Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution
Abstract
1. Introduction
2. Traditional Views on the Sources of Biological Variation
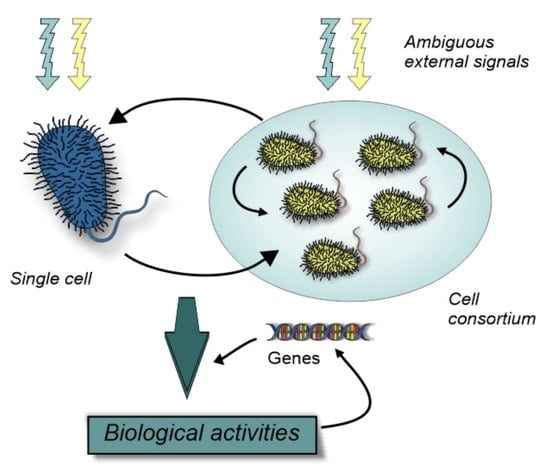
3. Unicellular Insights into Biological Variation
4. Variations in Multicellular Organisms
4.1. General Mechanisms
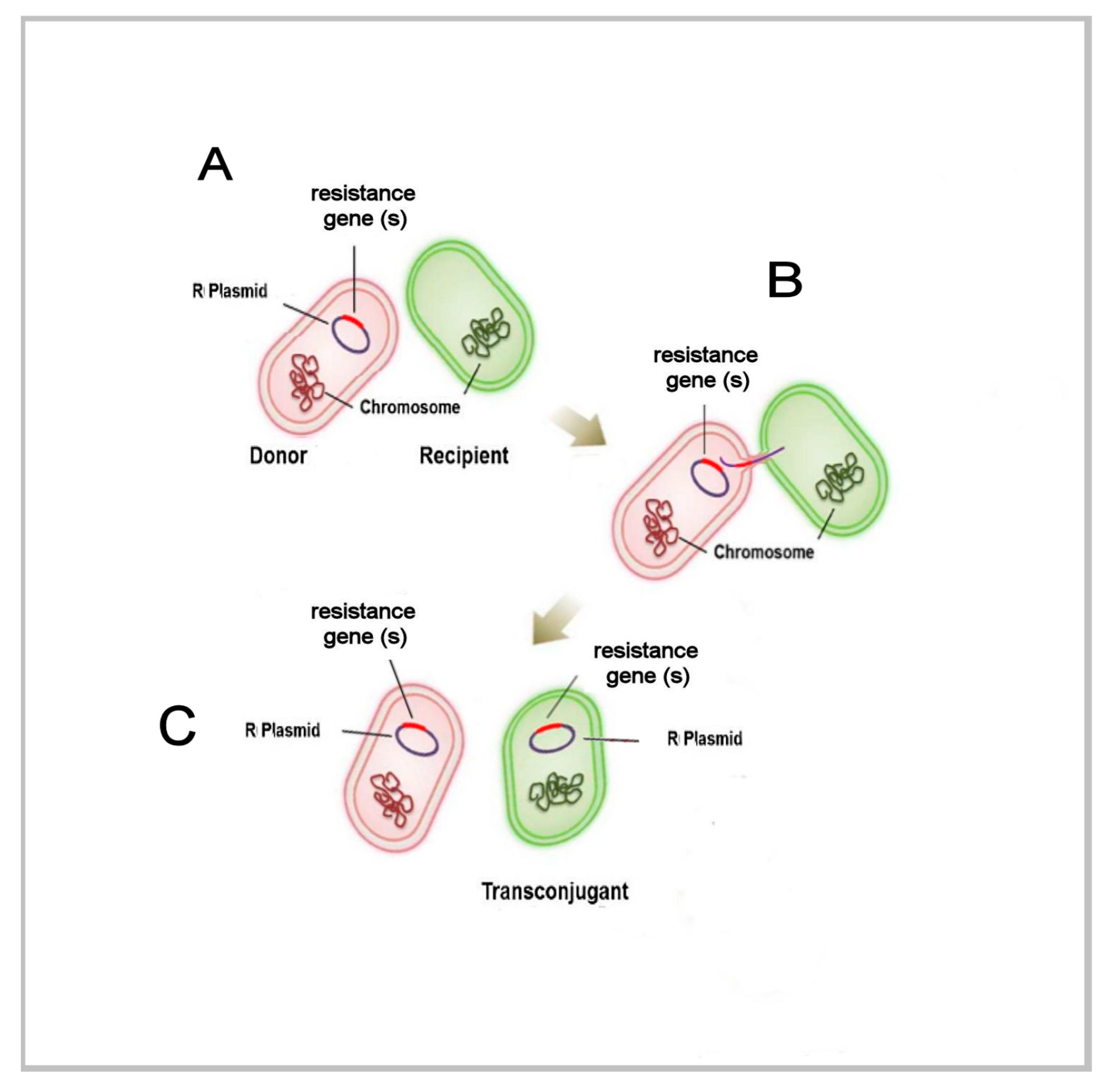
4.2. The Tools of Viral-Cellular Variation
5. The Origins and Extent of Natural Cellular Engineering
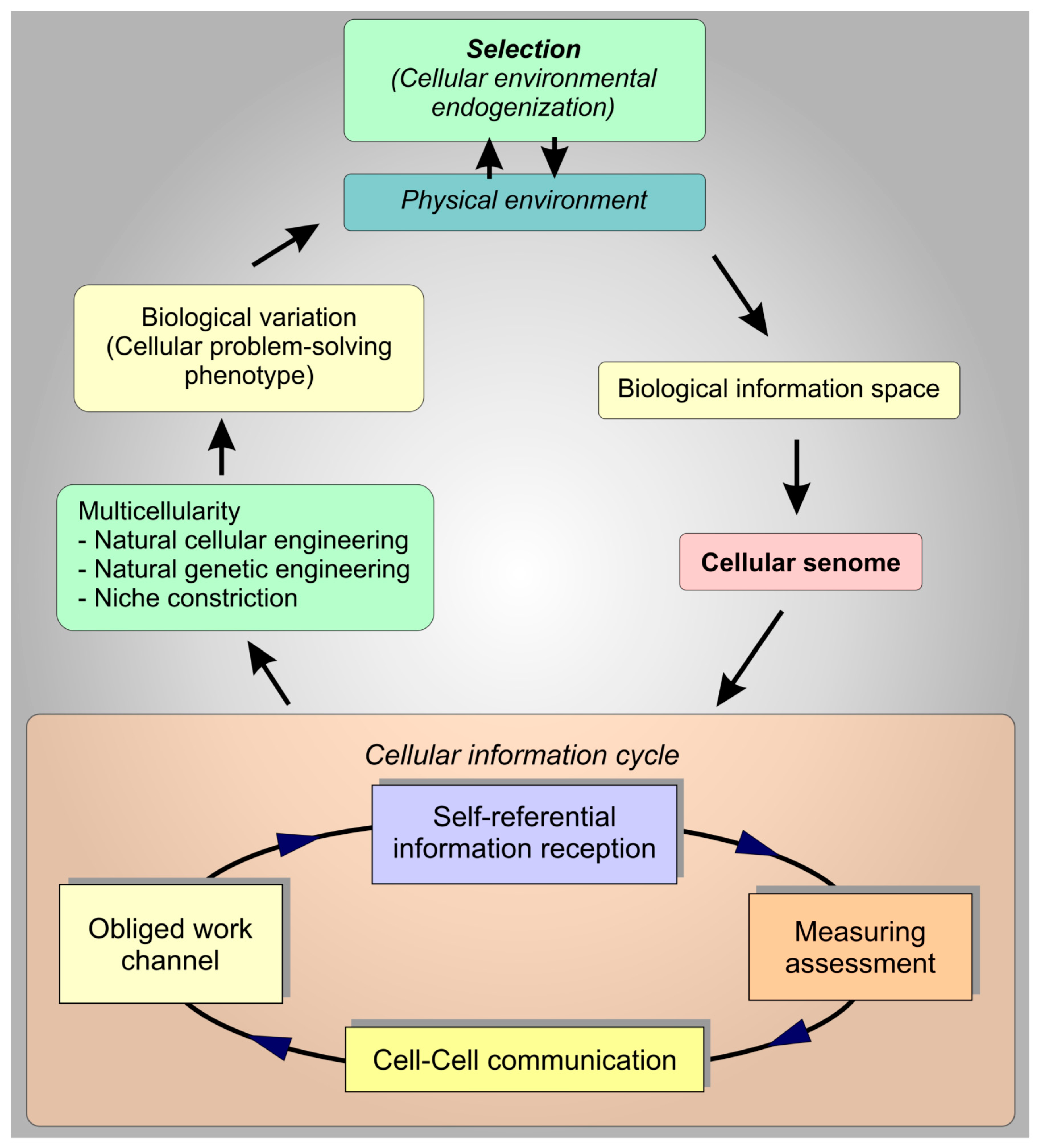
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Futuyma, D.J. Evolutionary Biology Today and the Call for an Extended Synthesis. Interface Focus 2017, 7, 20160145. [Google Scholar] [CrossRef]
- Koonin, E.V. The Logic of Chance: The Nature and Origin of Biological Evolution; Pearson Education: Upper Saddle River, NJ, USA, 2012; ISBN 978-0-13-254249-4. [Google Scholar]
- Miller, W.B. Cognition, Information Fields and Hologenomic Entanglement: Evolution in Light and Shadow. Biology 2016, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.B. Biological Information Systems: Evolution as Cognition-Based Information Management. Prog. Biophys. Mol. Biol. 2018, 134, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.B.; Torday, J.S. A Systematic Approach to Cancer: Evolution beyond Selection. Clin. Transl. Med. 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.B.; Torday, J.S. Four Domains: The Fundamental Unicell and Post-Darwinian Cognition-Based Evolution. Prog. Biophys. Mol. Biol. 2018, 140, 49–73. [Google Scholar] [CrossRef]
- Miller, W.B., Jr.; Torday, J.S.; Baluška, F. Biological Evolution as Defense of “Self”. Prog. Biophys. Mol. Biol. 2019, 142, 54–74. [Google Scholar] [CrossRef]
- Miller, W.; Torday, J.; Baluška, F. The N-Space Episenome Unifies Cellular Information Space-Time within Cognition-Based Evolution. Prog. Biophys. Mol. Biol. 2020, 150. [Google Scholar] [CrossRef]
- Miller, W.B.; Baluška, F.; Torday, J.S. Cellular Senomic Measurements in Cognition-Based Evolution. Prog. Biophys. Mol. Biol. 2020, 156, 20–33. [Google Scholar] [CrossRef]
- Torday, J.; Miller, W., Jr. Cellular-Molecular Mechanisms in Epigenetic Evolutionary Biology; Springer Nature: Cham, Switzerland, 2020. [Google Scholar]
- Leitão, A.L.; Costa, M.C.; Gabriel, A.F.; Enguita, F.J. Interspecies Communication in Holobionts by Non-Coding RNA Exchange. Int. J. Mol. Med. Sci. 2020, 21, 2333. [Google Scholar] [CrossRef]
- Shapiro, J.A. Evolution: A View from the 21st Century; FT Press Science: Saddle River, NJ, USA, 2011; ISBN 978-0-13-278093-3. [Google Scholar]
- Baluška, F.; Levin, M. On Having No Head: Cognition throughout Biological Systems. Front. Psychol. 2016, 7. [Google Scholar] [CrossRef]
- Reber, A.S. The First Minds: Caterpillars, Karyotes, and Consciousness; Oxford University Press: Oxford, UK, 2018; ISBN 978-0-1-085415-7. [Google Scholar]
- Slijepcevic, P. Natural Intelligence and Anthropic Reasoning. Biosemiotics 2020, 13, 285–307. [Google Scholar] [CrossRef]
- Ford, B.J. Are Cells Ingenious? Microscope 2004, 52, 135–144. [Google Scholar]
- Ford, B.J. On Intelligence in Cells: The Case for Whole Cell Biology. Interdiscip. Sci. Rev. 2009, 34, 350–365. [Google Scholar] [CrossRef]
- Ford, B.J. Cellular Intelligence: Microphenomenology and the Realities of Being. Prog. Biophys. Mol. Biol. 2017, 131, 273–287. [Google Scholar] [CrossRef]
- Dodig-Crnkovic, G. Modeling Life as Cognitive Info-Computation. In CiE 2014: Language, Life, Limits; Beckmann, A., Csuhaj-Varjú, E., Meer, K., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 153–162. [Google Scholar]
- Lyon, P. The Cognitive Cell: Bacterial Behavior Reconsidered. Front. Microbiol. 2015, 6, 264. [Google Scholar] [CrossRef]
- Baluška, F.; Reber, A. Sentience and Consciousness in Single Cells: How the First Minds Emerged in Unicellular Species. Bioessays 2019, 41, 1800229. [Google Scholar] [CrossRef]
- De Loof, A. From Darwin’s On the Origin of Species by Means of Natural Selection...to the Evolution of Life with Communication Activity as its Very Essence and Driving Force (=Mega-Evolution). Life Excit. Biol. 2015, 3, 153–187. [Google Scholar] [CrossRef]
- Zakirov, B.; Charalambous, G.; Aspalter, I.M.; van-Vuuren, K.; Mead, T.; Harrington, K.; Thuret, R.; Regan, E.R.; Herbert, S.P.; Bentley, K. Active Perception during Angiogenesis: Filopodia Speed Up Notch Selection of Tip Cells in Silico and in Vivo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing Cognition: Getting Down to Biological Basics. Phil. Trans. R. Soc. B. 2021, 376. [Google Scholar] [CrossRef] [PubMed]
- Shettleworth, S.J. Cognition, Evolution, and Behavior; Oxford University Press: Oxford, UK, 1998; ISBN 978-0-19-988638. [Google Scholar]
