MicroRNAs in Medullary Thyroid Carcinoma: A State of the Art Review of the Regulatory Mechanisms and Future Perspectives
Abstract
1. Introduction
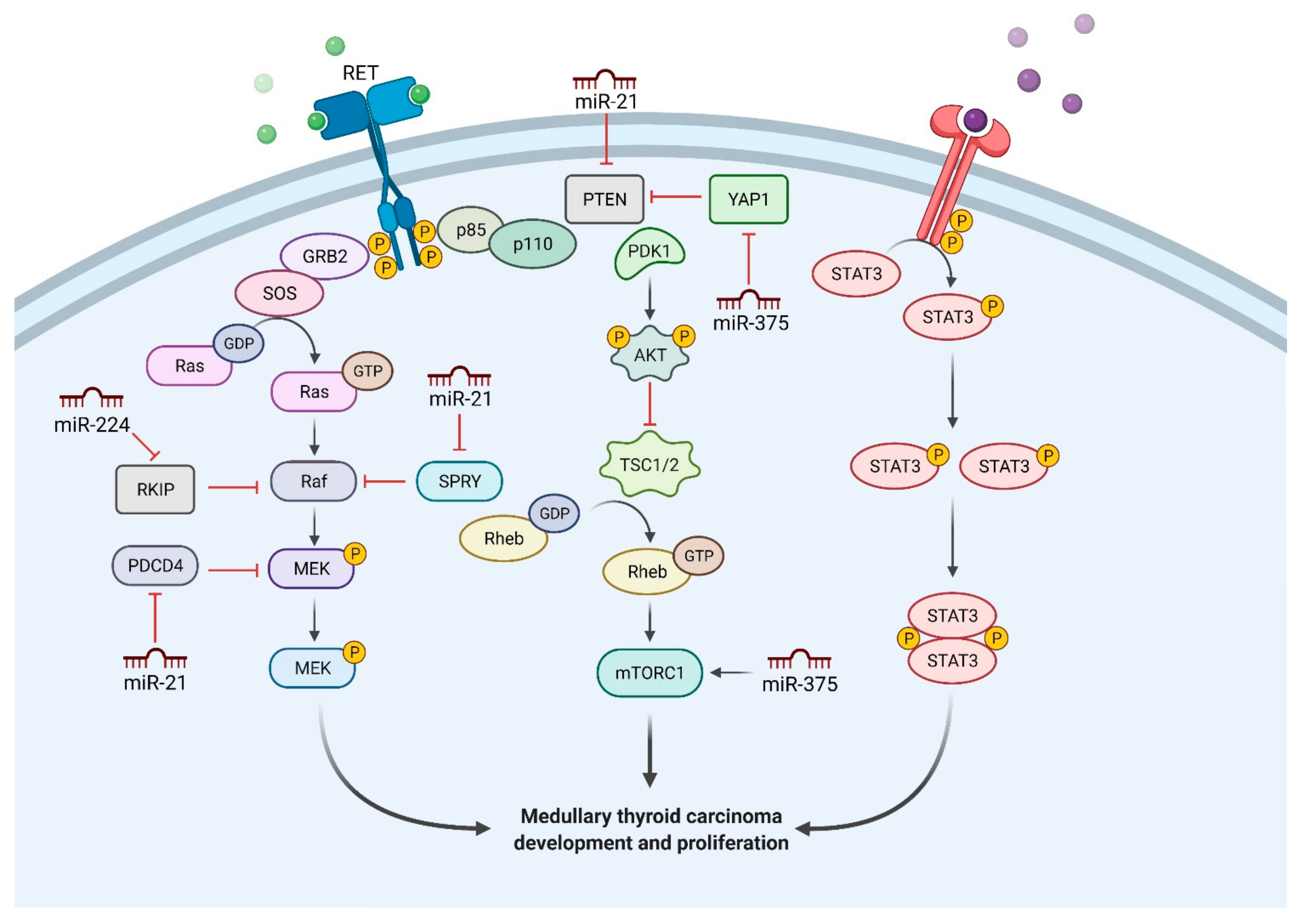
2. Deregulated miRNAs in MTC
2.1. MiR-21
2.2. MiR-375
2.3. MiR-224
2.4. MiR-183
2.5. MiR-127
2.6. MiR-153-3p
2.7. The Long-Non-Coding-RNA—MALAT1
2.8. MiR-31-3p
2.9. MiR-34a and miR-144
2.10. MiR-10a
2.11. The miR-200 Family
2.12. MiR-323
2.13. MiR-592
2.14. MiR-9-3p
2.15. MiR-129-5p
2.16. MiR-182
2.17. MiR-222-3p and miR-17-5p
3. MiRNAs as Biomarkers in MTC and Future Perspectives
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rosai, J.; DeLellis, R.A.; Carcangiu, M.L.; Frable, W.J.; Tallini, G. Classification of Thyroid Tumors; Silverberg, S.G., DeLellis, R.A., Sobin, L.H., Eds.; (4 Series); ARP PRESS: Silver Spring, MD, USA, 2014; pp. 61–65. [Google Scholar]
- Moo-Young, T.A.; Traugott, A.L.; Moley, J.F. Sporadic and familial medullary thyroid carcinoma: State of the art. Surg. Clin. N. Am. 2009, 89, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Takami, H.; Thosen, T.; Shirahama, S.; Ogura, K.; Hikiji, K. Does the syndrome of familial medullary thyroid carcinoma describe a distinct clinical entity? Eur. J. Cancer 1998, 34, 1639–1640. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Rosati, R.; Grieco, M.; Berlingieri, M.T.; D’Amato, G.L.; de Franciscis, V.; Fusco, A. The ret proto-oncogene is consistently expressed in human pheochromocytomas and thyroid medullary carcinomas. Oncogene 1990, 5, 1595–1598. [Google Scholar] [PubMed]
