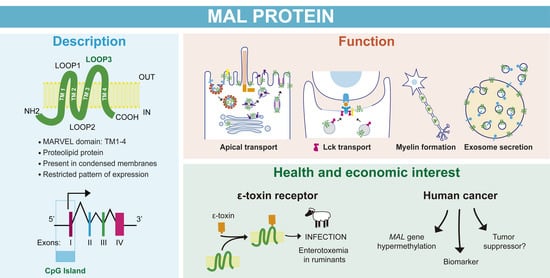
The MAL Protein, an Integral Component of Specialized Membranes, in Normal Cells and Cancer
Abstract
1. Introduction
2. Tissue Specific Expression, Intracellular Distribution, and Biochemical Properties of MAL
2.1. MAL Expression Is Restricted to Specific Tissues and Cell Types
2.2. MAL Is Mainly Found at the Plasma Membrane and Cytoplasmic Tubulovesicular Structures
2.3. MAL Has Unusual Biochemical Properties
2.3.1. MAL Is a Proteolipid Protein
2.3.2. MAL Selectively Partitions into Condensed Membranes
3. The Role of MAL in Epithelial Cells
3.1. MAL in Apical Transport in Cultured Cells
3.2. MAL in Apical Transport: KO Mice
3.3. MAL in Apical Endocytosis
3.4. MAL in Apical Retention of Membrane Proteins
3.5. MAL in Apical Lumen Formation
3.6. MAL and Primary Cilium Biogenesis
4. MAL in T Lymphocytes
4.1. MAL Expression in T Lymphocytes
4.2. MAL on Vesicular Traffic of the Tyrosine Kinase Lck
4.3. Subcompartmentalization of the Immunological Synapse
5. MAL in Myelin-Forming Cells
5.1. The Role of MAL in Myelin Proteolipid Protein (PLP) Targeting to Myelin in Cultured Cells
5.2. MAL in Myelin-Forming Cells in Mice
6. Role of MAL in Exosome Secretion
6.1. MAL in Prostasomes
6.2. MAL in Exosome Biogenesis by T Lymphocytes
7. MAL in Other Cell Systems and Processes
8. MAL and the Clostridial Epsilon Toxin
9. MAL as Organizer of Condensed Membranes for Specialized Processes
- (1)
- In polarized epithelial cells, HA is sorted to the apical surface and partitions into DRMs in control cells, whereas HA transport to the apical membrane is blocked and the association of HA with DRMs is decreased in MAL KD cells [59].
- (2)
- (3)
- The base of primary cilia is highly condensed in wild-type MDCK cells, whereas the degree of condensation is dramatically lower in MAL KD cells, resulting in fewer and shorter cilia [46].
- (4)
- Lck is transported to the plasma membrane and partitions into DRMs in control T cells, whereas Lck is retained at the Golgi and loses its association with DRMs in the absence of MAL expression [24]. Another effect of MAL depletion in T cells is that the biogenesis of exosomes carrying GPI-anchored proteins, which normally partition into DRMs, is much less extensive than in control cells [128]. Additionally, the mistargeting of MAL from the cSMAC to the pSMAC in T cell–APC conjugates in the presence of antigen reorganizes the structure of the IS in such a way that condensed membranes and associated proteins, such as Lck and LAT, change their distribution in the same manner as MAL [45].
- (5)
- In myelin-forming cells, neurofascin 55 distributes to the paranodal domain and is recovered from the DRM fraction in wild-type mice, but is excluded from the paranodal zone and from DRMs in MAL KO mice [119]. In oligodendrocytes, MAL overexpression interferes with the shift of PLP from a Triton X100-insoluble to a TritonX-100-soluble and CHAPS-insoluble form, impeding the switch of PLP targeting to myelin from transcytosis to lateral diffusion, which is associated with a conformational change in PLP [112,114].
- (6)
- (7)
- The expression of MAL in hepatic WIF-B cells, which lack endogenous MAL expression as do normal hepatocytes, increases the partitioning of specific apical proteins into DRMs and reroutes them from the indirect route to a direct pathway to the apical surface [64].
- (8)
- ETX, which associates with condensed plasma membrane regions, binds only to cells that express MAL, and the binding is lost upon MAL KD [152,153,154]. Reciprocally, the expression of MAL in cells that do not express the protein endogenously renders condensed membranes competent for ETX binding [152,153].
- (9)
- (10)
- Giant plasma membrane vesicles separate into coexisting ordered and disordered lipid phases [32,178]. From a set of 24 structurally and functionally diverse multipass membrane proteins, only MAL and PLP, which is a major myelin component, showed affinity for the ordered lipid phase [41]. It is of note that MAL overexpression out-competes PLP from these domains but PLP overexpression does not affect MAL partitioning. This observation was interpreted as meaning that MAL and PLP compete for a component of the ordered lipid phase present in limited amounts, and that MAL has a higher affinity for it than does PLP [41].
10. A Speculative Model of MAL Function in Membrane Trafficking
11. Structure of the MAL Gene and Promoter Region
11.1. The MAL Gene
11.2. The MAL Promoter
12. MAL in Cancer
12.1. MAL Gene Hypermethylation and Silencing Are Common Features in Many Carcinomas
12.2. MAL as a Cancer Biomarker
12.3. MAL as a Tumor Suppressor Protein
13. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alonso, M.A.; Weissman, S.M. cDNA Cloning and Sequence of MAL, a Hydrophobic Protein Associated with Human T-Cell Differentiation. Proc. Natl. Acad. Sci. USA 1987, 84, 1997–2001. [Google Scholar] [CrossRef]
- Kim, T.; Fiedler, K.; Madison, D.L.; Krueger, W.H.; Pfeiffer, S.E. Cloning and Characterization of MVP17: A Developmentally Regulated Myelin Protein in Oligodendrocytes. J. Neurosci. Res. 1995, 42, 413–422. [Google Scholar] [CrossRef]
- Schaeren-Wiemers, N.; Schaefer, C.; Valenzuela, D.M.; Yancopoulos, G.D.; Schwab, M.E. Identification of New Oligodendrocyte- and Myelin-Specific Genes by a Differential Screening Approach. J. Neurochem. 1995, 65, 10–22. [Google Scholar] [CrossRef]
- Zacchetti, D.; Peranen, J.; Murata, M.; Fiedler, K.; Simons, K. VIP17/MAL, a Proteolipid in Apical Transport Vesicles. FEBS Lett. 1995, 377, 465–469. [Google Scholar]
- Millán, J.; Puertollano, R.; Fan, L.; Alonso, M.A. Caveolin and MAL, Two Protein Components of Internal Detergent-Insoluble Membranes, Are in Distinct Lipid Microenvironments in MDCK Cells. Biochem. Biophys. Res. Commun. 1997, 233, 707–712. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Martin-Belmonte, F.; Valencia, A.; Alonso, M.A. MARVEL: A Conserved Domain Involved in Membrane Apposition Events. Trends Biochem. Sci. 2002, 27, 599–601. [Google Scholar] [CrossRef]
- Duan, H.-J.; Li, X.-Y.; Liu, C.; Deng, X.-L. Chemokine-like Factor-like MARVEL Transmembrane Domain-Containing Family in Autoimmune Diseases. Chin. Med. J. 2020, 133, 951–958. [Google Scholar] [CrossRef]
- Magyar, J.P.; Ebensperger, C.; Schaeren-Wiemers, N.; Suter, U. Myelin and Lymphocyte Protein (MAL/MVP17/VIP17) and Plasmolipin Are Members of an Extended Gene Family. Gene 1997, 189, 269–275. [Google Scholar] [CrossRef]
- Pérez, P.; Puertollano, R.; Alonso, M.A. Structural and Biochemical Similarities Reveal a Family of Proteins Related to the MAL Proteolipid, a Component of Detergent-Insoluble Membrane Microdomains. Biochem. Biophys. Res. Commun. 1997, 232, 618–621. [Google Scholar] [CrossRef]
- Janz, R.; Südhof, T.C.; Hammer, R.E.; Unni, V.; Siegelbaum, S.A.; Bolshakov, V.Y. Essential Roles in Synaptic Plasticity for Synaptogyrin I and Synaptophysin I. Neuron 1999, 24, 687–700. [Google Scholar] [CrossRef]
- Adams, D.J.; Arthur, C.P.; Stowell, M.H.B. Architecture of the Synaptophysin/Synaptobrevin Complex: Structural Evidence for an Entropic Clustering Function at the Synapse. Sci. Rep. 2015, 5, 13659. [Google Scholar] [CrossRef]
- Raleigh, D.R.; Marchiando, A.M.; Zhang, Y.; Shen, L.; Sasaki, H.; Wang, Y.; Long, M.; Turner, J.R. Tight Junction-Associated MARVEL Proteins Marveld3, Tricellulin, and Occludin Have Distinct but Overlapping Functions. Mol. Biol. Cell 2010, 21, 1200–1213. [Google Scholar] [CrossRef] [PubMed]
- Rehm, H.; Wiedenmann, B.; Betz, H. Molecular Characterization of Synaptophysin, a Major Calcium-Binding Protein of the Synaptic Vesicle Membrane. EMBO J. 1986, 5, 535–541. [Google Scholar] [CrossRef]
- Arthur, C.P.; Stowell, M.H.B. Structure of Synaptophysin: A Hexameric MARVEL-Domain Channel Protein. Structure 2007, 15, 707–714. [Google Scholar] [CrossRef]
- Pennuto, M.; Dunlap, D.; Contestabile, A.; Benfenati, F.; Valtorta, F. Fluorescence Resonance Energy Transfer Detection of Synaptophysin I and Vesicle-Associated Membrane Protein 2 Interactions during Exocytosis from Single Live Synapses. Mol. Biol. Cell 2002, 13, 2706–2717. [Google Scholar] [CrossRef]
- Valtorta, F.; Pennuto, M.; Bonanomi, D.; Benfenati, F. Synaptophysin: Leading Actor or Walk-on Role in Synaptic Vesicle Exocytosis? Bioessays 2004, 26, 445–453. [Google Scholar] [CrossRef]
- Marazuela, M.; Acevedo, A.; Adrados, M.; Garcia-Lopez, M.A.; Alonso, M.A. Expression of MAL, an Integral Protein Component of the Machinery for Raft-Mediated Pical Transport, in Human Epithelia. J. Histochem. Cytochem. 2003, 51, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Copie-Bergman, C.; Plonquet, A.; Alonso, M.A.; Boulland, M.-L.; Marquet, J.; Divine, M.; Moller, P.; Leroy, K.; Gaulard, P. MAL Expression in Lymphoid Cells: Further Evidence for MAL as a Distinct Molecular Marker of Primary Mediastinal Large B-Cell Lymphomas. Mod. Pathol. 2002, 15, 1172–1180. [Google Scholar] [CrossRef]
- Lacorre, D.-A.; Baekkevold, E.S.; Garrido, I.; Brandtzaeg, P.; Haraldsen, G.; Amalric, F.; Girard, J.-P. Plasticity of Endothelial Cells: Rapid Dedifferentiation of Freshly Isolated High Endothelial Venule Endothelial Cells Outside the Lymphoid Tissue Microenvironment. Blood 2004, 103, 4164–4172. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.; Schaeren-Wiemers, N.; Schneider, R.; Schwab, M.E. Developmental Expression Pattern of the Myelin Proteolipid MAL Indicates Different Functions of MAL for Immature Schwann Cells and in a Late Step of CNS Myelinogenesis. J. Neurochem. 1999, 73, 587–597. [Google Scholar] [CrossRef]
- Frank, M. MAL, a Proteolipid in Glycosphingolipid Enriched Domains: Functional Implications in Myelin and Beyond. Prog. Neurobiol. 2000, 60, 531–544. [Google Scholar] [CrossRef]
- Antón, O.; Batista, A.; Millan, J.; Andres-Delgado, L.; Puertollano, R.; Correas, I.; Alonso, M.A. An Essential Role for the MAL Protein in Targeting Lck to the Plasma Membrane of Human T Lymphocytes. J. Exp. Med. 2008, 205, 3201–3213. [Google Scholar] [CrossRef]
