; Kohda, D. Structural Basis of Presequence Recognition by the Mitochondrial Protein Import Receptor Tom. Cell 2000, 100, 551–560. [Google Scholar] [CrossRef]
- Brix, J.; Dietmeier, K.; Pfanner, N. Differential Recognition of Preproteins by the Purified Cytosolic Domains of the Mitochondrial Import Receptors Tom20, Tom22, and Tom. J. Biol. Chem. 1997, 272, 20730–20735. [Google Scholar] [CrossRef]
- Vardi-Oknin, D.; Arava, Y. Characterization of Factors Involved in Localized Translation Near Mitochondria by Ribosome-Proximity Labeling. Front. Cell Dev. Biol. 2019, 7, 305. [Google Scholar] [CrossRef]
- Backes, S.; Hess, S.; Boos, F.; Woellhaf, M.W.; Gödel, S.; Jung, M.; Mühlhaus, T.; Herrmann, J.M. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 2018, 217, 1369–1382. [Google Scholar] [CrossRef]
- Mills, R.D.; Trewhella, J.; Qiu, T.W.; Welte, T.; Ryan, T.M.; Hanley, T.; Knott, R.B.; Lithgow, T.; Mulhern, T.D. Domain Organization of the Monomeric Form of the Tom70 Mitochondrial Import Receptor. J. Mol. Biol. 2009, 388, 1043–1058. [Google Scholar] [CrossRef]
- Li, J.; Qian, X.; Hu, J.; Sha, B. Molecular Chaperone Hsp70/Hsp90 Prepares the Mitochondrial Outer Membrane Translocon Receptor Tom71 for Preprotein Loading. J. Biol. Chem. 2009, 284, 23852–23859. [Google Scholar] [CrossRef]
- Dutta, D.; Briere, L.C.; Kanca, O.; Marcogliese, P.C.; Walker, M.A.; High, F.A.; Vanderver, A.; Krier, J.; Carmichael, N.; Callahan, C.; et al. De novo mutations in TOMM70, a receptor of the mitochondrial import translocase, cause neurological impairment. Hum. Mol. Genet. 2020, 29, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Nugent, S.; Potvin, O.; Cunnane, S.C.; Chen, T.-H.; Duchesne, S. Associating Type 2 Diabetes Risk Factor Genes and FDG-PET Brain Metabolism in Normal Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 580633. [Google Scholar] [CrossRef]
- Wei, X.; Du, M.; Xie, J.; Luo, T.; Zhou, Y.; Zhang, K.; Li, J.; Chen, D.; Xu, P.; Jia, M.; et al. Mutations in TOMM70 lead to multi-OXPHOS deficiencies and cause severe anemia, lactic acidosis, and developmental delay. J. Hum. Genet. 2020, 65, 231–240. [Google Scholar] [CrossRef]
- Gornicka, A.; Bragoszewski, P.; Chroscicki, P.; Wenz, L.-S.; Schulz, C.; Rehling, P.; Chacinska, A. A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol. Biol. Cell 2014, 25, 3999–4009. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, H.; Shiota, T.; Ishizaka, N.; Kawano, S.; Tamura, Y.; Tan, K.S.; Imai, K.; Motono, C.; Hirokawa, T.; Taki, K.; et al. Porin Associates with Tom22 to Regulate the Mitochondrial Protein Gate Assembly. Mol. Cell 2019, 73, 1044–1055.e8. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.S.; Bildl, W.; Haupt, A.; Ellenrieder, L.; Becker, T.; Hunte, C.; Fakler, B.; Schulte, U. Cryo-slicing Blue Native-Mass Spectrometry (csBN-MS), a Novel Technology for High Resolution Complexome Profiling. Mol. Cell. Proteom. 2016, 15, 669–681. [Google Scholar] [CrossRef]
- Bausewein, T.; Mills, D.J.; Langer, J.D.; Nitschke, B.; Nussberger, S.; Kühlbrandt, W. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell 2017, 170, 693–700.e7. [Google Scholar] [CrossRef]
- Yang, W.; Shin, H.-Y.; Cho, H.; Chung, J.-Y.; Lee, E.-J.; Kim, J.-H.; Kang, E.-S. TOM40 Inhibits Ovarian Cancer Cell Growth by Modulating Mitochondrial Function Including Intracellular ATP and ROS Levels. Cancers 2020, 12, 1329. [Google Scholar] [CrossRef] [PubMed]