- Slijepcevic, P. Principles of Information Processing and Natural Learning in Biological Systems. J. Gen. Philos. Sci. 2019. [Google Scholar] [CrossRef]
- Pigliucci, M. Do We Need an Extended Evolutionary Synthesis? Evolution 2007, 61, 2743–2749. [Google Scholar] [CrossRef] [PubMed]
- Laland, K.; Uller, T.; Feldman, M.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; Odling Smee, J.; Wray, G.A.; Hoekstra, H.E.; et al. Does Evolutionary Theory Need a Rethink? Nature 2014, 514, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D.; Barton, N.H.; Charlesworth, B. The Sources of Adaptive Variation. Proc. R. Soc. B. 2017, 284. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.F. Too Much Eukaryote LGT. Bioessays 2017, 39. [Google Scholar] [CrossRef]
- Gould, S.J. The Return of Hopeful Monsters. Nat. Hist. 1977, 86, 23–30. Available online: http://www.somosbacteriasyvirus.com/monsters.pdf (accessed on 21 February 2021).
- Gould, S.J.; Eldredge, N. Punctuated Equilibria: The Tempo and Mode of Evolution Reconsidered. Paleobiology 1977, 3, 115–151. Available online: https://www.jstor.org/stable/2400177 (accessed on 14 January 2021). [CrossRef]
- Laland, K.N.; Uller, T.; Feldman, M.W.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; Odling-Smee, J. The Extended Evolutionary Synthesis: Its Structure, Assumptions and Predictions. Proc. R. Soc. B. 2015, 282. [Google Scholar] [CrossRef]
- Moczek, A.P. An Evolutionary Biology for the 21st Century. In Perspectives on Evolutionary and Developmental Biology: Essays for Alessandro Minelli; Giuseppe, F., Ed.; Padova University Press: Padova, Italy, 2019; pp. 23–27. ISBN 978-88-6938-140-9. [Google Scholar]
- Trerotola, M.; Relli, V.; Simeone, P.; Alberti, S. Epigenetic Inheritance and the Missing Heritability. Hum. Genom. 2015, 9, 17. [Google Scholar] [CrossRef]
- Asfahl, K.L.; Schuster, M. Social Interactions in Bacterial Cell-Cell Signaling. FEMS Microbiol. Rev. 2017, 41, 92–107. [Google Scholar] [CrossRef]
- Brown-Jaque, M.; Calero-Cáceres, W.; Muniesa, M. Transfer of Antibiotic-Resistance Genes via Phage-Related Mobile Elements. Plasmid 2015, 79, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Balcazar, J.L. Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef]
- Popat, R.; Cornforth, D.M.; McNally, L.; Brown, S.P. Collective Sensing and Collective Responses in Quorum-Sensing Bacteria. J. R. Soc. Interface 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, M.J.; Bassler, B.L. SnapShot: Bacterial quorum sensing. Cell 2018, 174, 1328–1328e1. [Google Scholar] [CrossRef]
- Goldenfeld, N.; Woese, C. Life is Physics: Evolution as a Collective Phenomenon Far from Equilibrium. Annu. Rev. Condens. Matter Phys. 2011, 2, 375–399. [Google Scholar] [CrossRef]
- Zatyka, M.; Thomas, C.M. Control of Genes for Conjugative Transfer of Plasmids and Other Mobile Elements. FEMS Microbiol. Rev. 1998, 21, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Pathways of DNA Transfer to Plants from Agrobacterium Tumefaciens and Related Bacterial Species. Annu. Rev. Phytopathol. 2019, 57, 231–251. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Otten, L. Widespread Occurrence of Natural Genetic Transformation of Plants by Agrobacterium. Plant Mol. Biol. 2019, 101, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.B.; Mulley, G.; Dills, A.H.; Alsohim, A.S.; McGuffin, L.J.; Studholme, D.J.; Silby, M.W.; Brockhurst, M.A.; Johnson, L.J.; Jackson, R.W. Evolutionary Resurrection of Flagellar Motility via Rewiring of the Nitrogen Regulation System. Science 2015, 347, 1014–1017. [Google Scholar] [CrossRef]
- Ponder, R.G.; Fonville, N.C.; Rosenberg, S.M. A Switch from High-Fidelity to Error-Prone DNA Double-Strand Break Repair Underlies Stress-Induced Mutation. Mol. Cell 2005, 19, 791–804. [Google Scholar] [CrossRef]
- Shee, C.; Gibson, J.L.; Darrow, M.C.; Gonzalez, C.; Rosenberg, S.M. Impact of a Stress-Inducible Switch to Mutagenic Repair of DNA Breaks on Mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 13659–13664. [Google Scholar] [CrossRef] [PubMed]
- Wlodek, A.; Kendrew, S.G.; Coates, N.J.; Hold, A.; Pogwizd, J.; Rudder, S.; Sheehan, L.S.; Higginbotham, S.J.; Stanley-Smith, A.E.; Warneck, T.; et al. Diversity Oriented Biosynthesis via Accelerated Evolution of Modular Gene Clusters. Nat. Commun. 2017, 8, 1206. [Google Scholar] [CrossRef]
- Peng, H.; Ishida, K.; Sugimoto, Y.; Jenke-Kodama, H.; Hertweck, C. Emulating Evolutionary Processes to Morph Aureothin-Type Modular Polyketide Synthases and Associated Oxygenases. Nat. Commun. 2019, 10, 3918. [Google Scholar] [CrossRef] [PubMed]