- Puertollano, R.; Martinez-Menarguez, J.A.; Batista, A.; Ballesta, J.; Alonso, M.A. An Intact Dilysine-like Motif in the Carboxyl Terminus of MAL Is Required for Normal Apical Transport of the Influenza Virus Hemagglutinin Cargo Protein in Epithelial Madin-Darby Canine Kidney Cells. Mol. Biol. Cell 2001, 12, 1869–1883. [Google Scholar] [CrossRef]
- Puertollano, R.; Alonso, M.A. MAL, an Integral Element of the Apical Sorting Machinery, Is an Itinerant Protein That Cycles between the Trans-Golgi Network and the Plasma Membrane. Mol. Biol. Cell 1999, 10, 3435–3447. [Google Scholar] [CrossRef][Green Version]
- Folch, J.; Lees, M. Proteolipides, a New Type of Tissue Lipoproteins; Their Isolation from Brain. J. Biol. Chem. 1951, 191, 807–817. [Google Scholar] [CrossRef]
- Schlesinger, M.J. Proteolipids. Annu. Rev. Biochem. 1981, 50, 193–206. [Google Scholar] [CrossRef]
- Rancaño, C.; Rubio, T.; Correas, I.; Alonso, M.A. Genomic Structure and Subcellular Localization of MAL, a Human T-Cell-Specific Proteolipid Protein. J. Biol. Chem. 1994, 269, 8159–8164. [Google Scholar] [CrossRef]
- Korlach, J.; Schwille, P.; Webb, W.W.; Feigenson, G.W. Characterization of Lipid Bilayer Phases by Confocal Microscopy and Fluorescence Correlation Spectroscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 8461–8466. [Google Scholar] [CrossRef]
- Dietrich, C.; Bagatolli, L.A.; Volovyk, Z.N.; Thompson, N.L.; Levi, M.; Jacobson, K.; Gratton, E. Lipid Rafts Reconstituted in Model Membranes. Biophys. J. 2001, 80, 1417–1428. [Google Scholar] [CrossRef]
- Veatch, S.L.; Keller, S.L. Organization in Ipid Membranes Containing Cholesterol. Phys. Rev. Lett. 2002, 89, 268101. [Google Scholar] [CrossRef]
- Simons, K.; Ikonen, E. Functional Rafts in Cell Membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid Rafts as a Membrane-Organizing Principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef]
- Brown, D.A.; London, E. Structure and Origin of Ordered Lipid Domains in Biological Membranes. J. Membr. Biol. 1998, 164, 103–114. [Google Scholar] [CrossRef]
- Schroeder, R.J.; Ahmed, S.N.; Zhu, Y.; London, E.; Brown, D.A. Cholesterol and Sphingolipid Enhance the Triton X-100 Insolubility of Glycosylphosphatidylinositol-Anchored Proteins by Promoting the Formation of Detergent-Insoluble Ordered Membrane Domains. J. Biol. Chem. 1998, 273, 1150–1157. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, D.; Goñi, F.M.; Heerklotz, H. Detergent-Resistant Membranes Should Not Be Identified with Membrane Rafts. Trends Biochem. Sci. 2005, 30, 430–436. [Google Scholar] [CrossRef]
- Puertollano, R.; Alonso, M.A. A Short Peptide Motif at the Carboxyl Terminus Is Required for Incorporation of the Integral Membrane MAL Protein to Glycolipid-Enriched Membranes. J. Biol. Chem. 1998, 273, 12740–12745. [Google Scholar] [CrossRef]
- Puertollano, R.; Alonso, M.A. Targeting of MAL, a Putative Element of the Apical Sorting Machinery, to Glycolipid-Enriched Membranes Requires a Pre-Golgi Sorting Event. Biochem. Biophys. Res. Commun. 1999, 254, 689–692. [Google Scholar] [CrossRef]
- Puertollano, R.; Menéndez, M.; Alonso, M.A. Incorporation of MAL, an Integral Protein Element of the Machinery for the Glycolipid and Cholesterol-Mediated Apical Pathway of Transport, into Artificial Membranes Requires Neither of These Lipid Species. Biochem. Biophys. Res. Commun. 1999, 266, 330–333. [Google Scholar] [CrossRef]
- Castello-Serrano, I.; Lorent, J.H.; Ippolito, R.; Levental, K.R.; Levental, I. Myelin-Associated MAL and PLP Are Unusual among Multipass Transmembrane Proteins in Preferring Ordered Membrane Domains. J. Phys. Chem. B 2020, 124, 5930–5939. [Google Scholar] [CrossRef]
- Gaus, K.; Gratton, E.; Kable, E.P.W.; Jones, A.S.; Gelissen, I.; Kritharides, L.; Jessup, W. Visualizing Lipid Structure and Raft Domains in Living Cells with Two-Photon Microscopy. Proc. Natl. Acad. Sci. USA 2003, 100, 15554–15559. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.A.; Tricerri, M.A.; Gratton, E. Laurdan Generalized Polarization Fluctuations Measures Membrane Packing Micro-Heterogeneity In Vivo. Proc. Nat. Acad. Sci. USA 2012, 109, 7314–7319. [Google Scholar] [CrossRef] [PubMed]
- Magal, L.G.; Yaffe, Y.; Shepshelovich, J.; Aranda, J.F.; de Marco, M.d.C.; Gaus, K.; Alonso, M.A.; Hirschberg, K. Clustering and Lateral Concentration of Raft Lipids by the MAL Protein. Mol. Biol. Cell 2009, 20, 3751–3762. [Google Scholar] [CrossRef] [PubMed]
- Antón, O.M.; Andrés-Delgado, L.; Reglero-Real, N.; Batista, A.; Alonso, M.A. MAL Protein Controls Protein Sorting at the Supramolecular Activation Cluster of Human T Lymphocytes. J. Immunol. 2011, 186, 6345–6356. [Google Scholar] [CrossRef] [PubMed]
- Reales, E.; Bernabé-Rubio, M.; Casares-Arias, J.; Rentero, C.; Fernández-Barrera, J.; Rangel, L.; Correas, I.; Enrich, C.; Andrés, G.; Alonso, M.A. The MAL Protein Is Crucial for Proper Membrane Condensation at the Ciliary Base, Which Is Required for Primary Cilium Elongation. J. Cell Sci. 2015, 128, 2261–2270. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodriguez-Boulan, E.; Kreitzer, G.; Musch, A. Organization of Vesicular Trafficking in Epithelia. Nat. Rev. Mol. Cell. Biol. 2005, 6, 233–247. [Google Scholar] [CrossRef]
- Weisz, O.A.; Rodriguez-Boulan, E. Apical Trafficking in Epithelial Cells: Signals, Clusters and Motors. J. Cell Sci. 2009, 122, 4253–4266. [Google Scholar] [CrossRef]
- van Meer, G.; Stelzer, E.H.; Wijnaendts-van-Resandt, R.W.; Simons, K. Sorting of Sphingolipids in Epithelial (Madin-Darby Canine Kidney) Cells. J. Cell Biol. 1987, 105, 1623–1635. [Google Scholar] [CrossRef]
- Simons, K.; Van Meer, G. Lipid Sorting in Epithelial Cells. Biochemistry 1988, 27, 6197–6202. [Google Scholar] [CrossRef]
- Hagmann, J.; Fishman, P.H. Detergent Extraction of Cholera Toxin and Gangliosides from Cultured Cells and Isolated Membranes. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1982, 720, 181–187. [Google Scholar] [CrossRef]
- Skibbens, J.E.; Roth, M.G.; Matlin, K.S. Differential Extractability of Influenza Virus Hemagglutinin during Intracellular Transport in Polarized Epithelial Cells and Nonpolar Fibroblasts. J. Cell Biol. 1989, 108, 821–832. [Google Scholar] [CrossRef]
- Brown, D.A.; Rose, J.K. Sorting of GPI-Anchored Proteins to Glycolipid-Enriched Membrane Subdomains during Transport to the Apical Cell Surface. Cell 1992, 68, 533–544. [Google Scholar] [CrossRef]
- Simons, K.; Wandinger-Ness, A. Polarized Sorting in Epithelia. Cell 1990, 62, 207–210. [Google Scholar] [CrossRef]
- Kurzchalia, T.V.; Dupree, P.; Parton, R.G.; Kellner, R.; Virta, H.; Lehnert, M.; Simons, K. VIP21, a 21-KD Membrane Protein Is an Integral Component of Trans-Golgi-Network-Derived Transport Vesicles. J. Cell Biol. 1992, 118, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, K.; Parton, R.G.; Kellner, R.; Etzold, T.; Simons, K. VIP36, a Novel Component of Glycolipid Rafts and Exocytic Carrier Vesicles in Epithelial Cells. EMBO J. 1994, 13, 1729–1740. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, K.; Lafont, F.; Parton, R.G.; Simons, K. Annexin XIIIb: A Novel Epithelial Specific Annexin Is Implicated in Vesicular Traffic to the Apical Plasma Membrane. J. Cell Biol. 1995, 128, 1043–1053. [Google Scholar] [CrossRef]
- Cheong, K.H.; Zacchetti, D.; Schneeberger, E.E.; Simons, K. VIP17/MAL, a Lipid Raft-Associated Protein, Is Involved in Apical Transport in MDCK Cells. Proc. Natl. Acad. Sci. USA 1999, 96, 6241–6248. [Google Scholar] [CrossRef]
- Puertollano, R.; Martin-Belmonte, F.; Millan, J.; de Marco, M.C.; Albar, J.P.; Kremer, L.; Alonso, M.A. The MAL Proteolipid Is Necessary for Normal Apical Transport and Accurate Sorting of the Influenza Virus Hemagglutinin in Madin-Darby Canine Kidney Cells. J. Cell Biol. 1999, 145, 141–151. [Google Scholar] [CrossRef]
- Martin-Belmonte, F.; Puertollano, R.; Millan, J.; Alonso, M.A. The MAL Proteolipid Is Necessary for the Overall Apical Delivery of Membrane Proteins in the Polarized Epithelial Madin-Darby Canine Kidney and Fischer Rat Thyroid Cell Lines. Mol. Biol. Cell 2000, 11, 2033–2045. [Google Scholar] [CrossRef]
- Martin-Belmonte, F.; Arvan, P.; Alonso, M.A. MAL Mediates Apical Transport of Secretory Proteins in Polarized Epithelial Madin-Darby Canine Kidney Cells. J. Biol. Chem. 2001, 276, 49337–49342. [Google Scholar] [CrossRef] [PubMed]
- Mostov, K.E.; Verges, M.; Altschuler, Y. Membrane Traffic in Polarized Epithelial Cells. Curr. Opin. Cell Biol. 2000, 12, 483–490. [Google Scholar] [CrossRef]
- de Marco, M.C.; Martin-Belmonte, F.; Kremer, L.; Albar, J.P.; Correas, I.; Vaerman, J.P.; Marazuela, M.; Byrne, J.A.; Alonso, M.A. MAL2, a Novel Raft Protein of the MAL Family, Is an Essential Component of the Machinery for Transcytosis in Hepatoma HepG2 Cells. J. Cell Biol. 2002, 159, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Ramnarayanan, S.P.; Cheng, C.A.; Bastaki, M.; Tuma, P.L. Exogenous MAL Reroutes Selected Hepatic Apical Proteins into the Direct Pathway in WIF-B Cells. Mol. Biol. Cell 2007, 18, 2707–2715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kachar, B.; Liang, F.; Lins, U.; Ding, M.; Wu, X.-R.; Stoffler, D.; Aebi, U.; Sun, T.-T. Three-Dimensional Analysis of the 16 Nm Urothelial Plaque Particle: Luminal Surface Exposure, Preferential Head-to-Head Interaction, and Hinge Formation 1 1Edited by W. Baumeisser. J. Mol. Biol. 1999, 285, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-R.; Kong, X.-P.; Pellicer, A.; Kreibich, G.; Sun, T.-T. Uroplakins in Urothelial Biology, Function, and Disease. Kidney Int. 2009, 75, 1153–1165. [Google Scholar] [CrossRef]
- Liang, F.; Kachar, B.; Ding, M.; Zhai, Z.; Wu, X.-R.; Sun, T.-T. Urothelial Hinge as a Highly Specialized Membrane: Detergent-Insolubility, Urohingin Association, and in Vitro Formation. Differentiation 1999, 65, 59–69. [Google Scholar] [CrossRef]
- Hudoklin, S.; Jezernik, K.; Neumüller, J.; Pavelka, M.; Romih, R. Electron Tomography of Fusiform Vesicles and Their Organization in Urothelial Cells. PLoS ONE 2012, 7, e32935. [Google Scholar] [CrossRef]