- Domańska, G.; Motz, C.; Meinecke, M.; Harsman, A.; Papatheodorou, P.; Reljic, B.; Dian-Lothrop, E.A.; Galmiche, A.; Kepp, O.; Becker, L.; et al. Helicobacter pylori VacA Toxin/Subunit p34: Targeting of an Anion Channel to the Inner Mitochondrial Membrane. PLoS Pathog. 2010, 6, e1000878. [Google Scholar] [CrossRef]
- Kang, B.H.; Xia, F.; Pop, R.; Dohi, T.; Socolovsky, M.; Altieri, D.C. Developmental Control of Apoptosis by the Immunophilin Aryl Hydrocarbon Receptor-interacting Protein (AIP) Involves Mitochondrial Import of the Survivin Protein. J. Biol. Chem. 2011, 286, 16758–16767. [Google Scholar] [CrossRef] [PubMed]
- Künkele, K.-P.; Heins, S.; Dembowski, M.E.; Nargang, F.; Benz, R.; Thieffry, M.; Walz, J.; Lill, R.; Nussberger, S.; Neupert, W. The Preprotein Translocation Channel of the Outer Membrane of Mitochondria. Cell 1998, 93, 1009–1019. [Google Scholar] [CrossRef]
- Model, K.; Meisinger, C.; Kühlbrandt, W. Cryo-Electron Microscopy Structure of a Yeast Mitochondrial Preprotein Translocase. J. Mol. Biol. 2008, 383, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Model, K.; Prinz, T.; Ruiz, T.; Radermacher, M.; Krimmer, T.; Kühlbrandt, W.; Pfanner, N.; Meisinger, C. Protein translocase of the outer mitochondrial membrane: Role of import receptors in the structural organization of the TOM complex. J. Mol. Biol. 2002, 316, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Shiota, T.; Imai, K.; Qiu, J.; Hewitt, V.L.; Tan, K.; Shen, H.-H.; Sakiyama, N.; Fukasawa, Y.; Hayat, S.; Kamiya, M.; et al. Molecular architecture of the active mitochondrial protein gate. Science 2015, 349, 1544–1548. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, J.-Y.; Mihara, K.; Oka, T. Identification and Characterization of a New Tom40 Isoform, a Central Component of Mitochondrial Outer Membrane Translocase. J. Biochem. 2007, 141, 897–906. [Google Scholar] [CrossRef]
- Mager, F.; Gessmann, D.; Nussberger, S.; Zeth, K. Functional Refolding and Characterization of Two Tom40 Isoforms from Human Mitochondria. J. Membr. Biol. 2011, 242, 11–21. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N.; Ryan, M.T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001, 20, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, T.; Ichinohe, T.; Sasaki, O.; Otera, H.; Kawabata, S.-I.; Mihara, K.; Koshiba, T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014, 5, 4713. [Google Scholar] [CrossRef]
- Loo, Y.-M.; Gale, M. Immune Signaling by RIG-I-like Receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Zara, V.; Ferramosca, A.; Günnewig, K.; Kreimendahl, S.; Schwichtenberg, J.; Sträter, D.; Çakar, M.; Emmrich, K.; Guidato, P.; Palmieri, F.; et al. Mimivirus-Encoded Nucleotide Translocator VMC1 Targets the Mitochondrial Inner Membrane. J. Mol. Biol. 2018, 430, 5233–5245. [Google Scholar] [CrossRef]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef]
- Tang, Z.; Takahashi, Y.; He, H.; Hattori, T.; Chen, C.; Liang, X.; Chen, H.; Young, M.M.; Wang, H.-G. TOM40 Targets Atg2 to Mitochondria-Associated ER Membranes for Phagophore Expansion. Cell Rep. 2019, 28, 1744–1757.e5. [Google Scholar] [CrossRef] [PubMed]
- Namba, T. BAP31 regulates mitochondrial function via interaction with Tom40 within ER-mitochondria contact sites. Sci. Adv. 2019, 5, eaaw1386. [Google Scholar] [CrossRef] [PubMed]