- Van Gestel, J.; Vlamakis, H.; Kolter, R. From Cell Differentiation to Cell Collectives: Bacillus Subtilis Uses Division of Labor to Migrate. PLoS Biol. 2015, 13, e1002141. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jacob, E.; Levine, H. Self-Engineering Capabilities of Bacteria. J. R. Soc. Interface 2006, 3, 197–214. [Google Scholar] [CrossRef]
- Ben-Jacob, E. Social Behavior of Bacteria: From Physics to Complex Organization. Eur. Phys. J. B 2008, 65, 315–322. [Google Scholar] [CrossRef]
- Ben-Jacob, E. Learning from Bacteria about Natural Information Processing. Ann. N. Y. Acad. Sci. 2009, 1178, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Baluška, F.; Miller, W.B., Jr. Senomic View of the Cell: Senome versus Genome. Commun. Integr. Biol. 2018, 11, 1–9. [Google Scholar] [CrossRef]
- Villarreal, L.P.; Ryan, F. Viruses in the Origin of Life and its Subsequent Diversification. In Handbook of Astrobiology; Kolb, V.M., Ed.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar] [CrossRef]
- Broecker, F.; Moelling, K. Evolution of Immune Systems from Viruses and Transposable Elements. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration Site Selection by Retroviruses and Transposable Elements in Eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, R.N. Functional Dualism of Transposon Transcripts in Evolution of Eukaryotic Genomes. Russ. J. Dev. Biol. 2018, 49, 339–355. [Google Scholar] [CrossRef]
- Caporale, L.H.; Doyle, J. In Darwinian Evolution, Feedback from Natural Selection Leads to Biased Mutations. Ann. N. Y. Acad. Sci. 2013, 1305, 18–28. [Google Scholar] [CrossRef]
- Gerhart, J.; Kirschner, M. The Theory of Facilitated Variation. Pnas 2007, 104, 8582–8589. [Google Scholar] [CrossRef]
- Shapiro, J.A. How Life Changes Itself: The Read-Write (RW) Genome. Phys. Life Rev. 2013, 10, 287–323. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. Living Organisms Author Their Read-Write Genomes in Evolution. Biology 2017, 6, 42. [Google Scholar] [CrossRef]
- Witzany, G. Natural Genome Editing from a Biocommunicative Perspective. Biosemiotics 2011, 4, 349–368. [Google Scholar] [CrossRef]
- De Loof, A. The Evolution of “Life”: A Metadarwinian Integrative Approach. Commun. Integr. Biol. 2017, 10, e1301335. [Google Scholar] [CrossRef] [PubMed]
- Lupien, L.E.; Bloch, K.; Dehairs, J.; Feng, W.W.; Davis, W.L.; Dennis, T.; Swinnen, J.V.; Wells, W.A.; Smits, N.C.; Kuemmerle, N.B.; et al. Endocytosis of Very Low-Density Lipoprotein Particles: An Unexpected Mechanism for Lipid Acquisition by Breast Cancer Cells. bioRxiv 2019. [Google Scholar] [CrossRef]
- Sobhy, H. A Comparative Review of Viral Entry and Attachment during Large and Giant DsDNA Virus Infections. Arch. Virol. 2017, 162, 3567–3585. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.F.; Garg, S.; Zimorski, V. Endosymbiotic Theories for Eukaryote Origin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140330. [Google Scholar] [CrossRef]
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, 911–921. [Google Scholar] [CrossRef]
- Nowack, E.C.M.; Price, D.C.; Bhattacharya, D.; Singer, A.; Melkonian, M.; Grossman, A.R. Gene Transfers from Diverse Bacteria Compensate for Reductive Genome Evolution in the Chromatophore of Paulinella chromatophora. Proc. Natl. Acad. Sci. USA 2016, 113, 12214–12219. [Google Scholar] [CrossRef]
- Overholtzer, M.; Mailleux, A.A.; Mouneimne, G.; Normand, G.; Schnitt, S.J.; King, R.W.; Cibas, E.S.; Brugge, J.S. A Nonapoptotic Cell Death Process, Entosis, That Occurs by Cell-in-Cell Invasion. Cell 2007, 131, 966–979. [Google Scholar] [CrossRef]
- Janssen, A.; Medema, R.H. Entosis: Aneuploidy by Invasion. Nat. Cell. Biol. 2011, 13, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Vogel, D.; Dussutour, A. Direct Transfer of Learned Behaviour via Cell Fusion in Non-Neural Organisms. Proc. R. Soc. B 2016, 283. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Yushenova, I.A. Giant Transposons in Eukaryotes: Is Bigger Better? Genome Biol. Evol. 2019, 11, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Cordaux, R. Viruses as Vectors of Horizontal Transfer of Genetic Material in Eukaryotes. Curr. Opin. Virol. 2017, 25, 16–22. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Gilbert, C. Endogenous Viruses: Insights into Viral Evolution and Impact on Host Biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef]
- Villarreal, L.P. Viruses and the Placenta: The Essential Virus First View. APMIS 2016, 124, 20–30. [Google Scholar] [CrossRef]
- Roberts, R.M.; Ezashi, T.; Schulz, L.C.; Sugimoto, J.; Schust, D.J.; Khan, T.; Zhou, J. Syncytins Expressed in Human Placental Trophoblast. Placenta 2021. [Google Scholar] [CrossRef]
- Koonin, E.V. Darwinian Evolution in the Light of Genomics. Nucleic Acids Res. 2009, 37, 1011–1034. [Google Scholar] [CrossRef]