- Liebert, M.; Yuan, T.Y.; Grossman, H.B.; Hubbel, A.; Chung, M.; Wedemeyer, G.; Brozovich, M.; Lomax, M.I.; Hegeman, A.; Wheelock, M.J. Expression of Mal Is Associated with Urothelial Differentiation In Vitro: Identification by Differential Display Reverse-Transcriptase Polymerase Chain Reaction. Differentiation 1997, 61, 177–185. [Google Scholar] [CrossRef]
- Zhou, G.; Liang, F.-X.; Romih, R.; Wang, Z.; Liao, Y.; Ghiso, J.; Luque-Garcia, J.L.; Neubert, T.A.; Kreibich, G.; Alonso, M.A.; et al. MAL Facilitates the Incorporation of Exocytic Uroplakin-Delivering Vesicles into the Apical Membrane of Urothelial Umbrella Cells. Mol. Biol. Cell 2012, 23, 1354–1366. [Google Scholar] [CrossRef]
- Wankel, B.; Ouyang, J.; Guo, X.; Hadjiolova, K.; Miller, J.; Liao, Y.; Tham, D.K.L.; Romih, R.; Andrade, L.R.; Gumper, I.; et al. Sequential and Compartmentalized Action of Rabs, SNAREs, and MAL in the Apical Delivery of Fusiform Vesicles in Urothelial Umbrella Cells. Mol. Biol. Cell 2016, 27, 1621–1634. [Google Scholar] [CrossRef]
- Martin-Belmonte, F.; Martinez-Menarguez, J.A.; Aranda, J.F.; Ballesta, J.; de Marco, M.C.; Alonso, M.A. MAL Regulates Clathrin-Mediated Endocytosis at the Apical Surface of Madin-Darby Canine Kidney Cells. J. Cell Biol. 2003, 163, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.; Frøkiær, J.; Marples, D.; Kwon, T.-H.; Agre, P.; Knepper, M.A. Aquaporins in the Kidney: From Molecules to Medicine. Physiol. Rev. 2002, 82, 205–244. [Google Scholar] [CrossRef]
- Nielsen, S.; Kwon, T.-H.; Frøkiær, J.; Agre, P. Regulation and Dysregulation of Aquaporins in Water Balance Disorders. J. Intern. Med. 2007, 261, 53–64. [Google Scholar] [CrossRef]
- Kamsteeg, E.-J.; Duffield, A.S.; Konings, I.B.M.; Spencer, J.; Pagel, P.; Deen, P.M.T.; Caplan, M.J. MAL Decreases the Internalization of the Aquaporin-2 Water Channel. Proc. Natl. Acad. Sci. USA 2007, 104, 16696–16701. [Google Scholar] [CrossRef]
- Carmosino, M.; Rizzo, F.; Procino, G.; Basco, D.; Valenti, G.; Forbush, B.; Schaeren-Wiemers, N.; Caplan, M.J.; Svelto, M. MAL/VIP17, a New Player in the Regulation of NKCC2 in the Kidney. Mol. Biol. Cell 2010, 21, 3985–3997. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Roignot, J.; Datta, A.; Overeem, A.W.; Kim, M.; Yu, W.; Peng, X.; Eastburn, D.J.; Ewald, A.J.; Werb, Z.; et al. A Molecular Switch for the Orientation of Epithelial Cell Polarization. Dev. Cell 2014, 31, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Boulan, E.; Macara, I.G. Organization and Execution of the Epithelial Polarity Programme. Nat. Rev. Mol. Cell. Biol. 2014, 15, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Bryant, D.M.; Mostov, K.E. Molecular Regulation of Lumen Morphogenesis. Curr. Biol. 2011, 21, R126–R136. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.; Atanasoski, S.; Sancho, S.; Magyar, J.P.; Rülicke, T.; Schwab, M.E.; Suter, U. Progressive Segregation of Unmyelinated Axons in Peripheral Nerves, Myelin Alterations in the CNS, and Cyst Formation in the Kidneys of Myelin and Lymphocyte Protein-Overexpressing Mice. J. Neurochem. 2000, 75, 1927–1939. [Google Scholar] [CrossRef]
- Wang, A.Z.; Ojakian, G.K.; Nelson, W.J. Steps in the Morphogenesis of a Polarized Epithelium. I. Uncoupling the Roles of Cell-Cell and Cell-Substratum Contact in Establishing Plasma Membrane Polarity in Multicellular Epithelial (MDCK) Cysts. J. Cell Sci. 1990, 95, 137–151. [Google Scholar] [CrossRef]
- Torkko, J.M.; Manninen, A.; Schuck, S.; Simons, K. Depletion of Apical Transport Proteins Perturbs Epithelial Cyst Formation and Ciliogenesis. J. Cell Sci. 2008, 121, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Takiar, V.; Mistry, K.; Carmosino, M.; Schaeren-Wiemers, N.; Caplan, M.J. VIP17/MAL Expression Modulates Epithelial Cyst Formation and Ciliogenesis. Am. J. Physiol. Cell Physiol. 2012, 303, C862–C871. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bernabe-Rubio, M.; Alonso, M.A. Routes and Machinery of Primary Cilium Biogenesis. Cell. Mol. Life Sci. 2017, 74, 4077–4095. [Google Scholar] [CrossRef]
- Nachury, M.V.; Mick, D.U. Establishing and Regulating the Composition of Cilia for Signal Transduction. Nat. Rev. Mol. Cell. Biol. 2019, 20, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Hildebrandt, F. Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028191. [Google Scholar] [CrossRef]
- Reiter, J.F.; Leroux, M.R. Genes and Molecular Pathways Underpinning Ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef]
- Bernabé-Rubio, M.; Bosch-Fortea, M.; García, E.; de la Serna, J.B.; Alonso, M.A. Adaptive Lipid Immiscibility and Membrane Remodeling Are Active Functional Determinants of Primary Ciliogenesis. Small Methods 2020, 5, 2000711. [Google Scholar] [CrossRef]
- Labat-de-Hoz, L.; Rubio-Ramos, A.; Casares-Arias, J.; Bernabé-Rubio, M.; Correas, I.; Alonso, M.A. A Model for Primary Cilium Biogenesis by Polarized Epithelial Cells: Role of the Midbody Remnant and Associated Specialized Membranes. Front. Cell Dev. Biol. 2021, 8, 622918. [Google Scholar] [CrossRef]
- Taniuchi, I. CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annu. Rev. Immunol. 2018, 36, 579–601. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Freeman, G.J. The B7-CD28 Superfamily. Nat. Rev. Immunol. 2002, 2, 116–126. [Google Scholar] [CrossRef]
- Riley, J.L.; Mao, M.; Kobayashi, S.; Biery, M.; Burchard, J.; Cavet, G.; Gregson, B.P.; June, C.H.; Linsley, P.S. Modulation of TCR-Induced Transcriptional Profiles by Ligation of CD28, ICOS, and CTLA-4 Receptors. Proc. Nat. Acad. Sci. USA 2002, 99, 11790–11795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Gu, W.; Sun, B. Th1/Th2 Cell Differentiation and Molecular Signals. In T Helper Cell Differentiation and Their Function; Sun, B., Ed.; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2014; Volume 841, pp. 15–44. ISBN 978-94-017-9486-2. [Google Scholar]
- Matsumoto, Y.; Oshida, T.; Obayashi, I.; Imai, Y.; Matsui, K.; Yoshida, N.L.; Nagata, N.; Ogawa, K.; Obayashi, M.; Kashiwabara, T.; et al. Identification of Highly Expressed Genes in Peripheral Blood T Cells from Patients with Atopic Dermatitis. Int. Arch. Allergy Immunol. 2002, 129, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Yoshida, N.L.; Sugita, Y.; Arai, T.; Seki, Y.; Kubo, M.; Tsujimoto, G.; Akasawa, A.; Saito, H.; Oshida, T. Mite-Antigen Stimulates MAL Expression in Peripheral Blood T Cells of Mite-Sensitive Subjects. Allergol. Int. 2005, 54, 273–282. [Google Scholar] [CrossRef][Green Version]
- Bosco, A.; McKenna, K.L.; Devitt, C.J.; Firth, M.J.; Sly, P.D.; Holt, P.G. Identification of Novel Th2-Associated Genes in T Memory Responses to Allergens. J. Immunol. 2006, 176, 4766. [Google Scholar] [CrossRef] [PubMed]
- Millán, J.; Puertollano, R.; Fan, L.; Rancaño, C.; Alonso, M.A. The MAL Proteolipid Is a Component of the Detergent-Insoluble Membrane Subdomains of Human T-Lymphocytes. Biochem. J. 1997, 321, 247. [Google Scholar] [CrossRef]
- Millan, J.; Alonso, M.A. MAL, a Novel Integral Membrane Protein of Human T Lymphocytes, Associates with Glycosylphosphatidylinositol-Anchored Proteins and Src-like Tyrosine Kinases. Eur. J. Immunol. 1998, 28, 3675–3684. [Google Scholar] [CrossRef]
- Andres-Delgado, L.; Anton, O.M.; Madrid, R.; Byrne, J.A.; Alonso, M.A. Formin INF2 Regulates MAL-Mediated Transport of Lck to the Plasma Membrane of Human T Lymphocytes. Blood 2010, 116, 5919–5929. [Google Scholar] [CrossRef]
- Leitner, J.; Mahasongkram, K.; Schatzlmaier, P.; Pfisterer, K.; Leksa, V.; Pata, S.; Kasinrerk, W.; Stockinger, H.; Steinberger, P. Differentiation and Activation of Human CD4 T Cells Is Associated with a Gradual Loss of Myelin and Lymphocyte Protein. Eur. J. Immunol. 2021, 51, 848–863. [Google Scholar] [CrossRef]
- Silva, J.G.; Martins, N.P.; Henriques, R.; Soares, H. HIV-1 Nef Impairs the Formation of Calcium Membrane Territories Controlling the Signaling Nanoarchitecture at the Immunological Synapse. J. Immunol. 2016, 197, 4042. [Google Scholar] [CrossRef]
- Ventimiglia, L.N.; Alonso, M.A. The Role of Membrane Rafts in Lck Transport, Regulation and Signalling in T-Cells. Biochem. J. 2013, 454, 169–179. [Google Scholar] [CrossRef][Green Version]
- Bromley, S.K.; Burack, W.R.; Johnson, K.G.; Somersalo, K.; Sims, T.N.; Sumen, C.; Davis, M.M.; Shaw, A.S.; Allen, P.M.; Dustin, M.L. The Immunological Synapse. Annu. Rev. Immunol. 2001, 19, 375–396. [Google Scholar] [CrossRef] [PubMed]
- Soares, H.; Henriques, R.; Sachse, M.; Ventimiglia, L.; Alonso, M.A.; Zimmer, C.; Thoulouze, M.-I.; Alcover, A. Regulated Vesicle Fusion Generates Signaling Nanoterritories That Control T Cell Activation at the Immunological Synapse. J. Exp. Med. 2013, 210, 2415. [Google Scholar] [CrossRef] [PubMed]
- Baron, W.; Hoekstra, D. On the Biogenesis of Myelin Membranes: Sorting, Trafficking and Cell Polarity. FEBS Lett. 2010, 584, 1760–1770. [Google Scholar] [CrossRef] [PubMed]
- Stadelmann, C.; Timmler, S.; Barrantes-Freer, A.; Simons, M. Myelin in the Central Nervous System: Structure, Function, and Pathology. Physiol. Rev. 2019, 99, 1381–1431. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.; van der Haar, M.E.; Schaeren-Wiemers, N.; Schwab, M.E. RMAL Is a Glycosphingolipid-Associated Protein of Myelin and Apical Membranes of Epithelial Cells in Kidney and Stomach. J. Neurosci. 1998, 18, 4901. [Google Scholar] [CrossRef]
- Erne, B.; Sansano, S.; Frank, M.; Schaeren-Wiemers, N. Rafts in Adult Peripheral Nerve Myelin Contain Major Structural Myelin Proteins and Myelin and Lymphocyte Protein (MAL) and CD59 as Specific Markers. J. Neurochem. 2002, 82, 550–562. [Google Scholar] [CrossRef]
- Wakeman, J.A.; Heath, P.R.; Pearson, R.C.; Andrews, P.W. MAL mRNA Is Induced during the Differentiation of Human Embryonal Carcinoma Cells into Neurons and Is Also Localised within Specific Regions of the Human Brain. Differentiation 1997, 62, 97–105. [Google Scholar] [CrossRef]
- Baron, W.; de Jonge, J.C.; de Vries, H.; Hoekstra, D. Regulation of Oligodendrocyte Differentiation: Protein Kinase C Activation Prevents Differentiation of O2A Progenitor Cells toward Oligodendrocytes. Glia 1998, 22, 121–129. [Google Scholar] [CrossRef]