- Khachane, A.N.; Timmis, K.N.; Dos Santos, V.A.P.M. Dynamics of Reductive Genome Evolution in Mitochondria and Obligate Intracellular Microbes. Mol. Biol. Evol. 2006, 24, 449–456. [Google Scholar] [CrossRef]
- Jin, G.; Xu, C.; Zhang, X.; Long, J.; Rezaeian, A.H.; Liu, C.; Furth, M.E.; Kridel, S.; Pasche, B.; Bian, X.-W.; et al. Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat. Immunol. 2018, 19, 29–40. [Google Scholar] [CrossRef]
- Bender, A.; Desplats, P.; Spencer, B.; Rockenstein, E.; Adame, A.; Elstner, M.; Laub, C.; Mueller, S.; Koob, A.O.; Mante, M.; et al. TOM40 Mediates Mitochondrial Dysfunction Induced by α-Synuclein Accumulation in Parkinson’s Disease. PLoS ONE 2013, 8, e62277. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2019, 48, D1031–D1041. [Google Scholar] [CrossRef] [PubMed]
- Bellot, G.; Cartron, P.-F.; Er, E.; Oliver, L.; Juin, P.; Armstrong, L.C.; Bornstein, P.; Mihara, K.; Manon, S.; Vallette, F.M. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2006, 14, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Curado, S.; Ober, E.A.; Walsh, S.; Cortes-Hernandez, P.; Verkade, H.; Koehler, C.M.; Stainier, D.Y.R. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Dis. Model. Mech. 2010, 3, 486–495. [Google Scholar] [CrossRef]
- Rajapaksha, M.; Kaur, J.; Prasad, M.; Pawlak, K.J.; Marshall, B.; Perry, E.W.; Whittal, R.M.; Bose, H.S. An Outer Mitochondrial Translocase, Tom22, Is Crucial for Inner Mitochondrial Steroidogenic Regulation in Adrenal and Gonadal Tissues. Mol. Cell. Biol. 2016, 36, 1032–1047. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Tsuji, J.; Fu, S.-C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved Prediction of Mitochondrial Targeting Sequences and Their Cleavage Sites*. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef]
- Liu, Z.; Ding, Y.; Ye, N.; Wild, C.; Chen, H.; Zhou, J. Direct Activation of Bax Protein for Cancer Therapy. Med. Res. Rev. 2016, 36, 313–341. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.-F.; Bellot, G.; Oliver, L.; Grandier-Vazeille, X.; Manon, S.; Vallette, F.M. Bax inserts into the mitochondrial outer membrane by different mechanisms. FEBS Lett. 2008, 582, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Grosse, L.; Wurm, A.C.; Bruser, C.; Neumann, D.C.J.; Jakobs, S. Bax assembles into large ring-like structures remodeling the mitochondrial outer membrane in apoptosis. EMBO J. 2016, 35, 402–413. [Google Scholar]
- Renault, T.T.; Grandier-Vazeille, X.; Arokium, H.; Velours, G.; Camougrand, N.; Priault, M.; Teijido, O.; Dejean, L.M.; Manon, S. The cytosolic domain of human Tom22 modulates human Bax mitochondrial translocation and conformation in yeast. FEBS Lett. 2012, 586, 116–121. [Google Scholar] [CrossRef]
- Rumlová, M.; Křížová, I.; Keprová, A.; Hadravová, R.; Doležal, M.; Strohalmová, K.; Pichová, I.; Hájek, M.; Ruml, T. HIV-1 protease-induced apoptosis. Retrovirology 2014, 11, 37. [Google Scholar] [CrossRef]
- Veresov, V.G.; Davidovskii, A.I. Structural insights into proapoptotic signaling mediated by MTCH2, VDAC2, TOM40 and TOM22. Cell. Signal. 2014, 26, 370–382. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, D.; Fan, C.; Ma, C.; Yang, W.; Meng, Y.; Wu, W.; Guan, S.; Jiang, B.; Yang, M.; et al. ER stress-mediated apoptosis induced by celastrol in cancer cells and important role of glycogen synthase kinase-3beta in the signal network. Cell Death Dis. 2013, 4, e715. [Google Scholar] [CrossRef]