- Ryan, F. Virolution; Harper-Collins: New York, NY, USA, 2009; ISBN 978-0-00-731512-3. [Google Scholar]
- Black, S.G.; Arnaud, F.; Palmarini, M.; Spencer, T.E. Endogenous Retroviruses in Trophoblast Differentiation and Placental Development. Am. J. Reprod. Immunol. 2010, 64, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Enard, D.; Cai, L.; Gwennap, C.; Petrov, D.A. Viruses Are a Dominant Driver of Protein Adaptation in Mammals. Elife 2016, 5, e12469. [Google Scholar] [CrossRef] [PubMed]
- Popov, M.; Kolotova, T.; Davidenko, M. Endogenous Retroviruses as Genetic Modules that Shape the Genome Regulatory Networks during Evolution. J. VN Karazin Kharkiv Natl. Univ. Ser. Med. 2019, 36, 80–95. [Google Scholar]
- Martinez, G.; Castellano, M.; Tortosa, M.; Pallas, V.; Gomez, G. A Pathogenic Non-Coding RNA Induces Changes in Dynamic DNA Methylation of Ribosomal RNA Genes in Host Plants. Nucleic Acids Res. 2013, 42, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional Activation of Retrotransposons Alters the Expression of Adjacent Genes in Wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B. Retroviruses Facilitate the Rapid Evolution of the Mammalian Placenta. Bioessays 2013, 35, 853–861. [Google Scholar] [CrossRef]
- Moon, C.; Baldridge, M.T.; Wallace, M.A.; Burnham, C.-A.; Virgin, H.W.; Stappenbeck, T.S. Vertically Transmitted Faecal IgA Levels Determine Extra-Chromosomal Phenotypic Variation. Nature 2015, 521, 90–93. [Google Scholar] [CrossRef]
- Wilke, C.O.; Sawyer, S.L. At the Mercy of Viruses. eLife 2016, 5, e16758. [Google Scholar] [CrossRef]
- Wallau, G.L.; Vieira, C.; Loreto, É.L.S. Genetic Exchange in Eukaryotes through Horizontal Transfer: Connected by the Mobilome. Mob. DNA 2018, 9, 6. [Google Scholar] [CrossRef]
- Wei, B.; Liu, H.; Liu, X.; Xiao, Q.; Wang, Y.; Zhang, J.; Hu, Y.; Liu, Y.; Yu, G.; Huang, Y. Genome-Wide Characterization of Non-Reference Transposons in Crops Suggests Non-Random Insertion. BMC Genom. 2016, 17, 536. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten Things You Should Know about Transposable Elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Witzany, G. Noncoding RNAs: Persistent Viral Agents as Modular Tools for Cellular Needs. Ann. N. Y. Acad. Sci. 2009, 1178, 244–267. [Google Scholar] [CrossRef]
- Brandt, J.; Schrauth, S.; Veith, A.-M.; Froschauer, A.; Haneke, T.; Schultheis, C.; Gessler, M.; Leimeister, C.; Volff, J.-N. Transposable Elements as a Source of Genetic Innovation: Expression and Evolution of a Family of Retrotransposon-Derived Neogenes in Mammals. Gene 2005, 345, 101–111. [Google Scholar] [CrossRef]
- Schaack, S.; Gilbert, C.; Feschotte, C. Promiscuous DNA: Horizontal Transfer of Transposable Elements and Why It Matters for Eukaryotic Evolution. Trends Ecol. Evol. 2010, 25, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Kukita, C.; Zhang, Z. Transposon-Mediated Directed Mutation in Bacteria and Eukaryotes. Front. Biosci. 2017, 22, 1458–1468. [Google Scholar] [CrossRef]
- Schrader, L.; Schmitz, J. The Impact of Transposable Elements in Adaptive Evolution. Mol. Ecol. 2019, 28, 1537–1549. [Google Scholar] [CrossRef]
- Vicient, C.M.; Casacuberta, J.M. Impact of Transposable Elements on Polyploid Plant Genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Rodriguez, F.; Arkhipova, I.R. Transposable Elements and Polyploid Evolution in Animals. Curr. Opin. Genet. Dev. 2018, 49, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Transposable Elements Contribute to Fungal Genes and Impact Fungal Lifestyle. Sci. Rep. 2019, 9, 4307. [Google Scholar] [CrossRef]
- Jangam, D.; Feschotte, C.; Betrán, E. Transposable Element Domestication as an Adaptation to Evolutionary Conflicts. Trends Genet. 2017, 33, 817–831. [Google Scholar] [CrossRef]
- Belyayev, A. Bursts of Transposable Elements as an Evolutionary Driving Force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R. Neutral Theory, Transposable Elements, and Eukaryotic Genome Evolution. Mol. Biol. Evol. 2018, 35, 1332–1337. [Google Scholar] [CrossRef]
- Oliver, K.R.; Greene, W.K. Mobile DNA and the TE-Thrust Hypothesis: Supporting Evidence from the Primates. Mob. DNA 2011, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Stapley, J.; Santure, A.W.; Dennis, S.R. Transposable Elements as Agents of Rapid Adaptation May Explain the Genetic Paradox of Invasive Species. Mol. Ecol. 2015, 24, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Fadloun, A.; Le Gras, S.; Jost, B.; Ziegler-Birling, C.; Takahashi, H.; Gorab, E.; Carninci, P.; Torres-Padilla, M.-E. Chromatin Signatures and Retrotransposon Profiling in Mouse Embryos Reveal Regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. Nat. 2013, 20, 332–338. [Google Scholar] [CrossRef]