- de Vries, H.; Schrage, C.; Hoekstra, D. An Apical-Type Trafficking Pathway Is Present in Cultured Oligodendrocytes but the Sphingolipid-Enriched Myelin Membrane Is the Target of a Basolateral-Type Pathway. Mol. Biol. Cell 1998, 9, 599–609. [Google Scholar] [CrossRef]
- Bijlard, M.; de Jonge, J.C.; Klunder, B.; Nomden, A.; Hoekstra, D.; Baron, W. MAL Is a Regulator of the Recruitment of Myelin Protein PLP to Membrane Microdomains. PLoS ONE 2016, 11, e0155317. [Google Scholar] [CrossRef] [PubMed]
- Marazuela, M.; Alonso, M.A. Expression of MAL and MAL2, Two Elements of the Protein Machinery for Raft-Mediated Transport, in Normal and Neoplastic Human Tissue. Histol. Histopathol. 2004, 19, 925–933. [Google Scholar]
- Baron, W.; Ozgen, H.; Klunder, B.; de Jonge, J.C.; Nomden, A.; Plat, A.; Trifilieff, E.; de Vries, H.; Hoekstra, D. The Major Myelin-Resident Protein PLP Is Transported to Myelin Membranes via a Transcytotic Mechanism: Involvement of Sulfatide. Mol. Cell. Biol. 2015, 35, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Woods, B.L.; Gladfelter, A.S. The State of the Septin Cytoskeleton from Assembly to Function. Curr. Opin. Cell Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef]
- Buser, A.M.; Erne, B.; Werner, H.B.; Nave, K.A.; Schaeren-Wiemers, N. The Septin Cytoskeleton in Myelinating Glia. Mol. Cell. Neurosci. 2009, 40, 156–166. [Google Scholar] [CrossRef]
- Shaimardanova, A.A.; Chulpanova, D.S.; Solovyeva, V.V.; Mullagulova, A.I.; Kitaeva, K.V.; Allegrucci, C.; Rizvanov, A.A. Metachromatic Leukodystrophy: Diagnosis, Modeling, and Treatment Approaches. Front. Med. 2020, 7, 576221. [Google Scholar] [CrossRef]
- Saravanan, K.; Schaeren-Wiemers, N.; Klein, D.; Sandhoff, R.; Schwarz, A.; Yaghootfam, A.; Gieselmann, V.; Franken, S. Specific Downregulation and Mistargeting of the Lipid Raft-Associated Protein MAL in a Glycolipid Storage Disorder. Neurobiol. Dis. 2004, 16, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Schaeren-Wiemers, N.; Bonnet, A.; Erb, M.; Erne, B.; Bartsch, U.; Kern, F.; Mantei, N.; Sherman, D.; Suter, U. The Raft-Associated Protein MAL Is Required for Maintenance of Proper Axon-Glia Interactions in the Central Nervous System. J. Cell Biol. 2004, 166, 731–742. [Google Scholar] [CrossRef]
- Buser, A.M.; Schmid, D.; Kern, F.; Erne, B.; Lazzati, T.; Schaeren-Wiemers, N. The Myelin Protein MAL Affects Peripheral Nerve Myelination: A New Player Influencing P75 Neurotrophin Receptor Expression. Eur. J. Neurosci. 2009, 29, 2276–2290. [Google Scholar] [CrossRef]
- Schmid, D.; Zeis, T.; Sobrio, M.; Schaeren-Wiemers, N. MAL Overexpression Leads to Disturbed Expression of Genes That Influence Cytoskeletal Organization and Differentiation of Schwann Cells. ASN Neuro 2014, 6. [Google Scholar] [CrossRef]
- Arancibia-Carcamo, I.L.; Attwell, D. The Node of Ranvier in CNS Pathology. Acta Neuropathol. 2014, 128, 161–175. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell. Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Sánchez-Madrid, F. Intercellular Communication: Diverse Structures for Exchange of Genetic Information. Nat. Rev. Mol. Cell. Biol. 2012, 13, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, G. Prostasomes Are Mediators of Intercellular Communication: From Basic Research to Clinical Implications: Review: Prostasomes Are Mediators of Intercellular Communication. J. Int. Med. 2012, 271, 400–413. [Google Scholar] [CrossRef]
- Llorente, A.; de Marco, M.C.; Alonso, M.A. Caveolin-1 and MAL Are Located on Prostasomes Secreted by the Prostate Cancer PC-3 Cell Line. J. Cell Sci. 2004, 117, 5343–5351. [Google Scholar] [CrossRef]
- de Gassart, A.; Géminard, C.; Février, B.; Raposo, G.; Vidal, M. Lipid Raft-Associated Protein Sorting in Exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef]
- Ventimiglia, L.N.; Fernández-Martín, L.; Martínez-Alonso, E.; Antón, O.M.; Guerra, M.; Martínez-Menárguez, J.A.; Andrés, G.; Alonso, M.A. Cutting Edge: Regulation of Exosome Secretion by the Integral MAL Protein in T Cells. J. Immunol. 2015, 195, 810. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional Transfer of MicroRNA-Loaded Exosomes from T Cells to Antigen-Presenting Cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Ventimiglia, L.N.; Alonso, M.A. Biogenesis and Function of T Cell-Derived Exosomes. Front. Cell Dev. Biol. 2016, 4. [Google Scholar] [CrossRef]
- Jones, S.A.; Jenkins, B.J. Recent Insights into Targeting the IL-6 Cytokine Family in Inflammatory Diseases and Cancer. Nat. Rev. Immunol. 2018, 18, 773–789. [Google Scholar] [CrossRef]
- Dalous, J.; Pansiot, J.; Pham, H.; Chatel, P.; Nadaradja, C.; D’Agostino, I.; Vottier, G.; Schwendimann, L.; Vanneaux, V.; Charriaut-Marlangue, C.; et al. Use of Human Umbilical Cord Blood Mononuclear Cells to Prevent Perinatal Brain Injury: A Preclinical Study. Stem Cells Dev. 2013, 22, 169–179. [Google Scholar] [CrossRef]
- Henning, R.J.; Abu-Ali, H.; Balis, J.U.; Morgan, M.B.; Willing, A.E.; Sanberg, P.R. Human Umbilical Cord Blood Mononuclear Cells for the Treatment of Acute Myocardial Infarction. Cell Transplant. 2004, 13, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Ireland, R.C.; Iovene, C.; Wagner, E.F.; McInnis, R.; Oblon, D.; Alonso, M.A.; Paul, S.R. Use of Messenger RNA Differential Display to Identify Interleukin-11-Responsive Genes in Human Umbilical Cord Blood Mononuclear Cells: IL-11 Upregulates the Expression of the HMAL Gene. J. Interf. Cytok. Res. 1996, 16, 829–834. [Google Scholar] [CrossRef]
- Itoh, Y. Membrane-Type Matrix Metalloproteinases: Their Functions and Regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef]
- Schröder, H.M.; Hoffmann, S.C.; Hecker, M.; Korff, T.; Ludwig, T. The Tetraspanin Network Modulates MT1-MMP Cell Surface Trafficking. Int. J. Biochem. Cell Biol. 2013, 45, 1133–1144. [Google Scholar] [CrossRef]
- Barker, D.J. The Fetal and Infant Origins of Adult Disease. BMJ 1990, 301, 1111. [Google Scholar] [CrossRef] [PubMed]
- Van Campen, H.; Bishop, J.V.; Abrahams, V.M.; Bielefeldt-Ohmann, H.; Mathiason, C.K.; Bouma, G.J.; Winger, Q.A.; Mayo, C.E.; Bowen, R.A.; Hansen, T.R. Maternal Influenza A Virus Infection Restricts Eetal and Placental Growth and Adversely Affects the Fetal Thymic Transcriptome. Viruses 2020, 12, 1003. [Google Scholar] [CrossRef] [PubMed]
- Bello-Morales, R.; Crespillo, A.J.; García, B.; Dorado, L.Á.; Martín, B.; Tabarés, E.; Krummenacher, C.; de Castro, F.; López-Guerrero, J.A. The Effect of Cellular Differentiation on HSV-1 Infection of Oligodendrocytic Cells. PLoS ONE 2014, 9, e89141. [Google Scholar] [CrossRef] [PubMed]
- López-Guerrero, J.A.; de la Nuez, C.; Praena, B.; Sánchez-León, E.; Krummenacher, C.; Bello-Morales, R. Herpes Simplex Virus 1 Spread in Oligodendrocytic Cells Is Highly Dependent on MAL Proteolipid. J. Virol. 2019, 94, e01739-19. [Google Scholar] [CrossRef]
- Popoff, M.R. Epsilon Toxin: A Fascinating Pore-Forming Toxin. FEBS J. 2011, 278, 4602–4615. [Google Scholar] [CrossRef]
- Rumah, K.R.; Linden, J.; Fischetti, V.A.; Vartanian, T. Isolation of Clostridium Perfringens Type B in an Individual at First Clinical Presentation of Multiple Sclerosis Provides Clues for Environmental Triggers of the Disease. PLoS ONE 2013, 8, e76359. [Google Scholar] [CrossRef]
- Compston, A.; Coles, A. Multiple Sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of Multiple Sclerosis. Nat. Rev. Immunol. 2015, 15, 545. [Google Scholar] [CrossRef]
- Petit, L.; Gibert, M.; Gillet, D.; Laurent-Winter, C.; Boquet, P.; Popoff, M.R. Clostridium Perfringens Epsilon-Toxin Acts on MDCK Cells by Forming a Large Membrane Complex. J. Bacteriol. 1997, 179, 6480–6487. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, M.; Ochi, S.; Sakurai, J. Assembly of Clostridium Perfringens Epsilon-Toxin on MDCK Cell Membrane. J. Natl. Toxins 1998, 7, 291–302. [Google Scholar]
- Türkcan, S.; Masson, J.-B.; Casanova, D.; Mialon, G.; Gacoin, T.; Boilot, J.-P.; Popoff, M.R.; Alexandrou, A. Observing the Confinement Potential of Bacterial Pore-Forming Toxin Receptors inside Rafts with Nonblinking Eu3+-Doped Oxide Nanoparticles. Biophys. J. 2012, 102, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, M.; Itohayashi, Y.; Hara, H.; Higashihara, M.; Fukatani, Y.; Takagishi, T.; Oda, M.; Kobayashi, K.; Nakagawa, I.; Sakurai, J. Cellular Vacuolation Induced by Clostridium Perfringens Epsilon-Toxin. FEBS J. 2011, 278, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Gil, C.; Dorca-Arévalo, J.; Blasi, J. Clostridium Perfringens Epsilon Toxin Binds to Membrane Lipids and Its Cytotoxic Action Depends on Sulfatide. PLoS ONE 2015, 10, e0140321. [Google Scholar] [CrossRef]
- Ferret-Sena, V.; Sena, A.; Besnard, F.; Fressinaud, C.; Rebel, G.; Sarliève, L.L. Comparison of the Mechanisms of Action of Insulin and Triiodothyronine on the Synthesis of Cerebroside Sulfotransferase in Cultures of Cells Dissociated from Brains of Embryonic Mice. Dev. Neurosci. 1990, 12, 89–105. [Google Scholar] [CrossRef]
- Pombo, P.M.G.; Ibarrola, N.; Alonso, M.A.; Rodríguez-Peña, A. Thyroid Hormone Regulates the Expression of the MAL Proteolipid, a Component of Glycolipid-Enriched Membranes, in Neonatal Rat Brain. J. Neurosci. Res. 1998, 52, 584–590. [Google Scholar] [CrossRef]
- Rumah, K.R.; Ma, Y.; Linden, J.R.; Oo, M.L.; Anrather, J.; Schaeren-Wiemers, N.; Alonso, M.A.; Fischetti, V.A.; McClain, M.S.; Vartanian, T. The Myelin and Lymphocyte Protein MAL Is Required for Binding and Activity of Clostridium Perfringens Epsilon-Toxin. PLoS Pathog. 2015, 11, e1004896. [Google Scholar] [CrossRef] [PubMed]
- Blanch, M.; Dorca-Arévalo, J.; Not, A.; Cases, M.; Gómez de Aranda, I.; Martínez-Yélamos, A.; Martínez-Yélamos, S.; Solsona, C.; Blasi, J. The Cytotoxicity of Epsilon Toxin from Clostridium Perfringenson Lymphocytes Is Mediated by MAL Protein Expression. Mol. Cell. Biol. 2018, 38, e00086-18. [Google Scholar] [CrossRef]