- Zeng, Y.; Pan, Q.; Wang, X.; Li, N.; Lin, Y.; Man, F.; Xiao, F.; Guo, L. Impaired Mitochondrial Fusion and Oxidative Phosphorylation Triggered by High Glucose Is Mediated by Tom22 in Endothelial Cells. Oxidative Med. Cell. Longev. 2019, 2019, 4508762. [Google Scholar] [CrossRef]
- Siemen, D.; Loupatatzisa, C.; Boreckyb, J.; Gulbinsa, E.; Langa, F. Ca2+-Activated K Channel of the BK-Type in the Inner Mitochondrial Membrane of a Human Glioma Cell Line. Biochem. Biophys. Res. Commun. 1999, 257, 549–554. [Google Scholar] [CrossRef]
- Zhang, J.; Li, M.; Zhang, Z.; Zhu, R.; Olcese, R.; Stefani, E.; Toro, L. The mitochondrial BKCa channel cardiac interactome reveals BKCa association with the mitochondrial import receptor subunit Tom22, and the adenine nucleotide translocator. Mitochondrion 2017, 33, 84–101. [Google Scholar] [CrossRef]
- Sokol, A.M.; Sztolsztener, M.E.; Wasilewski, M.; Heinz, E.; Chacinska, A. Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 2014, 588, 2484–2495. [Google Scholar] [CrossRef]
- Bajaj, R.; Jaremko, Ł.; Jaremko, M.; Becker, S.; Zweckstetter, M. Molecular Basis of the Dynamic Structure of the TIM23 Complex in the Mitochondrial Intermembrane Space. Structure 2014, 22, 1501–1511. [Google Scholar] [CrossRef] [PubMed]
- Waegemann, K.; Popov-Čeleketić, D.; Neupert, W.; Azem, A.; Mokranjac, D. Cooperation of TOM and TIM23 Complexes during Translocation of Proteins into Mitochondria. J. Mol. Biol. 2015, 427, 1075–1084. [Google Scholar] [CrossRef]
- Qian, X.; Gebert, M.; Höpker, J.; Yan, M.; Li, J.; Wiedemann, N.; Van Der Laan, M.; Pfanner, N.; Sha, B. Structural Basis for the Function of Tim50 in the Mitochondrial Presequence Translocase. J. Mol. Biol. 2011, 411, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Bose, H.S.; Whittal, R.M.; Marshall, B.; Rajapaksha, M.; Wang, N.P.; Bose, M.; Perry, E.W.; Zhao, Z.-O.; Miller, W.L. A novel mitochondrial complex of P450c11AS, StAR and Tom22 synthesizes aldosterone in the rat heart. J. Pharmacol. Exp. Ther. 2021. [Google Scholar] [CrossRef]
- Lazarou, M.; Jin, S.M.; Kane, L.A.; Youle, R.J. Role of PINK1 Binding to the TOM Complex and Alternate Intracellular Membranes in Recruitment and Activation of the E3 Ligase Parkin. Dev. Cell 2012, 22, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Fiesel, F.C.; Caulfield, T.R.; Moussaud-Lamodière, E.L.; Ogaki, K.; Dourado, D.F.; Flores, S.C.; Ross, O.A.; Springer, W. Structural and Functional Impact of Parkinson Disease-Associated Mutations in the E3 Ubiquitin Ligase Parkin. Hum. Mutat. 2015, 36, 774–786. [Google Scholar] [CrossRef]
- Geisler, S.; Holmström, K.M.; Treis, A.; Skujat, D.; Weber, S.S.; Fiesel, F.C.; Kahle, P.J.; Springer, W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 2010, 6, 871–878. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- Jacoupy, M.; Hamon-Keromen, E.; Ordureau, A.; Erpapazoglou, Z.; Coge, F.; Corvol, J.-C.; Nosjean, O.; La Cour, C.M.; Millan, M.J.; Boutin, J.A.; et al. The PINK1 kinase-driven ubiquitin ligase Parkin promotes mitochondrial protein import through the presequence pathway in living cells. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Wu, X.; Li, L.; Jiang, H. Mitochondrial inner-membrane protease Yme1 degrades outer-membrane proteins Tom22 and Om45. J. Cell Biol. 2017, 217, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Mårtensson, C.U.; Priesnitz, C.; Song, J.; Ellenrieder, L.; Doan, K.N.; Boos, F.; Floerchinger, A.; Zufall, N.; Oeljeklaus, S.; Warscheid, B.; et al. Mitochondrial protein translocation-associated degradation. Nat. Cell Biol. 2019, 569, 679–683. [Google Scholar] [CrossRef]