- Suarez, N.A.; Macia, A.; Muotri, A.R. LINE-1 Retrotransposons in Healthy and Diseased Human Brain. Dev. Neurobiol. 2018, 78, 434–455. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, R.N. Hypothesis on the Origin of Viruses from Transposons. Mol. Genet. Microbiol. Virol. 2018, 33, 223–232. [Google Scholar] [CrossRef]
- Sundaram, V.; Wysocka, J. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos. Trans. R. Soc. Lond. B 2020, 30, 375. [Google Scholar] [CrossRef]
- Moschetti, R.; Palazzo, A.; Lorusso, P.; Viggiano, L.; Massimiliano Marsano, R. “What You Need, Baby, I Got It”: Transposable Elements as Suppliers of Cis-Operating Sequences in Drosophila. Biology 2020, 9, 25. [Google Scholar] [CrossRef]
- Han, J.; Masonbrink, R.E.; Shan, W.; Song, F.; Zhang, J.; Yu, W.; Wang, K.; Wu, Y.; Tang, H.; Wendel, J.F.; et al. Rapid Proliferation and Nucleolar Organizer Targeting Centromeric Retrotransposons in Cotton. Plant J. 2016, 88, 992–1005. [Google Scholar] [CrossRef]
- Villarreal, L.; Witzany, G. That is Life: Communicating RNA Networks from Viruses and Cells in Continuous Interaction. Ann. N. Y. Acad. Sci. 2019, 1447, 5–20. [Google Scholar] [CrossRef]
- Smalheiser, N.R. The RNA-Centred View of the Synapse: Non-Coding RNAs and Synaptic Plasticity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef]
- Palmer, W.H.; Hadfield, J.D.; Obbard, D.J. RNA-Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates. Genetics 2018, 208, 1585–1599. [Google Scholar] [CrossRef]
- Villarreal, L.P.; Witzany, G. Editorial: Genome Invading RNA Networks. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Shapiro, J.A. Nothing in Evolution Makes Sense Except in the Light of Genomics: Read–Write Genome Evolution as an Active Biological Process. Biology 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Zamai, L. Unveiling Human Non-Random Genome Editing Mechanisms Activated in Response to Chronic Environmental Changes: I. Where Might These Mechanisms Come from and What Might They Have Led To? Cells 2020, 9, 2362. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, T.; Shibata, Y.; Kumar, P.; Dillon, L.; Dutta, A. Extrachromosomal Circular DNA, MicroDNA, without Canonical Promoters Produce Short Regulatory RNAs that Suppress Gene Expression. bioRxiv 2019. [Google Scholar] [CrossRef]
- Lanciano, S.; Carpentier, M.C.; Llauro, C.; Jobet, E.; Robakowska-Hyzorek, D.; Lasserre, E.; Ghesquière, A.; Panaud, O.; Mirouze, M. Sequencing the Extrachromosomal Circular Mobilome Reveals Retrotransposon Activity in Plants. PLoS Genet. 2017, 13, e1006630. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of Form and Function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-Coding RNA Regulatory Networks. Biochim. Biophys. Acta Gene. Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs Function as CeRNAs to Regulate and Control Human Cancer Progression. Mole. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Ecco, G.; Imbeault, M.; Trono, D. KRAB Zinc Finger Proteins. Development 2017, 144, 2719–2729. [Google Scholar] [CrossRef]
- Lupo, A.; Cesaro, E.; Montano, G.; Zurlo, D.; Izzo, P.; Costanzo, P. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr. Genom. 2013, 14, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Petrie, K.L.; Palmer, N.D.; Johnson, D.T.; Medina, S.J.; Yan, S.J.; Li, V.; Burmeister, A.R.; Meyer, J.R. Destabilizing Mutations Encode Nongenetic Variation that Drives Evolutionary Innovation. Science 2018, 359, 1542–1545. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Jung, I.; Selvaraj, S.; Shen, Y.; Antosiewicz-Bourget, J.E.; Lee, A.Y.; Ye, Z.; Kim, A.; Rajagopal, N.; Xie, W.; et al. Chromatin Architecture Reorganization during Stem Cell Differentiation. Nature 2015, 518, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Waszak, S.M.; Delaneau, O.; Gschwind, A.R.; Kilpinen, H.; Raghav, S.K.; Witwicki, R.M.; Orioli, A.; Wiederkehr, M.; Panousis, N.I.; Yurovsky, A.; et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell 2015, 162, 1039–1050. [Google Scholar] [CrossRef]
- Wellenreuther, M.; Mérot, C.; Berdan, E.; Bernatchez, L. Going beyond SNPs: The Role of Structural Genomic Variants in Adaptive Evolution and Species Diversification. Mol. Ecol. 2019, 28, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Shorter, J.; Lindquist, S. Prions as Adaptive Conduits of Memory and Inheritance. Nat. Rev. Genet. 2005, 6, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Jarosz, D.F.; Jones, S.K.; Chang, A.; Lancaster, A.K.; Lindquist, S. Prions Are a Common Mechanism for Phenotypic Inheritance in Wild Yeasts. Nature 2012, 482, 363–368. [Google Scholar] [CrossRef]