- Dorca-Arévalo, J.; Blanch, M.; Pradas, M.; Blasi, J. Epsilon Toxin from Clostridium Perfringens Induces Cytotoxicity in FRT Thyroid Epithelial Cells. Anaerobe 2018, 53, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Morcrette, H.; Bokori-Brown, M.; Ong, S.; Bennett, L.; Wren, B.W.; Lewis, N.; Titball, R.W. Clostridium Perfringens Epsilon Toxin Vaccine Candidate Lacking Toxicity to Cells Expressing Myelin and Lymphocyte Protein. NPJ Vaccines 2019, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Linden, J.R.; Ma, Y.; Zhao, B.; Harris, J.M.; Rumah, K.R.; Schaeren-Wiemers, N.; Vartanian, T. Clostridium Perfringens Epsilon Toxin Causes Selective Death of Mature Oligodendrocytes and Central Nervous System Demyelination. mBio 2015, 6, e02513-14. [Google Scholar] [CrossRef]
- Adler, D.; Linden, J.R.; Shetty, S.V.; Ma, Y.; Bokori-Brown, M.; Titball, R.W.; Vartanian, T. Clostridium Perfringens Epsilon Toxin Compromises the Blood-Brain Barrier in a Humanized Zebrafish Model. iScience 2019, 15, 39–54. [Google Scholar] [CrossRef]
- Dorca-Arévalo, J.; Dorca, E.; Torrejón-Escribano, B.; Blanch, M.; Martín-Satué, M.; Blasi, J. Lung Endothelial Cells Are Sensitive to Epsilon Toxin from Clostridium Perfringens. Vet. Res. 2020, 51, 27. [Google Scholar] [CrossRef]
- Linden, J.R.; Flores, C.; Schmidt, E.F.; Uzal, F.A.; Michel, A.O.; Valenzuela, M.; Dobrow, S.; Vartanian, T. Clostridium Perfringens Epsilon Toxin Induces Blood Brain Barrier Permeability via Caveolae-Dependent Transcytosis and Requires Expression of MAL. PLoS Pathog. 2019, 15, e1008014. [Google Scholar] [CrossRef]
- van Bunderen, C.C.; Bomers, M.K.; Wesdorp, E.; Peerbooms, P.; Veenstra, J. Clostridium Perfringens Septicaemia with Massive Intravascular Haemolysis: A Case Report and Review of the Literature. Neth. J. Med. 2010, 68, 343–346. [Google Scholar] [PubMed]
- Simon, T.G.; Bradley, J.; Jones, A.; Carino, G. Massive Intravascular Hemolysis From Clostridium Perfringens Septicemia: A Review. J. Inten. Care Med. 2014, 29, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Hashiba, M.; Tomino, A.; Takenaka, N.; Hattori, T.; Kano, H.; Tsuda, M.; Takeyama, N. Clostridium Perfringens Infection in a Febrile Patient with Severe Hemolytic Anemia. Am. J. Case Rep. 2016, 17, 219–223. [Google Scholar] [CrossRef]
- Gao, J.; Xin, W.; Huang, J.; Ji, B.; Gao, S.; Chen, L.; Kang, L.; Yang, H.; Shen, X.; Zhao, B.; et al. Hemolysis in Human Erythrocytes by Clostridium perfringens Epsilon Toxin Requires Activation of P2 Receptors. Virulence 2018, 9, 1601–1614. [Google Scholar] [CrossRef]
- Geng, Z.; Huang, J.; Kang, L.; Gao, S.; Yuan, Y.; Li, Y.; Wang, J.; Xin, W.; Wang, J. Clostridium Perfringens Epsilon Toxin Binds to Erythrocyte MAL Receptors and Triggers Phosphatidylserine Exposure. J. Cell. Mol. Med. 2020, 24, 7341–7352. [Google Scholar] [CrossRef]
- Rumah, K.R.; Eleso, O.E.; Fischetti, V.A. Human Blood Exposure to Clostridium Perfringens Epsilon Toxin May Shed Light on Erythrocyte Fragility during Active Multiple Sclerosis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Bryk, A.H.; Wiśniewski, J.R. Quantitative Analysis of Human Red Blood Cell Proteome. J. Proteome Res. 2017, 16, 2752–2761. [Google Scholar] [CrossRef]
- Alves, G.G.; Machado de Ávila, R.A.; Chávez-Olórtegui, C.D.; Lobato, F.C.F. Clostridium Perfringens Epsilon Toxin: The Third Most Potent Bacterial Toxin Known. Anaerobe 2014, 30, 102–107. [Google Scholar] [CrossRef]
- Binz, T.; Sikorra, S.; Mahrhold, S. Clostridial Neurotoxins: Mechanism of SNARE Cleavage and Outlook on Potential Substrate Specificity Reengineering. Toxins 2010, 2, 665–682. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, O.; Montecucco, C. Tetanus and botulinum neurotoxins. In Microbial Toxins; Gopalakrishnakone, P., Stiles, B., Alape-Girón, A., Dubreuil, J.D., Mandal, M., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 1–16. ISBN 978-94-007-6725-6. [Google Scholar]
- Jahn, R.; Scheller, R.H. SNAREs-Engines for Membrane Fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631. [Google Scholar] [CrossRef] [PubMed]
- Rizo, J.; Südhof, T. The Membrane Fusion Enigma: SNAREs, Sec1/Munc18 Proteins, and Their Accomplices-Guilty as Charged? Ann. Rev. Cell Dev. Biol. 2012, 28, 279–308. [Google Scholar] [CrossRef] [PubMed]
- Martin-Belmonte, F.; Kremer, L.; Albar, J.P.; Marazuela, M.; Alonso, M.A. Expression of the MAL Gene in the Thyroid: The MAL Proteolipid, a Component of Glycolipid-Enriched Membranes, Is Apically Distributed in Thyroid Follicles. Endocrinology 1998, 139, 2077–2084. [Google Scholar] [CrossRef][Green Version]
- Schuck, S.; Honsho, M.; Ekroos, K.; Shevchenko, A.; Simons, K. Resistance of Cell Membranes to Different Detergents. Proc. Natl. Acad. Sci. USA 2003, 100, 5795–5800. [Google Scholar] [CrossRef]
- Lajoie, P.; Goetz, J.G.; Dennis, J.W.; Nabi, I.R. Lattices, Rafts, and Scaffolds: Domain Regulation of Receptor Signaling at the Plasma Membrane. J. Cell Biol. 2009, 185, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ariotti, N.; Rae, J.; Liang, H.; Tillu, V.; Tee, S.; Bastiani, M.; Bademosi, A.T.; Collins, B.M.; Meunier, F.A.; et al. Caveolin-1 and Cavin1 Act Synergistically to Generate a Unique Lipid Environment in Caveolae. J. Cell Biol. 2021, 220, e202005138. [Google Scholar] [CrossRef] [PubMed]
- Tall, R.D.; Alonso, M.A.; Roth, M.G. Features of Influenza HA Required for Apical Sorting Differ from Those Required for Association with DRMs or MAL. Traffic 2003, 4, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Dukhovny, A.; Goldstein Magal, L.; Hirschberg, K. The MAL Proteolipid Restricts Detergent-Mediated Membrane Pore Expansion and Percolation. Mol. Membr. Biol. 2006, 23, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Heberle, F.A.; Feigenson, G.W. Phase Separation in Lipid Membranes. Cold Spring Harb. Perspect. Biol. 2011, 3, a004630. [Google Scholar] [CrossRef]
- Puertollano, R.; Li, S.; Lisanti, M.P.; Alonso, M.A. Recombinant Expression of the MAL Proteolipid, a Component of Glycolipid-Enriched Membrane Microdomains, Induces the Formation of Vesicular Structures in Insect Cells. J. Biol. Chem. 1997, 272, 18311–18315. [Google Scholar] [CrossRef]
- Li, S.; Song, K.S.; Koh, S.S.; Kikuchi, A.; Lisanti, M.P. Baculovirus-Based Expression of Mammalian Caveolin in Sf21 Insect Cells. J. Biol. Chem. 1996, 271, 28647–28654. [Google Scholar] [CrossRef]
- Jackson, M.R.; Nilsson, T.; Peterson, P.A. Identification of a Consensus Motif for Retention of Transmembrane Proteins in the Endoplasmic Reticulum. EMBO J. 1990, 9, 3153–3162. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.R.; Nilsson, T.; Peterson, P.A. Retrieval of Transmembrane Proteins to the Endoplasmic Reticulum. J. Cell Biol. 1993, 121, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Itin, C.; Kappeler, F.; Linstedt, A.D.; Hauri, H.P. A Novel Endocytosis Signal Related to the KKXX ER-Retrieval Signal. EMBO J. 1995, 14, 2250–2256. [Google Scholar] [CrossRef] [PubMed]
- Puertollano, R.; Alonso, M.A. Substitution of the Two Carboxyl-Terminal Serines by Alanine Causes Retention of MAL, a Component of the Apical Sorting Machinery, in the Endoplasmic Reticulum. Biochem. Biophys. Res. Commun. 1999, 260, 188–192. [Google Scholar] [CrossRef]
- Alonso, M.A.; Barton, D.E.; Francke, U. Assignment of the T-Cell Differentiation Gene MAL to Human Chromosome 2, Region Cen→Q13. Immunogenetics 1988, 27, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Rancaño, C.; Rubio, T.; Alonso, M.A. Alternative Splicing of Human T-Cell-Specific MAL mRNA and Its Correlation with the Exon/Intron Organization of the Gene. Genomics 1994, 21, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG Islands and the Regulation of Transcription. Gene Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The Molecular Hallmarks of Epigenetic Control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in Epigenetics Link Genetics to the Environment and Disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA Methylation and DNA Methyltransferases. Epigenet. Chrom. 2017, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Esteller, M. DNA Methylation and Cancer. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2010; Volume 70, pp. 27–56. ISBN 978-0-12-380866-0. [Google Scholar]
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic Modulators, Modifiers and Mediators in Cancer Aetiology and Progression. Nat. Rev. Genet. 2016, 17, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic Plasticity and the Hallmarks of Cancer. Science 2017, 357. [Google Scholar] [CrossRef]
- Smith, J.; Sen, S.; Weeks, R.J.; Eccles, M.R.; Chatterjee, A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer 2020, 6, 392–406. [Google Scholar] [CrossRef]
- Tugores, A.; Rubio, T.; Rancano, C.; Alonso, M.A. A Tandem Array of Sp-1 Sites and a Reverse Initiator Element Are Both Required for Synergistic Transcriptional Activation of the T-Cell-Specific MAL Gene. DNA Cell Biol. 1997, 16. [Google Scholar] [CrossRef]
- Lind, G.E.; Ahlquist, T.; Lothe, R.A. DNA Hypermethylation of MAL: A Promising Diagnostic Biomarker for Colorectal Tumors. Gastroenterology 2007, 132, 1631–1632. [Google Scholar] [CrossRef]
- Lind, G.E.; Ahlquist, T.; Kolberg, M.; Berg, M.; Ekneas, M.; Alonso, M.A.; Kallioniemi, A.; Meling, G.I.; Skotheim, R.I.; Rognum, T.O.; et al. Hypermethylated MAL Gene—A Silent Marker of Early Colon Tumorigenesis. J. Transl. Med. 2008, 6, 13. [Google Scholar] [CrossRef]
- Ahlquist, T.; Lind, G.E.; Costa, V.L.; Meling, G.I.; Vatn, M.; Hoff, G.S.; Rognum, T.O.; Skotheim, R.I.; Thiis-Evensen, E.; Lothe, R.A. Gene Methylation Profiles of Normal Mucosa, and Benign and Malignant Colorectal Tumors Identify Early Onset Markers. Mol. Cancer 2008, 7, 94. [Google Scholar] [CrossRef]
- Buffart, T.E.; Overmeer, R.M.; Steenbergen, R.D.M.; Tijssen, M.; van Grieken, N.C.T.; Snijders, P.J.F.; Grabsch, H.I.; van de Velde, C.J.H.; Carvalho, B.; Meijer, G.A. MAL Promoter Hypermethylation as a Novel Prognostic Marker in Gastric Cancer. Br. J. Cancer 2008, 99, 1802–1807. [Google Scholar] [CrossRef]