- Schmidt, O.; Harbauer, A.B.; Rao, S.; Eyrich, B.; Zahedi, R.P.; Stojanovski, D.; Schönfisch, B.; Guiard, B.; Sickmann, A.; Pfanner, N.; et al. Regulation of Mitochondrial Protein Import by Cytosolic Kinases. Cell 2011, 144, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Kravic, B.; Harbauer, A.B.; Romanello, V.; Simeone, L.; Vögtle, F.-N.; Kaiser, T.; Straubinger, M.; Huraskin, D.; Böttcher, M.; Cerqua, C.; et al. In mammalian skeletal muscle, phosphorylation of TOMM22 by protein kinase CSNK2/CK2 controls mitophagy. Autophagy 2018, 14, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Horie, C.; Suzuki, H.; Sakaguchi, M.; Mihara, K. Targeting and Assembly of Mitochondrial Tail-anchored Protein Tom5 to the TOM Complex Depend on a Signal Distinct from That of Tail-anchored Proteins Dispersed in the Membrane. J. Biol. Chem. 2003, 278, 41462–41471. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brandner, K.; Mick, D.U.; Frazier, A.E.; Taylor, R.D.; Meisinger, C.; Rehling, P. Taz1, an Outer Mitochondrial Membrane Protein, Affects Stability and Assembly of Inner Membrane Protein Complexes: Implications for Barth Syndrome. Mol. Biol. Cell 2005, 16, 5202–5214. [Google Scholar] [CrossRef]
- Becker, T.; Guiard, B.; Thornton, N.; Zufall, N.; Stroud, D.A.; Wiedemann, N.; Pfanner, N. Assembly of the mitochondrial protein import channel: Role of Tom5 in two-stage interaction of Tom40 with the SAM complex. Mol. Biol. Cell. 2010, 21, 3106–3113. [Google Scholar] [CrossRef] [PubMed]
- Umemoto, T.; Asai, T.; Hirashima, K.; Shimizu, K.; Mihara, M.; Yamamoto, K.; Kubota, K.; Miyagawa, S.-I.; Oku, N. Proapoptotic Action of p53-Tom5 in p53-Resistant A549 Human Non-small Cell Lung Cancer Cells through Direct Mitochondrial Dysfunction. Biol. Pharm. Bull. 2011, 34, 551–554. [Google Scholar] [CrossRef][Green Version]
- Dukanovic, J.; Dimmer, K.S.; Bonnefoy, N.; Krumpe, K.; Rapaport, D. Genetic and Functional Interactions between the Mitochondrial Outer Membrane Proteins Tom6 and Sam37. Mol. Cell. Biol. 2009, 29, 5975–5988. [Google Scholar] [CrossRef]
- Turcu, A.L.; Versini, A.; Khene, N.; Gaillet, C.; Cañeque, T.; Müller, S.; Rodriguez, R. DMT1 Inhibitors Kill Cancer Stem Cells by Blocking Lysosomal Iron Translocation. Chem. A Eur. J. 2020, 26, 7369–7373. [Google Scholar] [CrossRef]
- Wolff, N.A.; Ghio, A.J.; Garrick, L.M.; Garrick, M.D.; Zhao, L.A.; Fenton, R.; Thévenod, F. Evidence for mitochondrial localization of divalent metal transporter 1 (DMT1). FASEB J. 2014, 28, 2134–2145. [Google Scholar] [CrossRef]
- Johnston, A.J.; Hoogenraad, J.; Dougan, D.A.; Truscott, K.N.; Yano, M.; Mori, M.; Hoogenraad, N.J.; Ryan, M.T. Insertion and Assembly of Human Tom7 into the Preprotein Translocase Complex of the Outer Mitochondrial Membrane. J. Biol. Chem. 2002, 277, 42197–42204. [Google Scholar] [CrossRef]
- Sekine, S.; Wang, C.; Sideris, D.P.; Bunker, E.; Zhang, Z.; Youle, R.J. Reciprocal Roles of Tom7 and OMA1 during Mitochondrial Import and Activation of PINK1. Mol. Cell 2019, 73, 1028–1043.e5. [Google Scholar] [CrossRef]