- Miller, W.B., Jr. The Eukaryotic Microbiome: Origins and Implications for Fetal and Neonatal Life. Front. Pediatr. 2016, 4, 96. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Hoffmann, A.R.; Proctor, L.M.; Surette, M.G.; Suchodolski, J.S. The Microbiome: The Trillions of Microorganisms that Maintain Health and Cause Disease in Humans and Companion Animals. Vet. Pathol. 2016, 53, 10–21. [Google Scholar] [CrossRef]
- Gilbert, S.F.; Bosch, T.C.G.; Ledón-Rettig, C. Eco-Evo-Devo: Developmental Symbiosis and Developmental Plasticity as Evolutionary Agents. Nat. Rev. Genet. 2015, 16, 611–622. [Google Scholar] [CrossRef]
- Malik, S.S.; Azem-e-Zahra, S.; Kim, K.M.; Caetano-Anollés, G.; Nasir, A. Do Viruses Exchange Genes across Superkingdoms of Life? Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Jobson, M.A.; Jordan, J.M.; Sandrof, M.A.; Hibshman, J.D.; Lennox, A.L.; Baugh, L.R. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis Elegans. Genetics 2015, 201, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Rupert, J.L. Hypoxia and Environmental Epigenetics. High Alt. Med. Biol. 2014, 15, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, R.; Buchner, D.A. Transgenerational Inheritance of Metabolic Disease. Semin. Cell Dev. Biol. 2015, 43, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Environmental Stress and Epigenetic Transgenerational Inheritance. BMC Med. 2014, 12, 153. [Google Scholar] [CrossRef]
- Milstein, J.N.; Meiners, J.-C. On the Role of DNA Biomechanics in the Regulation of Gene Expression. J. R. Soc. Interface 2011, 8, 1673–1681. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. Exploring the Read-Write Genome: Mobile DNA and Mammalian Adaptation. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable Elements, Epigenetics, and Genome Evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.; Greene, W. Transposable Elements: Powerful Facilitators of Evolution. Bioessays 2009, 31, 703–714. [Google Scholar] [CrossRef]
- Schrader, L.; Kim, J.W.; Ence, D.; Zimin, A.; Klein, A.; Wyschetzki, K.; Weichselgartner, T.; Kemena, C.; Stökl, J.; Schultner, E.; et al. Transposable Element Islands Facilitate Adaptation to Novel Environments in an Invasive Species. Nat. Commun. 2014, 5, 5495. [Google Scholar] [CrossRef]
- Mita, P.; Boeke, J.D. How Retrotransposons Shape Genome Regulation. Curr. Opin. Genet. Dev. 2016, 37, 90–100. [Google Scholar] [CrossRef]
- Torday, J.S.; Miller, W.B., Jr. The Resolution of Ambiguity as the Basis for Life: A Cellular Bridge Between Western Reductionism and Eastern Holism. Prog. Biophys. Mol. Biol. 2017, 131, 288–297. [Google Scholar] [CrossRef]
- Baverstock, K. The Role of Information in Cell Regulation. Prog. Biophys. Mol. Biol. 2013, 111, 141–143. [Google Scholar] [CrossRef]
- Torday, J.S.; Rehan, V.K. Lung Evolution as a Cipher for Physiology. Physiol. Genom. 2009, 38, 1–6. [Google Scholar] [CrossRef]
- Cárdenas-García, J.F. The Process of Info-Autopoiesis—The Source of All Information. Biosemiotics 2020, 13, 199–221. [Google Scholar] [CrossRef]
- Torday, J.S.; Miller, W.B. Phenotype as Agent for Epigenetic Inheritance. Biology 2016, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.I.; Kim, H.; Davies, P.C.W. The Informational Architecture of the Cell. Phil. Trans. R. Soc. A 2016, 374, 20150057. [Google Scholar] [CrossRef]
- Martinez, J.; Klasson, L.; Welch, J.J.; Jiggins, F.M. Life and Death of Selfish Genes: Comparative Genomics Reveals the Dynamic Evolution of Cytoplasmic Incompatibility. Mol. Biol. Evol. 2021, 38, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Chain, E.B. Social Responsibility and the Scientist in Modern Western Society. Perspect. Biol. Med. 1971, 14, 347–369. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Central Dogma or Central Debate? Physiology 2018, 33, 246–249. [Google Scholar] [CrossRef]
- Turner, P.; Nottale, L.; Zhao, J.; Pesquet, E. New Insights into the Physical Processes that Underpin Cell Division and the Emergence of Different Cellular and Multicellular Structures. Prog. Biophys. Mol. Biol. 2020, 150, 13–42. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Evolution Viewed from Physics, Physiology and Medicine. Interface Focus 2017, 7. [Google Scholar] [CrossRef]
- Rudkin, D.M.; Young, G.A. Horseshoe Crabs—An Ancient Ancestry Revealed. In Biology and Conservation of Horseshoe Crabs; Tanacredi, J.T., Botton, M.L., Smith, D., Eds.; Springer US: Boston, MA, USA, 2009; pp. 25–44. ISBN 978-0-387-89959-6. [Google Scholar]
- Stockdale, M.T.; Benton, M.J. Environmental Drivers of Body Size Evolution in Crocodile-Line Archosaurs. Commun. Bio. 2021, 4, 38. [Google Scholar] [CrossRef]