- Horne, H.N.; Lee, P.S.; Murphy, S.K.; Alonso, M.A.; Olson, J.A.; Marks, J.R. Inactivation of the MAL Gene in Breast Cancer Is a Common Event That Predicts Benefit from Adjuvant Chemotherapy. Mol. Cancer Res. 2009, 7, 199. [Google Scholar] [CrossRef]
- Overmeer, R.M.; Henken, F.E.; Bierkens, M.; Wilting, S.M.; Timmerman, I.; Meijer, C.J.L.M.; Snijders, P.J.F.; Steenbergen, R.D.M. Repression of MAL Tumour Suppressor Activity by Promoter Methylation during Cervical Carcinogenesis. J. Pathol. 2009, 219, 327–336. [Google Scholar] [CrossRef]
- Cao, W.; Zhang, Z.; Xu, Q.; Sun, Q.; Yan, M.; Zhang, J.; Zhang, P.; Han, Z.; Chen, W. Epigenetic Silencing of MAL, a Putative Tumor Suppressor Gene, Can Contribute to Human Epithelium Cell Carcinoma. Mol. Cancer 2010, 9, 296. [Google Scholar] [CrossRef]
- Jin, Z.; Wang, L.; Zhang, Y.; Cheng, Y.; Gao, Y.; Feng, X.; Dong, M.; Cao, Z.; Chen, S.; Yu, H.; et al. MAL Hypermethylation Is a Tissue-Specific Event That Correlates with MAL mRNA Expression in Esophageal Carcinoma. Sci. Rep. 2013, 3, 2838. [Google Scholar] [CrossRef]
- Suzuki, M.; Shiraishi, K.; Eguchi, A.; Ikeda, K.; Mori, T.; Yoshimoto, K.; Ohba, Y.; Yamada, T.; Ito, T.; Baba, Y.; et al. Aberrant Methylation of LINE-1, SLIT2, MAL and IGFBP7 in Non-Small Cell Lung Cancer. Oncol. Rep. 2013, 29, 1308–1314. [Google Scholar] [CrossRef]
- Vasiljevic, N.; Scibior-Bentkowska, D.; Brentnall, A.R.; Cuzick, J.; Lorincz, A.T. Credentialing of DNA Methylation Assays for Human Genes as Diagnostic Biomarkers of Cervical Intraepithelial Neoplasia in High-Risk HPV Positive Women. Gynecol. Oncol. 2014, 132, 709–714. [Google Scholar] [CrossRef]
- Ahmad, A.S.; Vasiljevic, N.; Carter, P.; Berney, D.M.; Moller, H.; Foster, C.S.; Cuzick, J.; Lorincz, A.T. A Novel DNA Methylation Score Accurately Predicts Death from Prostate Cancer in Men with Low to Intermediate Clinical Risk Factors. Oncotarget 2015, 7, 71833–71840. [Google Scholar] [CrossRef]
- Huang, K.T.; Mikeska, T.; Li, J.; Takano, E.A.; Millar, E.K.A.; Graham, P.H.; Boyle, S.E.; Campbell, I.G.; Speed, T.P.; Dobrovic, A.; et al. Assessment of DNA Methylation Profiling and Copy Number Variation as Indications of Clonal Relationship in Bilateral and Contralateral Breast Cancers to Distinguish Recurrent Breast Cancer from a Second Primary Tumour. BMC Cancer 2015, 15, 669. [Google Scholar] [CrossRef][Green Version]
- Kalmár, A.; Péterfia, B.; Hollósi, P.; Galamb, O.; Spisák, S.; Wichmann, B.; Bodor, A.; Tóth, K.; Patai, Ä.V.; Valcz, G.; et al. DNA Hypermethylation and Decreased mRNA Expression of MAL, PRIMA1, PTGDR and SFRP1 in Colorectal Adenoma and Cancer. BMC Cancer 2015, 15, 736. [Google Scholar] [CrossRef]
- Patai, A.V.; Valcz, G.; Hollösi, P.; Kalmár, A.; Péterfia, B.; Patai, Ä.; Wichmann, B.; Spisák, S.; Barták, B.K.; Leiszter, K.; et al. Comprehensive DNA Methylation Analysis Reveals a Common Ten-Gene Methylation Signature in Colorectal Adenomas and Carcinomas. PLoS ONE 2015, 10, e0133836. [Google Scholar] [CrossRef]
- Teneng, I.; Tellez, C.S.; Picchi, M.A.; Klinge, D.M.; Yingling, C.M.; Snider, A.M.; Liu, Y.; Belinsky, S.A. Global Identification of Genes Targeted by DNMT3b for Epigenetic Silencing in Lung Cancer. Oncogene 2015, 34, 621–630. [Google Scholar] [CrossRef]
- Choi, B.; Han, T.-S.; Min, J.; Hur, K.; Lee, S.-M.; Lee, H.-J.; Kim, Y.-J.; Yang, H.-K. MAL and TMEM220 Are Novel DNA Methylation Markers in Human Gastric Cancer. Biomarkers 2016, 22, 35–44. [Google Scholar] [CrossRef]
- Hassan, Z.K.; Hafez, M.M.; Kamel, M.M.; Zekri, A.R.N. Human Papillomavirus Genotypes and Methylation of CADM1, PAX1, MAL and ADCYAP1 Genes in Epithelial Ovarian Cancer Patients. Asian Pac. J. Cancer Prev. 2017, 18, 169–176. [Google Scholar] [CrossRef]
- Sambuudash, O.; Kim, H.-S.; Cho, M.Y. Lack of Aberrant Methylation in an Adjacent Area of Left-Sided Colorectal Cancer. Yonsei Med. J. 2017, 58, 749–755. [Google Scholar] [CrossRef]
- Verhoef, V.M.J.; Bosgraaf, R.P.; van Kemenade, F.J.; Rozendaal, L.; Heideman, D.A.M.; Hesselink, A.T.; Bekkers, R.L.M.; Steenbergen, R.D.M.; Massuger, L.F.A.G.; Melchers, W.J.G.; et al. Triage by Methylation-Marker Testing versus Cytology in Women Who Test HPV-Positive on Self-Collected Cervicovaginal Specimens (PROHTECT-3): A Randomised Controlled Non-Inferiority Trial. Lancet Oncol. 2017, 15, 315–322. [Google Scholar] [CrossRef]
- Holubekova, V.; Mersakova, S.; Grendar, M.; Snahnicanova, Z.; Kudela, E.; Kalman, M.; Lasabova, Z.; Danko, J.; Zubor, P. The Role of CADM1 and MAL Promoter Methylation in Inflammation and Cervical Intraepithelial Neoplasia. Genet. Test. Mol. Biomark. 2020, 24, 256–263. [Google Scholar] [CrossRef]
- Olkhov-Mitsel, E.; Bapat, B. Strategies for Discovery and Validation of Methylated and Hydroxymethylated DNA Biomarkers. Cancer Med. 2012, 1, 237–260. [Google Scholar] [CrossRef] [PubMed]
- Kurdyukov, S.; Bullock, M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel) 2016, 5, 3. [Google Scholar] [CrossRef]
- Galardi, F.; De Luca, F.; Romagnoli, D.; Biagioni, C.; Moretti, E.; Biganzoli, L.; Di Leo, A.; Migliaccio, I.; Malorni, L.; Benelli, M. Cell-Free DNA-Methylation-Based Methods and Applications in Oncology. Biomolecules 2020, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Mimori, K.; Shiraishi, T.; Mashino, K.; Sonoda, H.; Yamashita, K.; Yoshinaga, K.; Masuda, T.; Utsunomiya, T.; Alonso, M.A.; Inoue, H.; et al. MAL Gene Expression in Esophageal Cancer Suppresses Motility, Invasion and Tumorigenicity and Enhances Apoptosis through the Fas Pathway. Oncogene 2003, 22, 3463–3471. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beder, L.B.; Gunduz, M.; Hotomi, M.; Fujihara, K.; Shimada, J.; Tamura, S.; Gunduz, E.; Fukushima, K.; Yaykasli, K.; Grenman, R.; et al. T-Lymphocyte Maturation-Associated Protein Gene as a Candidate Metastasis Suppressor for Head and Neck Squamous Cell Carcinomas. Cancer Sci. 2009, 100, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Teaberry, V.S.; Bland, A.E.; Huang, Z.; Whitaker, R.S.; Baba, T.; Fujii, S.; Secord, A.A.; Berchuck, A.; Murphy, S.K. Elevated MAL Expression Is Accompanied by Promoter Hypomethylation and Platinum Resistance in Epithelial Ovarian Cancer. Int. J. Cancer 2010, 126, 1378–1389. [Google Scholar] [CrossRef]
- Copie-Bergman, C.; Gaulard, P.; Maouche-Chretien, L.; Briere, J.; Haioun, C.; Alonso, M.A.; Romeo, P.-H.; Leroy, K. The MAL Gene Is Expressed in Primary Mediastinal Large B-Cell Lymphoma. Blood 1999, 94, 3567–3575. [Google Scholar] [CrossRef]
- Tracey, L.; Villuendas, R.; Ortiz, P.; Dopazo, A.; Spiteri, I.; Lombardia, L.; Rodríguez-Peralto, J.L.; Fernández-Herrera, J.; Hernández, A.; Fraga, J.; et al. Identification of Genes Involved in Resistance to Interferon-Alpha in Cutaneous T-Cell Lymphoma. Am. J. Pathol. 2002, 161, 1825–1837. [Google Scholar] [CrossRef]
- Pileri, S.A.; Gaidano, G.; Zinzani, P.L.; Falini, B.; Gaulard, P.; Zucca, E.; Pieri, F.; Berra, E.; Sabattini, E.; Ascani, S.; et al. Primary Mediastinal B-Cell Lymphoma: High Frequency of BCL-6 Mutations and Consistent Expression of the Transcription Factors OCT-2, BOB.1, and PU.1 in the Absence of Immunoglobulins. Am. J. Pathol. 2003, 162, 243–253. [Google Scholar] [CrossRef]
- Traverse-Glehen, A.; Pittaluga, S.; Gaulard, P.; Sorbara, L.; Alonso, M.A.; Raffeld, M.; Jaffe, E.S. Mediastinal Gray Zone Lymphoma: The Missing Link between Classic Hodgkin’s Lymphoma and Mediastinal Large B-Cell Lymphoma. Am. J. Surg. Pathol. 2005, 29, 1411–1421. [Google Scholar] [CrossRef]
- Hsi, E.D.; Sup, S.J.; Alemany, C.; Tso, E.; Skacel, M.; Elson, P.; Alonso, M.A.; Pohlman, B. MAL Is Expressed in a Subset of Hodgkin Lymphoma and Identifies a Population of Patients with Poor Prognosis. Am. J. Clin. Pathol. 2006, 125, 776. [Google Scholar] [CrossRef]
- Berchuck, A.; Iversen, E.S.; Luo, J.; Clarke, J.P.; Horne, H.; Levine, D.A.; Boyd, J.; Alonso, M.A.; Secord, A.A.; Bernardini, M.Q.; et al. Microarray Analysis of Early Stage Serous Ovarian Cancers Shows Profiles Predictive of Favorable Outcome. Clin. Cancer Res. 2009, 15, 2448. [Google Scholar] [CrossRef]
- Fiches, G.N.; Zhou, D.; Kong, W.; Biswas, A.; Ahmed, E.H.; Baiocchi, R.A.; Zhu, J.; Santoso, N. Profiling of Immune Related Genes Silenced in EBV-Positive Gastric Carcinoma Identified Novel Restriction Factors of Human Gammaherpesviruses. PLoS Pathog. 2020, 16, e1008778. [Google Scholar] [CrossRef]
- Vasiljević, N.; Ahmad, A.S.; Thorat, M.A.; Fisher, G.; Berney, D.M.; Møller, H.; Foster, C.S.; Cuzick, J.; Lorincz, A.T. DNA Methylation Gene-Based Models Indicating Independent Poor Outcome in Prostate Cancer. BMC Cancer 2014, 14, 655. [Google Scholar] [CrossRef]
- Maruya, S.; Kim, H.-W.; Weber, R.S.; Lee, J.J.; Kies, M.; Luna, M.A.; Batsakis, J.G.; El-Naggar, A.K. Gene Expression Screening of Salivary Gland Neoplasms: Molecular Markers of Potential Histogenetic and Clinical Significance. J. Mol. Diagnost. 2004, 6, 180–190. [Google Scholar] [CrossRef]
- Pal, S.K.; Noguchi, S.; Yamamoto, G.; Yamada, A.; Isobe, T.; Hayashi, S.; Tanaka, J.-I.; Tanaka, Y.; Kamijo, R.; Yamane, G.-Y.; et al. Expression of Myelin and Lymphocyte Protein (MAL) in Oral Carcinogenesis. Med. Mol. Morphol. 2012, 45, 222–228. [Google Scholar] [CrossRef]
- Bhosale, P.G.; Cristea, S.; Ambatipudi, S.; Desai, R.S.; Kumar, R.; Patil, A.; Kane, S.; Borges, A.M.; Schäffer, A.A.; Beerenwinkel, N.; et al. Chromosomal Alerations and Gene Expression Changes Associated with the Progression of Leukoplakia to Advanced Gingivobuccal Cancer. Transl. Oncol. 2017, 10, 396–409. [Google Scholar] [CrossRef]
- Mori, Y.; Cai, K.; Cheng, Y.; Wang, S.; Paun, B.; Hamilton, J.P.; Jin, Z.; Sato, F.; Berki, A.T.; Kan, T.; et al. A Genome-Wide Search Identifies Epigenetic Silencing of Somatostatin, Tachykinin-1, and 5 Other Genes in Colon Cancer. Gastroenterology 2006, 131, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bi, H.; Ge, A.; Xia, T.; Fu, J.; Liu, Y.; Sun, H.; Li, D.; Zhao, Y. DNA Hypermethylation of MAL Gene May Act as an Independent Predictor of Favorable Prognosis in Patients with Colorectal Cancer. Transl. Cancer Res. 2019, 8, 1985–1996. [Google Scholar] [CrossRef]