- Okatsu, K.; Uno, M.; Koyano, F.; Go, E.; Kimura, M.; Oka, T.; Tanaka, K.; Matsuda, N. A Dimeric PINK1-containing Complex on Depolarized Mitochondria Stimulates Parkin Recruitment. J. Biol. Chem. 2013, 288, 36372–36384. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, R.; Liu, Z.; Bunker, E.; Lee, S.; Giuntini, M.; Chapnick, D.; Liu, X. The plant triterpenoid celastrol blocks PINK1-dependent mitophagy by disrupting PINK1’s association with the mitochondrial protein TOM20. J. Biol. Chem. 2019, 294, 7472–7487. [Google Scholar] [CrossRef]
- Bertolin, G.; Ferrando-Miguel, R.; Jacoupy, M.; Traver, S.; Grenier, K.; Greene, A.W.; Dauphin, A.; Waharte, F.; Bayot, A.; Salamero, J.; et al. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy 2013, 9, 1801–1817. [Google Scholar] [CrossRef]
- Kato, H.; Lu, Q.; Rapaport, D.; Kozjak-Pavlovic, V. Tom70 Is Essential for PINK1 Import into Mitochondria. PLoS ONE 2013, 8, e58435. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.C.; Morimoto, R.I. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996, 15, 2969–2979. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Raman, M.; Guarani-Pereira, V.; Sowa, M.E.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nat. Cell Biol. 2013, 496, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Kotrasová, V.; Keresztesová, B.; Ondrovičová, G.; Bauer, J.; Havalová, H.; Pevala, V.; Kutejová, E.; Kunová, N. Mitochondrial Kinases and the Role of Mitochondrial Protein Phosphorylation in Health and Disease. Life 2021, 11, 82. [Google Scholar] [CrossRef]
- Zhang, M.; Yu, Q.; Liu, Z.; Liang, C.; Zhang, B.; Li, M. UBX domain-containing proteins are involved in lipid homeostasis and stress responses in Pichia pastoris. Int. J. Biochem. Cell Biol. 2017, 90, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.O.; Dengjel, J.; Wilfling, F.; Kozjak-Pavlovic, V.; Häcker, G.; Weber, A. The Pro-Apoptotic BH3-Only Protein Bim Interacts with Components of the Translocase of the Outer Mitochondrial Membrane (TOM). PLoS ONE 2015, 10, e0123341. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Kohara, M.; Kasama, Y.; Nishimura, T.; Tsukiyama-Kohara, K.; Saito, M.; Kai, C. Translocase of outer mitochondrial membrane 70 expression is induced by hepatitis C virus and is related to the apoptotic response. J. Med. Virol. 2011, 83, 801–809. [Google Scholar] [CrossRef]
- Chou, C.-H.; Lee, R.-S.; Yang-Yen, H.-F. An Internal EELD Domain Facilitates Mitochondrial Targeting of Mcl-1 via a Tom70-dependent Pathway. Mol. Biol. Cell 2006, 17, 3952–3963. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, X.-Y.; Wei, B.; Shi, H.-X.; Shan, Y.-F.; Wang, C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010, 20, 994–1011. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Cui, Y.; Huang, Y.; Liu, H.; Li, L.; Li, M.; Ruan, K.-C.; Zhou, Q.; Wang, C. Tom70 Mediates Sendai Virus-Induced Apoptosis on Mitochondria. J. Virol. 2015, 89, 3804–3818. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; O’Meara, M.J.; Guo, J.Z.; Swaney, D.L.; Tummino, T.A.; Huettenhain, R.; et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. BioRxiv 2020. [Google Scholar] [CrossRef]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef]
- Jiang, H.W.; Zhang, H.-N.; Meg, Q.-F.; Xie, J.; Li, Y.; Chen, H.; Zheng, Y.-X.; Wang, X.-N.; Qi, H.; Zhang, J.; et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020, 17, 998–1000. [Google Scholar] [CrossRef]
- Hira, S.; Packialakshmi, B.; Tang, E.; Zhou, X. Dexamethasone upregulates mitochondrial Tom20, Tom70, and MnSOD through SGK1 in the kidney cells. J. Physiol. Biochem. 2021, 77, 1–11. [Google Scholar] [CrossRef]