- Witzany, G. Evolution of Genetic Information without Error Replication. In Theoretical Information Studies; World Scientific Series in Information Studies: London, UK, 2018; Volume 11, pp. 295–320. ISBN 978-981-327-748-9. [Google Scholar]
- Kiliç, A.M.; Plasencia, P.; Ishida, K.; Guex, J.; Hirsch, F. Proteromorphosis of Neospathodus (Conodonta) during the Permian–Triassic Crisis and Recovery. Rev. Micropaléontol. 2016, 59, 33–39. [Google Scholar] [CrossRef]
- Guex, J. The Controversial Cope’s, Haeckel’s and Dollo’s Evolutionary Rules: The Role of Evolutionary Retrogradation; Springer International Publishing: Cham, Switzerland, 2020; pp. 13–22. ISBN 978-3-030-47279-5. [Google Scholar] [CrossRef]
- Guex, J. Retrograde Evolution during Major Extinction Crises; Springer Briefs in Evolutionary Biology; Springer International Publishing: Lausanne, Switzerland, 2016; ISBN 978-3-319-27916-9. [Google Scholar] [CrossRef]
- Torday, J.S.; Miller, W.B., Jr. Terminal Addition in a Cellular World. Prog. Biophys. Mol. Biol. 2018, 135. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. No Genome Is an Island: Toward a 21st Century Agenda for Evolution. Ann. N. Y. Acad. Sci. 2019, 1447, 21–52. [Google Scholar] [CrossRef]
- González Plaza, J.J. Small RNAs as Fundamental Players in the Transference of Information during Bacterial Infectious Diseases. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. Force for Ancient and Recent Life: Viral and Stem-Loop RNA Consortia Promote Life. Ann. N. Y. Acad. Sci. 2015, 1341, 25–34. [Google Scholar] [CrossRef]
- Villarreal, L.P. Viral Ancestors of Antiviral Systems. Viruses 2011, 3, 1933–1958. [Google Scholar] [CrossRef] [PubMed]
- Witzany, G. RNA Sociology: Group Behavioral Motifs of RNA Consortia. Life 2014, 4, 800–818. [Google Scholar] [CrossRef]
- Ambrosi, A.; Cattoglio, C.; Serio, C.D. Retroviral Integration Process in the Human Genome: Is It Really Non-Random? A New Statistical Approach. PLoS Comput. Biol. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Rabadan, R.; Levine, A.J.; Krasnitz, M. Non-Random Reassortment in Human Influenza A Viruses. Influenza Other Respir. Viruses 2008, 2, 9–22. [Google Scholar] [CrossRef]
- Schmitz, M.; Driesch, C.; Jansen, L.; Runnebaum, I.B.; Dürst, M. Non-Random Integration of the HPV Genome in Cervical Cancer. PLoS ONE 2012, 7, e39632. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory Activities of Transposable Elements: From Conflicts to Benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Klimenko, O.V. Small non-coding RNAs as regulators of structural evolution and carcinogenesis. Non-Coding RNA Res. 2017, 2, 88–92. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Bazán, E.R. Symbiosis: Viruses as Intimate Partners. Annu. Rev. Virol. 2017, 4, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Ryan, F. Virusphere: Explains the Science Behind the Coronavirus Outbreak; HarperCollins Publishers: New York, NY, USA, 2019; ISBN 978-0-00-829669-8. [Google Scholar]
- Burke, G.R.; Simmonds, T.J.; Sharanowski, B.J.; Geib, S.M. Rapid Viral Symbiogenesis via Changes in Parasitoid Wasp Genome Architecture. Mol. Biol. Evol. 2018, 35, 2463–2474. [Google Scholar] [CrossRef]
- Venner, S.; Feschotte, C.; Biémont, C. Dynamics of Transposable Elements: Towards a Community Ecology of the Genome. Trends Genet. 2009, 25, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Venner, S.; Miele, V.; Terzian, C.; Biémont, C.; Daubin, V.; Feschotte, C.; Pontier, D. Ecological Networks to Unravel the Routes to Horizontal Transposon Transfers. PLoS Biol. 2017, 15, e2001536. [Google Scholar] [CrossRef]
- Auboeuf, D. Physicochemical Foundations of Life That Direct Evolution: Chance and Natural Selection Are Not Evolutionary Driving Forces. Life 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.T.; Loreto, E.L.S. The Catenulida Flatworm Can Express Genes from its Microbiome or from the DNA It Ingests. Sci. Rep. 2019, 9, 19045. [Google Scholar] [CrossRef]
- Annila, A.; Baverstock, K. Genes without Prominence: A Reappraisal of the Foundations of Biology. J. R. Soc. Interface 2014, 11. [Google Scholar] [CrossRef]
- Torday, J.S. The Cell as the First Niche Construction. Biology 2016, 5, 19. [Google Scholar] [CrossRef]