- Bierkens, M.; Hesselink, A.T.; Meijer, C.J.L.M.; Heideman, D.A.M.; Wisman, G.B.A.; van der Zee, A.G.J.; Snijders, P.J.F.; Steenbergen, R.D.M. CADM1 and MAL Promoter Methylation Levels in HrHPV-Positive Cervical Scrapes Increase Proportional to Degree and Duration of Underlying Cervical Disease: DNA Methylation in HrHPV-Positive Cervical Scrapes. Int. J. Cancer 2013, 133, 1293–1299. [Google Scholar] [CrossRef]
- De Strooper, L.M.A.; van Zummeren, M.; Steenbergen, R.D.M.; Bleeker, M.C.G.; Hesselink, A.T.; Wisman, G.B.A.; Snijders, P.J.F.; Heideman, D.A.M.; Meijer, C.J.L.M. CADM1, MAL and miR124-2 Methylation Analysis in Cervical Scrapes to Detect Cervical and Endometrial Cancer. J. Clin. Pathol. 2014, 67, 1067–1071. [Google Scholar] [CrossRef]
- De Vuyst, H.; Franceschi, S.; Plummer, M.; Mugo, N.R.; Sakr, S.R.; Meijer, C.J.L.M.; Heideman, D.A.M.; Tenet, V.; Snijders, P.J.F.; Hesselink, A.T.; et al. Methylation Levels of CADM1, MAL, and mIR124-2 in Cervical Scrapes for Triage of HIV-Infected, High-Risk HPV-Positive Women in Kenya. J. Acquir. Immune Defic. Syndr. 2015, 70, 311–318. [Google Scholar] [CrossRef]
- Ki, E.Y.; Lee, K.H.; Hur, S.Y.; Rhee, J.E.; Kee, M.K.; Kang, C.; Park, J.S. Methylation of Cervical Neoplastic Cells Infected With Human Papillomavirus 16. Int. J. Gynecol. Cancer 2016, 26, 176–183. [Google Scholar] [CrossRef]
- Kim, M.-K.; Lee, I.-H.; Lee, K.-H.; Lee, Y.K.; So, K.A.; Hong, S.R.; Hwang, C.-S.; Kee, M.-K.; Rhee, J.E.; Kang, C.; et al. DNA Methylation in Human Papillomavirus-Infected Cervical Cells Is Elevated in High-Grade Squamous Intraepithelial Lesions and Cancer. J. Gynecol. Oncol. 2016, 27, e14. [Google Scholar] [CrossRef]
- Uijterwaal, M.H.; van Zummeren, M.; Kocken, M.; Luttmer, R.; Berkhof, J.; Witte, B.I.; van Baal, W.M.; Graziosi, G.C.M.; Verheijen, R.H.M.; Helmerhorst, T.J.M.; et al. Performance of CADM1/MAL-Methylation Analysis for Monitoring of Women Treated for High-Grade CIN. Gynecol. Oncol. 2016, 143, 135–142. [Google Scholar] [CrossRef]
- van Baars, R.; van der Marel, J.; Snijders, P.J.F.; Rodriquez-Manfredi, A.; ter Harmsel, B.; van den Munckhof, H.A.M.; Ordi, J.; del Pino, M.; van de Sandt, M.M.; Wentzensen, N.; et al. CADM1 and MAL Methylation Status in Cervical Scrapes Is Representative of the Most Severe Underlying Lesion in Women with Multiple Cervical Biopsies: CADM1 and MAL Methylation on Lesion Level. Int. J. Cancer 2016, 138, 463–471. [Google Scholar] [CrossRef]
- Clarke, M.A.; Luhn, P.; Gage, J.C.; Bodelon, C.; Dunn, S.T.; Walker, J.; Zuna, R.; Hewitt, S.; Killian, J.K.; Yan, L.; et al. Discovery and Validation of Candidate Host DNA Methylation Markers for Detection of Cervical Precancer and Cancer. Int. J. Cancer 2017, 141, 701–710. [Google Scholar] [CrossRef]
- Fiano, V.; Trevisan, M.; Fasanelli, F.; Grasso, C.; Marabese, F.; da Graça Bicalho, M.; de Carvalho, N.S.; Maestri, C.A.; Merletti, F.; Sacerdote, C.; et al. Methylation in Host and Viral Genes as Marker of Aggressiveness in Cervical Lesions: Analysis in 543 Unscreened Women. Gynecol. Oncol. 2018, 151, 319–326. [Google Scholar] [CrossRef]
- Meršaková, S.; Holubeková, V.; Grendár, M.; Višňovsky, J.; Ňachajová, M.; Kalman, M.; Kúdela, E.; Žúbor, P.; Bielik, T.; Lasabová, Z.; et al. Methylation of CADM1 and MAL Together with HPV Status in Cytological Cervical Specimens Serves an Important Role in the Progression of Cervical Intraepithelial Neoplasia. Oncol. Lett. 2018. [Google Scholar] [CrossRef]
- van Zummeren, M.; Kremer, W.W.; Leeman, A.; Bleeker, M.C.G.; Jenkins, D.; van de Sandt, M.; Doorbar, J.; Heideman, D.A.M.; Steenbergen, R.D.M.; Snijders, P.J.F.; et al. HPV E4 Expression and DNA Hypermethylation of CADM1, MAL, and miR124-2 Genes in Cervical Cancer and Precursor Lesions. Mod. Pathol. 2018, 31, 1842–1850. [Google Scholar] [CrossRef]
- del Pino, M.; Sierra, A.; Marimon, L.; Martí Delgado, C.; Rodriguez-Trujillo, A.; Barnadas, E.; Saco, A.; Torné, A.; Ordi, J. CADM1, MAL, and miR124 Promoter Methylation as Biomarkers of Transforming Cervical Intraepithelial Lesions. Int. J. Mol. Sci. 2019, 20, 2262. [Google Scholar] [CrossRef]
- Blaveri, E.; Simko, J.P.; Korkola, J.E.; Brewer, J.L.; Baehner, F.; Mehta, K.; DeVries, S.; Koppie, T.; Pejavar, S.; Carroll, P.; et al. Bladder Cancer Outcome and Subtype Classification by Gene Expression. Clin. Cancer Res. 2005, 11, 4044–4055. [Google Scholar] [CrossRef]
- Iwasaki, T.; Matsushita, M.; Nonaka, D.; Nagata, K.; Kato, M.; Kuwamoto, S.; Murakami, I.; Hayashi, K. Lower Expression of CADM1 and Higher Expression of MAL in Merkel Cell Carcinomas Are Associated with Merkel Cell Polyomavirus Infection and Better Prognosis. Hum. Pathol. 2016, 48, 1–8. [Google Scholar] [CrossRef]
- Dasari, S.; Bernard Tchounwou, P. Cisplatin in Cancer Therapy: Molecular Mechanisms of Action. Eur J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P.B. Resistance to Cis- and Carboplatin Initiated by Epigenetic Changes in Ovarian Cancer Patients. Cancer Drug Resist. 2019. [Google Scholar] [CrossRef]
- Citron, F.; Fabris, L. Targeting Epigenetic Dependencies in Solid Tumors: Evolutionary Landscape Beyond Germ Layers Origin. Cancers 2020, 12, 682. [Google Scholar] [CrossRef]
- Zanotti, L.; Romani, C.; Tassone, L.; Todeschini, P.; Tassi, R.A.; Bandiera, E.; Damia, G.; Ricci, F.; Ardighieri, L.; Calza, S.; et al. MAL Gene Overexpression as a Marker of High-Grade Serous Ovarian Carcinoma Stem-like Cells That Predicts Chemoresistance and Poor Prognosis. BMC Cancer 2017, 17, 366. [Google Scholar] [CrossRef]
- Isaacson, P.G.; Norton, A.J.; Addis, B.J. The Human Thymus Contains a Novel Population of B Cells. Lancet 1987, 330, 1488–1491. [Google Scholar] [CrossRef]
- Hofmann, W.J.; Momburg, F.; Möller, P. Thymic Medullary Cells Expressing B Lymphocyte Antigens. Hum. Pathol. 1988, 19, 1280–1287. [Google Scholar] [CrossRef]
- Perera, J.; Huang, H. The Development and Function of Thymic B Cells. Cell. Mol. Life Sci. 2015, 72, 2657–2663. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.J. The Molecular Signature of Mediastinal Large B-Cell Lymphoma Differs from That of Other Diffuse Large B-Cell Lymphomas and Shares Features with Classical Hodgkin Lymphoma. Blood 2003, 102, 3871–3879. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chew, M.H.; Tham, C.K.; Tang, C.L.; Ong, S.Y.K.; Zhao, Y. Methylation of Serum SST Gene Is an Independent Prognostic Marker in Colorectal Cancer. Am. J. Cancer Res. 2016, 6, 2098–2108. [Google Scholar]
- Sherr, C.J. Principles of Tumor Suppression. Cell 2004, 116, 235–246. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetic Gene Silencing in Cancer: The DNA Hypermethylome. Hum. Mol. Genet. 2007, 16, R50–R59. [Google Scholar] [CrossRef]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic Silencing of Tumor Suppressor Genes: Paradigms, Puzzles, and Potential. Biochim. Biophys. Acta (BBA) Rev. Cancer 2016, 1865, 275–288. [Google Scholar] [CrossRef]
- Agostini, M.; Enzo, M.V.; Bedin, C.; Belardinelli, V.; Goldin, E.; Del Bianco, P.; Maschietto, E.; D’Angelo, E.; Izzi, L.; Saccani, A.; et al. Circulating Cell-Free DNA: A Promising Marker of Regional Lymphonode Metastasis in Breast Cancer Patients. Cancer Biomarkers 2012, 11, 89–98. [Google Scholar] [CrossRef]
- Mimori, K.; Nishida, K.; Nakamura, Y.; Ieta, K.; Yoshikawa, Y.; Sasaki, A.; Ishii, H.; Alonso, M.A.; Mori, M. Loss of MAL Expression in Precancerous Lesions of the Esophagus. Ann. Surg. Oncol. 2007, 14, 1670–1677. [Google Scholar] [CrossRef]
- Visser, E.; Franken, I.A.; Brosens, L.A.A.; Ruurda, J.P.; van Hillegersberg, R. Prognostic Gene Expression Profiling in Esophageal Cancer: A Systematic Review. Oncotarget 2017, 8, 5566–5577. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, M.; Cai, Y. MAL gene is down-regulated substantially in human esophageal cancer. Zhonghua Zhong Liu Za Zhi 1999, 21, 250–252. [Google Scholar] [PubMed]
- Wang, Z.; Wang, M.; Xu, X.; Xu, Z.; Han, Y.; Cai, Y.; Sun, Y.; Wu, M. Studies of MAL gene in human esophageal cancer by RNA in situ hybridization. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2000, 17, 329–331. [Google Scholar]
- Yue, Y.; Song, M.; Qiao, Y.; Li, P.; Yuan, Y.; Lian, J.; Wang, S.; Zhang, Y. Gene Function Analysis and Underlying Mechanism of Esophagus Cancer Based on Microarray Gene Expression Profiling. Oncotarget 2017, 8, 105222–105237. [Google Scholar] [CrossRef]
- Wang, X.; Li, G.; Luo, Q.; Gan, C. Identification of Crucial Genes Associated with Esophageal Squamous Cell Carcinoma by Gene Expression Profile Analysis. Oncol. Lett. 2018. [Google Scholar] [CrossRef]
- Kurashige, J.; Sawada, G.; Takahashi, Y.; Eguchi, H.; Sudo, T.; Ikegami, T.; Yoshizumi, T.; Soejima, Y.; Ikeda, T.; Kawanaka, H.; et al. Suppression of MAL Gene Expression in Gastric Cancer Correlates with Metastasis and Mortality. Fukuoka Igaku Zasshi 2013, 104, 344–349. [Google Scholar] [PubMed]
- Ma, R.; Xu, Y.E.; Wang, M.; Peng, W.E.I. Suppression of MAL Gene Expression Is Associated with Colorectal Cancer Metastasis. Oncol. Lett. 2016, 10, 957–961. [Google Scholar] [CrossRef]
- Lind, G.E.; Danielsen, S.A.; Ahlquist, T.; Merok, M.A.; Andresen, K.; Skotheim, R.I.; Hektoen, M.; Rognum, T.O.; Meling, G.I.; Hoff, G.; et al. Identification of an Epigenetic Biomarker Panel with High Sensitivity and Specificity for Colorectal Cancer and Adenomas. Mol. Cancer 2011, 10, 85. [Google Scholar] [CrossRef]
- Schwartz, D.R.; Kardia, S.L.R.; Shedden, K.A.; Kuick, R.; Michailidis, G.; Taylor, J.M.G.; Misek, D.E.; Wu, R.; Zhai, Y.; Darrah, D.M.; et al. Gene Expression in Ovarian Cancer Reflects Both Morphology and Biological Behavior, Distinguishing Clear Cell from Other Poor-Prognosis Ovarian Carcinomas. Cancer Res. 2002, 62, 4722–4729. [Google Scholar]
- Kulbe, H.; Otto, R.; Darb-Esfahani, S.; Lammert, H.; Abobaker, S.; Welsch, G.; Chekerov, R.; Schäfer, R.; Dragun, D.; Hummel, M.; et al. Discovery and Validation of Novel Biomarkers for Detection of Epithelial Ovarian Cancer. Cells 2019, 8, 713. [Google Scholar] [CrossRef]