- Recovery Collaborative Group; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar]
- Filadi, R.; Leal, N.S.; Schreiner, B.; Rossi, A.; Dentoni, G.; Pinho, C.M.; Wiehager, B.; Cieri, D.; Calì, T.; Pizzo, P.; et al. TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca2+ Transfer. Curr. Biol. 2018, 28, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Bartok, A.; Weaver, D.; Golenár, T.; Nichtova, Z.; Katona, M.; Bánsághi, S.; Alzayady, K.J.; Thomas, V.K.; Ando, H.; Mikoshiba, K.; et al. IP3 receptor isoforms differently regulate ER-mitochondrial contacts and local calcium transfer. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Li, J.; Qi, M.; Li, C.; Shi, D.; Zhang, D.; Xie, D.; Yuan, T.; Feng, J.; Liu, Y.; Liang, D.; et al. Tom70 serves as a molecular switch to determine pathological cardiac hypertrophy. Cell Res. 2014, 24, 977–993. [Google Scholar] [CrossRef] [PubMed]
- Herkenne, S.; Ek, O.; Zamberlan, M.; Pellattiero, A.; Chergova, M.; Chivite, I.; Novotná, E.; Rigoni, G.; Fonseca, T.B.; Samardzic, D.; et al. Developmental and Tumor Angiogenesis Requires the Mitochondria-Shaping Protein Opa1. Cell Metab. 2020, 31, 987–1003.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, D.; Yang, Y.; Hou, J.; Wan, J.; Ran, F.; Dai, X.; Zhou, P.; Yang, Y. Tom70 protects against diabetic cardiomyopathy through its antioxidant and antiapoptotic properties. Hypertens. Res. 2020, 43, 1047–1056. [Google Scholar] [CrossRef]
- Pei, H.; Yang, Y.; Zhao, H.; Li, X.; Yang, D.; Li, D.; Yang, Y. The Role of Mitochondrial Functional Proteins in ROS Production in Ischemic Heart Diseases. Oxidative Med. Cell. Longev. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Xue, Q.; Pei, H.; Liu, Q.; Zhao, M.; Sun, J.; Gao, E.; Ma, X.; Tao, L. MICU1 protects against myocardial ischemia/reperfusion injury and its control by the importer receptor Tom70. Cell Death Dis. 2017, 8, e2923. [Google Scholar] [CrossRef] [PubMed]
- Boengler, K.; Gres, P.; Cabestrero, A.; Ruiz-Meana, M.; García-Dorado, D.; Heusch, G.; Schulz, R. Prevention of the ischemia-induced decrease in mitochondrial Tom20 content by ischemic preconditioning. J. Mol. Cell. Cardiol. 2006, 41, 426–430. [Google Scholar] [CrossRef]
- Di Maio, R.; Barrett, P.J.; Hoffman, E.K.; Barrett, C.W.; Zharikov, A.; Borah, A.; Hu, X.; McCoy, J.; Chu, C.T.; Burton, E.A.; et al. α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci. Transl. Med. 2016, 8, 342ra78. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, B.R.; Rocha, E.M.; Castro, S.L.; Greenamyre, J.T. Protection from α-Synuclein induced dopaminergic neurodegeneration by overexpression of the mitochondrial import receptor TOM20. NPJ Park. Dis. 2020, 6, 1–10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitt, A.S.; Buchanan, S.K. A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease. Cells 2021, 10, 1164. https://doi.org/10.3390/cells10051164
Pitt AS, Buchanan SK. A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease. Cells. 2021; 10(5):1164. https://doi.org/10.3390/cells10051164
Chicago/Turabian StylePitt, Ashley S., and Susan K. Buchanan. 2021. "A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease" Cells 10, no. 5: 1164. https://doi.org/10.3390/cells10051164
APA StylePitt, A. S., & Buchanan, S. K. (2021). A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease. Cells, 10(5), 1164. https://doi.org/10.3390/cells10051164