- ElMaghraby, M.F.; Andersen, P.R.; Pühringer, F.; Hohmann, U.; Meixner, K.; Lendl, T.; Tirian, L.; Brennecke, J. A Heterochromatin-Specific RNA Export Pathway Facilitates PiRNA Production. Cell 2019, 178, 964–979.e20. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1: Evidence of Cross-Kingdom Regulation by MicroRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Chen, W.L.; Kung, W.-H.; Huang, H.-D. Plant MiRNAs Found in Human Circulating System Provide Evidences of Cross Kingdom RNAi. BMC Genom. 2017, 18, 112. [Google Scholar] [CrossRef]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.-B.; Wang, B.; Zhang, L.; et al. Grape Exosome-like Nanoparticles Induce Intestinal Stem Cells and Protect Mice from DSS-Induced Colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Torday, J.S.; Miller, W.B. Life is determined by its environment. Int. J. Astrobiol. 2016, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Noble, R.; Noble, D. Harnessing Stochasticity: How Do Organisms Make Choices? Chaos 2018, 28, 106309. [Google Scholar] [CrossRef]
- Noble, R.; Noble, D. Was the Watchmaker Blind? Or Was She One-Eyed? Biology 2017, 6, 47. [Google Scholar] [CrossRef]
- Kitano, H. Biological Robustness. Nat. Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef] [PubMed]
- De Visser, J.A.G.M.; Hermisson, J.; Wagner, G.P.; Ancel Meyers, L.; Bagheri-Chaichian, H.; Blanchard, J.L.; Chao, L.; Cheverud, J.M.; Elena, S.F.; Fontana, W.; et al. Perspective: Evolution and Detection of Genetic Robustness. Evolution 2003, 57, 1959–1972. [Google Scholar] [CrossRef]
- Siegal, M.L.; Bergman, A. Waddington’s Canalization Revisited: Developmental Stability and Evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 10528–10532. [Google Scholar] [CrossRef]
- Loison, L. Canalization and Genetic Assimilation: Reassessing the Radicality of the Waddingtonian Concept of Inheritance of Acquired Characters. Semin. Cell Dev. Biol. 2019, 88, 4–13. [Google Scholar] [CrossRef]
- Flatt, T. The Evolutionary Genetics of Canalization. Q. Rev. Biol. 2005, 80, 287–316. [Google Scholar] [CrossRef] [PubMed]
- Hornstein, E.; Shomron, N. Canalization of Development by MicroRNAs. Nat. Genet. 2006, 38, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Stelling, J.; Sauer, U.; Szallasi, Z.; Doyle, F.J.; Doyle, J. Robustness of Cellular Functions. Cell 2004, 118, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Marshall, P. Evolution 2.0: Breaking the Deadlock between Darwin and Design; BenBella Books: Dallas, TX, USA, 2015; ISBN 978-1-940363-90-5. [Google Scholar]
- Shakhnovich, B.E.; Koonin, E.V. Origins and Impact of Constraints in Evolution of Gene Families. Genome Res. 2006, 16, 1529–1536. [Google Scholar] [CrossRef]
- Futuyma, D.J. Evolutionary Constraint and Ecological Consequences. Evolution 2010, 64, 1865–1884. [Google Scholar] [CrossRef] [PubMed]
- Murren, C.J.; Auld, J.R.; Callahan, H.; Ghalambor, C.K.; Handelsman, C.A.; Heskel, M.A.; Kingsolver, J.G.; Maclean, H.J.; Masel, J.; Maughan, H.; et al. Constraints on the Evolution of Phenotypic Plasticity: Limits and Costs of Phenotype and Plasticity. Heredity 2015, 115, 293–301. [Google Scholar] [CrossRef]
- Babloyantz, I.A.; Kaczmarek, L.K. Self-Organization in Biological Systems with Multiple Cellular Contacts. Bull. Math. Biol. 1979, 41, 193–201. [Google Scholar] [CrossRef]
- Vijver, G.; Salthe, S.N.; Delpos, M. (Eds.) Evolutionary Systems: Biological and Epistemological Perspectives on Selection and Self-Organization; Springer-Science+Business Media: Dordrecht, The Netherlands, 2013; ISBN 978-94-017-1510-2. [Google Scholar] [CrossRef]
- Heylighen, F. Stigmergy as a Universal Coordination Mechanism I: Definition and Components. Cognit. Syst. Res. 2016, 38, 4–13. [Google Scholar] [CrossRef]
- Kauffman, S.A. Investigations; Oxford University Press: New York, NY, USA, 2000; ISBN 978-0-19-972894-7. [Google Scholar]
- Coelho, M.T.P.; Diniz-Filho, J.A.; Rangel, T.F. A Parsimonious View of the Parsimony Principle in Ecology and Evolution. Ecography 2019, 42, 968–976. [Google Scholar] [CrossRef]
- Noble, D. A theory of biological relativity: No privileged level of causation. Interface Focus 2012, 2, 55–64. [Google Scholar] [CrossRef]
- Kassen, R. Experimental Evolution of Innovation and Novelty. Trends Ecol. Evol. 2019, 34, 712–722. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, W.B., Jr.; Enguita, F.J.; Leitão, A.L. Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells 2021, 10, 1125. https://doi.org/10.3390/cells10051125
Miller WB Jr., Enguita FJ, Leitão AL. Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells. 2021; 10(5):1125. https://doi.org/10.3390/cells10051125
Chicago/Turabian StyleMiller, William B., Jr., Francisco J. Enguita, and Ana Lúcia Leitão. 2021. "Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution" Cells 10, no. 5: 1125. https://doi.org/10.3390/cells10051125
APA StyleMiller, W. B., Jr., Enguita, F. J., & Leitão, A. L. (2021). Non-Random Genome Editing and Natural Cellular Engineering in Cognition-Based Evolution. Cells, 10(5), 1125. https://doi.org/10.3390/cells10051125