- Berchuck, A.; Iversen, E.S.; Lancaster, J.M.; Pittman, J.; Luo, J.; Lee, P.; Murphy, S.; Dressman, H.K.; Febbo, P.G.; West, M.; et al. Patterns of Gene Expression That Characterize Long-Term Survival in Advanced Stage Serous Ovarian Cancers. Clin. Cancer Res. 2005, 11, 3686. [Google Scholar] [CrossRef]
- Lorincz, A.T. Virtues and Weaknesses of DNA Methylation as a Test for Cervical Cancer Prevention. Acta. Cytol. 2016, 60, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Ondič, O.; Němcová, J.; Alaghehbandan, R.; Černá, K.; Gomolčáková, B.; Kinkorová-Luňáčková, I.; Chytra, J.; Šidlová, H.; Májek, O.; Bouda, J. The Detection of DNA Methylation of Tumour Suppressor Genes in Cervical High-grade Squamous Intraepithelial Lesion: A Prospective Cytological-histological Correlation Study of 70 Cases. Cytopathology 2019, 30, 426–431. [Google Scholar] [CrossRef]
- Wilting, S.M.; de Wilde, J.; Meijer, C.J.L.M.; Berkhof, J.; Yi, Y.; van Wieringen, W.N.; Braakhuis, B.J.M.; Meijer, G.A.; Ylstra, B.; Snijders, P.J.F.; et al. Integrated Genomic and Transcriptional Profiling Identifies Chromosomal Loci with Altered Gene Expression in Cervical Cancer. Genes Chromosom. Cancer 2008, 47, 890–905. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.; Nagai, H.; Okino, K.; Onda, M.; Yoneyama, K.; Ohta, Y.; Nakayama, H.; Araki, T.; Emi, M. Down-Regulation of Members of Glycolipid-Enriched Membrane Raft Gene Family, MAL and BENE, in Cervical Squamous Cell Cancers. Obstet. Gynaecol. Res. 2004, 30, 53–58. [Google Scholar] [CrossRef]
- Sok, J.C.; Kuriakose, M.A.; Mahajan, V.B.; Pearlman, A.N.; DeLacure, M.D.; Chen, F.-A. Tissue-Specific Gene Expression of Head and Neck Squamous Cell Carcinoma In Vivo by Complementary DNA Microarray Analysis. Arch. Otolaryngol. Head Neck Surg. 2003, 129, 760. [Google Scholar] [CrossRef] [PubMed]
- Schmalbach, C.E.; Chepeha, D.B.; Giordano, T.J.; Rubin, M.A.; Teknos, T.N.; Bradford, C.R.; Wolf, G.T.; Kuick, R.; Misek, D.E.; Trask, D.K.; et al. Molecular Profiling and the Identification of Genes Associated with Metastatic Oral Cavity/Pharynx Squamous Cell Carcinoma. Arch. Otolaryngol. Head Neck Surg. 2004, 130, 295. [Google Scholar] [CrossRef] [PubMed]
- Belbin, T.J.; Singh, B.; Smith, R.V.; Socci, N.D.; Wreesmann, V.B.; Sanchez-Carbayo, M.; Masterson, J.; Patel, S.; Cordon-Cardo, C.; Prystowsky, M.B.; et al. Molecular Profiling of Tumor Progression in Head and Neck Cancer. Arch. Otolaryngol. Head Neck Surg. 2005, 131, 10. [Google Scholar] [CrossRef]
- Choi, P.; Chen, C. Genetic Expression Profiles and Biologic Pathway Alterations in Head and Neck Squamous Cell Carcinoma. Cancer 2005, 104, 1113–1128. [Google Scholar] [CrossRef]
- Riva, G.; Biolatti, M.; Pecorari, G.; Dell’Oste, V.; Landolfo, S. PYHIN Proteins and HPV: Role in the Pathogenesis of Head and Neck Squamous Cell Carcinoma. Microorganisms 2019, 8, 14. [Google Scholar] [CrossRef]
- Kornberg, L.J.; Villaret, D.; Popp, M.; Lui, L.; McLaren, R.; Brown, H.; Cohen, D.; Yun, J.; McFadden, M. Gene Expression Profiling in Squamous Cell Carcinoma of the Oral Cavity Shows Abnormalities in Several Signaling Pathways. Laryngoscope 2005, 115, 690–698. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, Y.; Gao, L.; Ye, Q.; Alonso, M.A. Expression pattern of MAL in normal epithelial cells, benign tumor, and squamous cell carcinoma of larynx. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi (J. Clin. Otorhinolaryngol. Head Neck Surg.) 2009, 23, 451–453. [Google Scholar]
- Misawa, K.; Imai, A.; Matsui, H.; Kanai, A.; Misawa, Y.; Mochizuki, D.; Mima, M.; Yamada, S.; Kurokawa, T.; Nakagawa, T.; et al. Identification of Novel Methylation Markers in HPV-Associated Oropharyngeal Cancer: Genome-Wide Discovery, Tissue Verification and Validation Testing in CtDNA. Oncogene 2020, 39, 4741–4755. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, A.E.; Nieuwenhuijzen, J.A.; Bosschieter, J.; van Splunter, A.P.; Lissenberg-Witte, B.I.; van der Voorn, J.P.; Segerink, L.I.; van Moorselaar, R.J.A.; Steenbergen, R.D.M. Comparative Analysis of Urine Fractions for Optimal Bladder Cancer Detection Using DNA Methylation Markers. Cancers (Basel) 2020, 12, 859. [Google Scholar] [CrossRef]
- Bosschieter, J.; Nieuwenhuijzen, J.A.; Hentschel, A.; van Splunter, A.P.; Segerink, L.I.; Vis, A.N.; Wilting, S.M.; Lissenberg-Witte, B.I.; van Moorselaar, R.J.A.; Steenbergen, R.D. A Two-Gene Methylation Signature for the Diagnosis of Bladder Cancer in Urine. Epigenomics 2019, 11, 337–347. [Google Scholar] [CrossRef]
- Chen, B.-J.; Ruminy, P.; Roth, C.G.; Bisig, B.; Mankel, B.; Steinhilber, J.; Bohers, E.; Jardin, F.; Fend, F.; Swerdlow, S.H.; et al. Cyclin D1-Positive Mediastinal Large B-Cell Lymphoma with Copy Number Gains of CCND1 Gene: A Study of 3 Cases With Nonmediastinal Disease. Am. J. Surg. Pathol. 2019, 43, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Kelley, T.W.; Pohlman, B.; Elson, P.; Hsi, E.D. The Ratio of FOXP3+ Regulatory T Cells to Granzyme B+ Cytotoxic T/NK Cells Predicts Prognosis in Classical Hodgkin Lymphoma and Is Independent of Bcl-2 and MAL Expression. Am. J. Clin. Pathol. 2007, 128, 958. [Google Scholar] [CrossRef]
- Jacquier, A.; Syrykh, C.; Bedgedjian, I.; Monnien, F.; Laurent, C.; Valmary-Degano, S.; Brousset, P. Immunohistochemistry with Anti-MAL Antibody and RNAscope with MAL Probes Are Complementary Techniques for Diagnosis of Primary Mediastinal Large B-Cell Lymphoma. J. Clin. Pathol. 2020. [Google Scholar] [CrossRef]
- Duns, G.; Viganò, E.; Ennishi, D.; Sarkozy, C.; Hung, S.S.; Chavez, E.A.; Takata, K.; Rushton, C.; Jiang, A.; Ben-Neriah, S.; et al. Characterization of DLBCL with a PMBL Gene Expression Signature. Blood 2021. [Google Scholar] [CrossRef]
- Mangasarova, J.K.; Misuyrin, A.V.; Magomedova, A.U.; Kravchenko, S.K. Molecular Signature Distinguishes Between Primary Mediastinal B-Cell Lymphoma and Diffuse Large B-Cell Lymphoma with Primary Involvment of Mediastinal Lymph Nodes. Blood 2011, 118, 2654. [Google Scholar] [CrossRef]
- Lara-Lemus, R. On the Role of Myelin and Lymphocyte Protein (MAL) in Cancer: A Puzzle with Two Faces. J. Cancer 2019, 10, 2312–2318. [Google Scholar] [CrossRef]
- Zimmerberg, J. How Can Proteolipids Be Central Players in Membrane Fusion? Trends Cell Biol. 2001, 11, 233–235. [Google Scholar] [CrossRef]
- Zimmerberg, J.; Kozlov, M.M. How Proteins Produce Cellular Membrane Curvature. Nat. Rev. Mol. Cell. Biol. 2006, 7, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Shivapurkar, N.; Gazdar, A. DNA Methylation Based Biomarkers in Non-Invasive Cancer Screening. Curr. Mol. Med. 2010, 10, 123–132. [Google Scholar] [CrossRef]
- Leygo, C.; Williams, M.; Jin, H.C.; Chan, M.W.Y.; Chu, W.K.; Grusch, M.; Cheng, Y.Y. DNA Methylation as a Noninvasive Epigenetic Biomarker for the Detection of Cancer. Dis. Markers 2017, 2017, 13. [Google Scholar] [CrossRef]
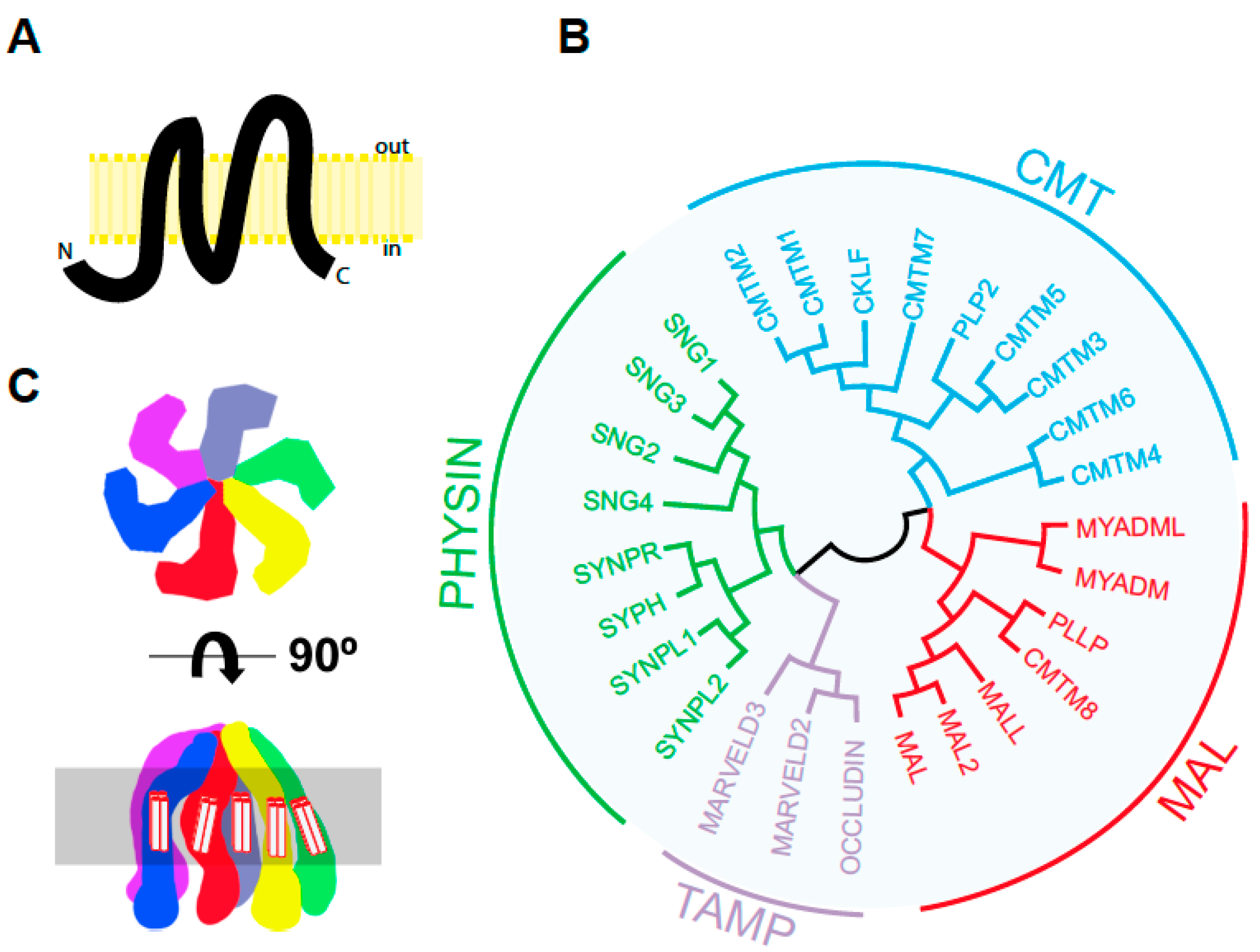
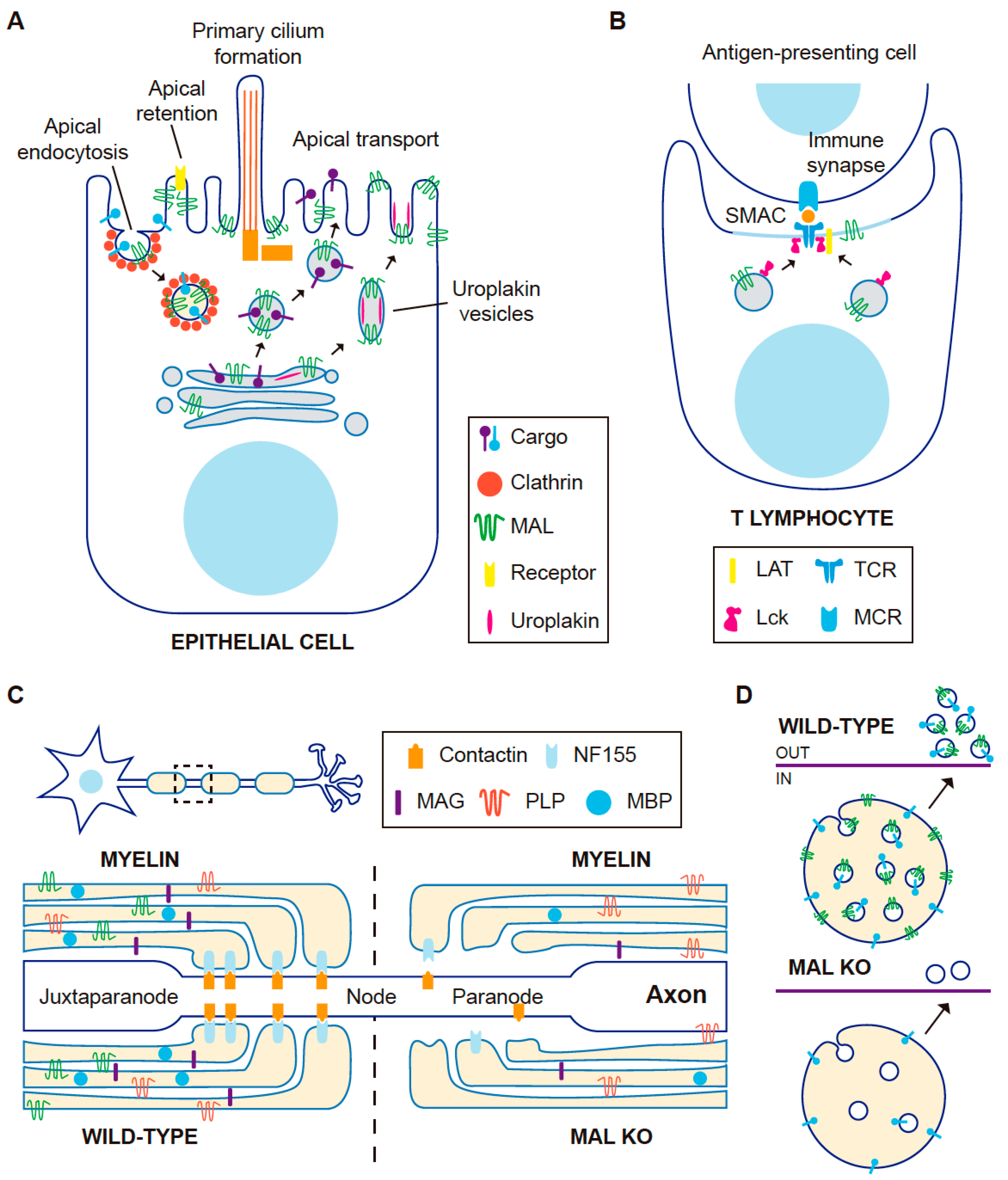
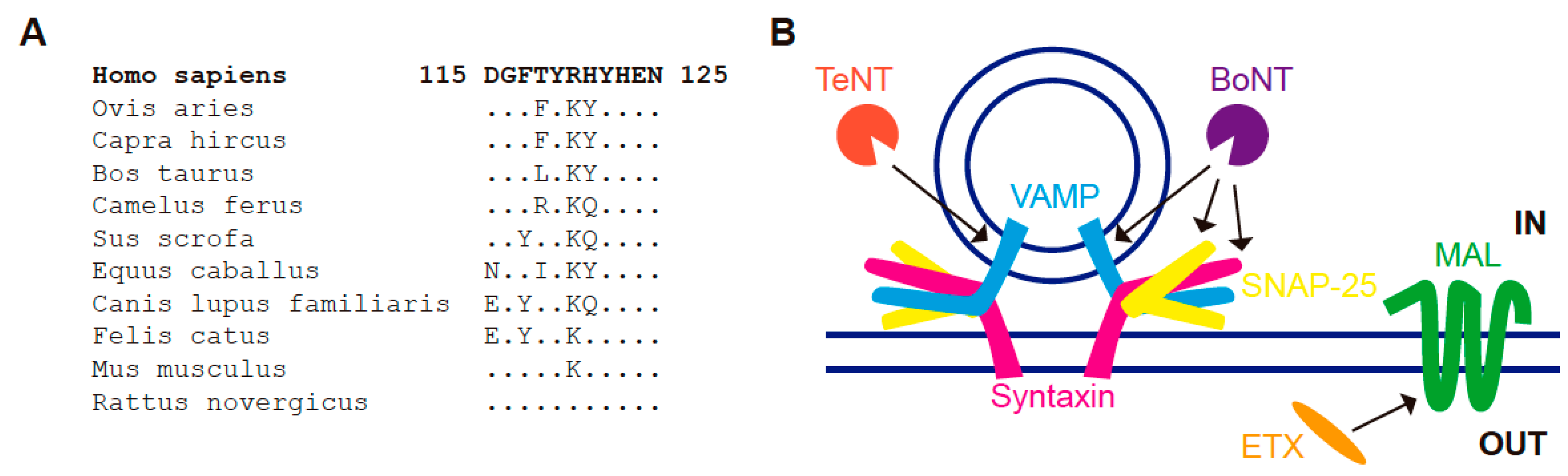
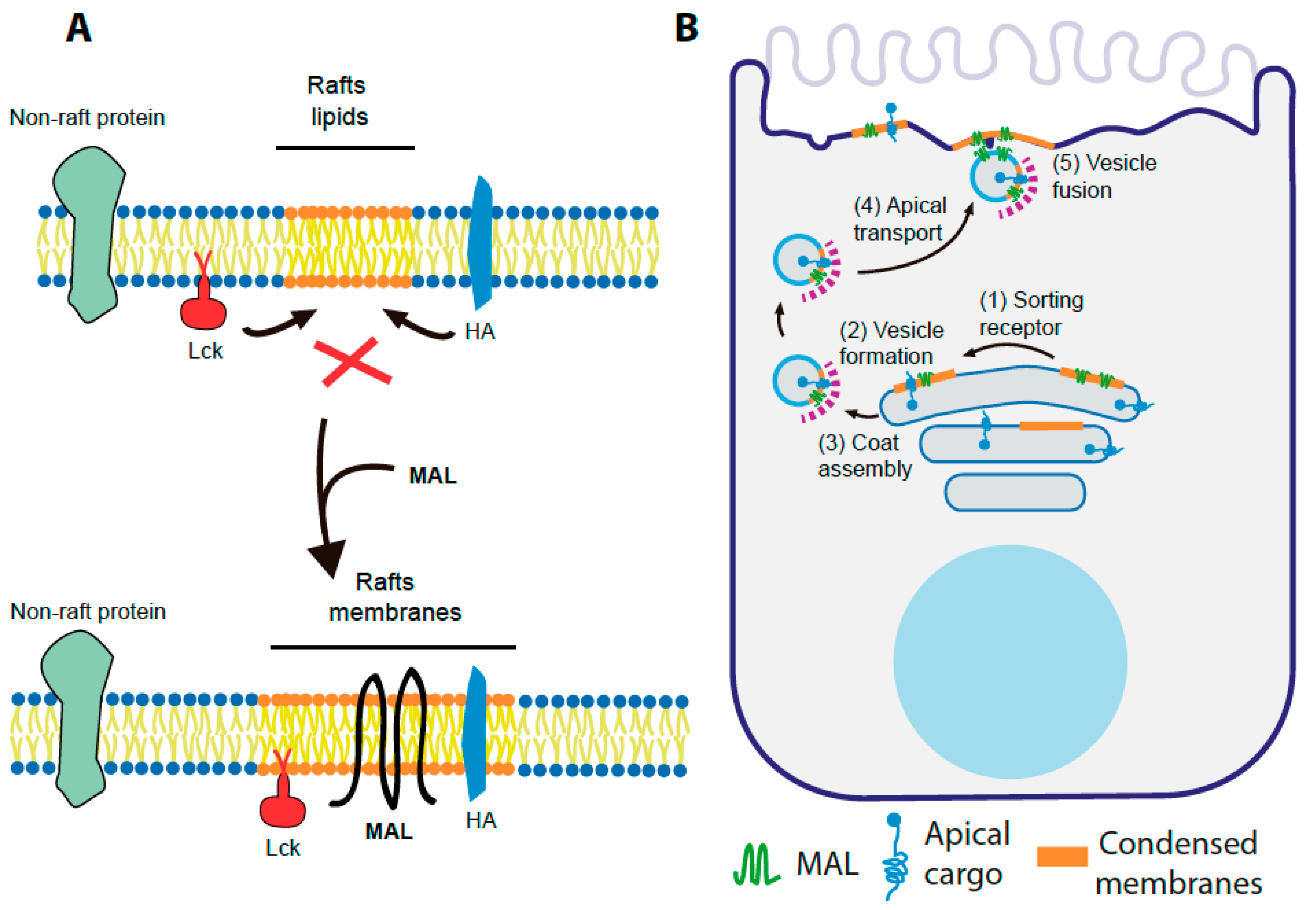

| Tissue/Organ | Cells Positive for MAL Expression |
|---|---|
| Esophagus | Stratified squamous epithelium |
| Stomach | Surface mucus cells, parietal cells, chief cells |
| Small intestine | Enterocytes, crypt cells, Paneth cells |
| Large intestine | Mucus cells |
| Pancreas | Acinar cells, ductal cells, endocrine cells |
| Kidney | Distal convoluted tubules, collecting tubules, loop of Henle |
| Bladder/ureter | Superficial cells from transitional epithelium |
| Prostate | Ductal and acinar cells |
| Thymus | Cortical thymocytes, medullary thymocytes, Hassall’s corpuscles |
| Lymph node | T cells, high endothelial cells venules endothelium |
| Bronchi/trachea | Respiratory epithelium, goblet cells |
| Lung | Type 2, pneumocytes, mucus cells |
| Thyroid | Thyrocytes |
| Testis | Leidy cells, Sertoli cells |
| Cancer | MAL Gene Hyper Methylation | Silenced MAL mRNA Expression | MAL mRNA Expression Rescue by DAC * Treatment | Down-Regulation of MAL Protein Expression (IHC ** Analysis) | Decreased Tumor Size after Exogenous Expression of MAL | Decreased Tumor Cell Functions after Exogenous Expression of MAL | References |
|---|---|---|---|---|---|---|---|
| Breast | Yes | Yes | Yes | Yes | Yes | [200,207,261] | |
| Esophagus | Yes | Yes | Yes | Yes | Yes | Yes | [203,219,262,263,264,265,266,267] |
| Stomach | Yes | Yes | Yes | [199,211,228,268] | |||
| Colon | Yes | Yes | Yes | Yes | [196,197,198,208,209,213,234,257,269,270] | ||
| Ovary | No | No | Yes | [212,221,227,250,252,271,272,273] | |||
| Cervix | Yes | Yes | Yes | Yes | [201,205,214,215,235,236,237,238,239,240,241,242,243,244,245,246,274,275,276,277] | ||
| Head and Neck | Yes | Yes | Yes | Yes | Yes | Yes | [202,220,230,231,232,278,279,280,281,282,283,284,285] |
| Lung | Yes | Yes | Yes | [204,210] | |||
| Bladder | Yes | [247,286,287] | |||||
| Prostate | Yes | Yes | [206,229] | ||||
| Kidney | Yes/No *** | [19] | |||||
| Skin | No | No | No | [248] | |||
| Cutaneous T-cell lymphoma | No | No | [223] | ||||
| B-cell lymphoma | No | No | [20,222,224,225,226,288,289,290,291,292] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubio-Ramos, A.; Labat-de-Hoz, L.; Correas, I.; Alonso, M.A. The MAL Protein, an Integral Component of Specialized Membranes, in Normal Cells and Cancer. Cells 2021, 10, 1065. https://doi.org/10.3390/cells10051065
Rubio-Ramos A, Labat-de-Hoz L, Correas I, Alonso MA. The MAL Protein, an Integral Component of Specialized Membranes, in Normal Cells and Cancer. Cells. 2021; 10(5):1065. https://doi.org/10.3390/cells10051065
Chicago/Turabian StyleRubio-Ramos, Armando, Leticia Labat-de-Hoz, Isabel Correas, and Miguel A. Alonso. 2021. "The MAL Protein, an Integral Component of Specialized Membranes, in Normal Cells and Cancer" Cells 10, no. 5: 1065. https://doi.org/10.3390/cells10051065
APA StyleRubio-Ramos, A., Labat-de-Hoz, L., Correas, I., & Alonso, M. A. (2021). The MAL Protein, an Integral Component of Specialized Membranes, in Normal Cells and Cancer. Cells, 10(5), 1065. https://doi.org/10.3390/cells10051065