- Lai, A.Z.; Gujral, T.S.; Mulligan, L.M. RET signaling in endocrine tumors: Delving deeper into molecular mechanisms. Endocr. Pathol. 2007, 18, 57–67. [Google Scholar] [CrossRef]
- Machens, A.; Holzhausen, H.J.; Thanh, P.N.; Dralle, H. Malignant progression from C-cell hyperplasia to medullary thyroid carcinoma in 167 carriers of RET germline mutations. Surgery 2003, 134, 425–431. [Google Scholar] [CrossRef]
- Santoro, M.; Carlomagno, F.; Romano, A.; Bottaro, D.P.; Dathan, N.A.; Grieco, M.; Fusco, A.; Vecchio, G.; Matoskova, B.; Kraus, M.H.; et al. Activation of RET as a dominant transforming gene by germline mutations of MEN2A and MEN2B. Science 1995, 267, 381–383. [Google Scholar] [CrossRef]
- Moura, M.M.; Cavaco, B.M.; Pinto, A.E.; Domingues, R.; Santos, J.R.; Cid, M.O.; Bugalho, M.J.; Leite, V. Correlation of RET somatic with clinicopathological features in sporadic medullary thyroid carcinomas. Br. J. Cancer 2009, 100, 1777–1783. [Google Scholar] [CrossRef]
- Moura, M.M.; Cavaco, B.M.; Pinto, A.E.; Leite, V. High prevalence of RAS mutations in RET-negative sporadic medullary thyroid carcinomas. J. Clin. Endocrinol. Metab. 2011, 96, E863–E868. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Pallante, P.; Visone, R.; Croce, C.M. Deregulation of microRNA expression in follicular-cell-derived human thyroid carcinomas. Endocr. Relat. Cancer 2010, 17, 91–104. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Abraham, D.; Jackson, N.E.; Gundara, J.S. MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis and potential therapeutic targets. Clin. Cancer Res. 2011, 17, 4772–4781. [Google Scholar] [CrossRef]
- Santarpia, L.; Calin, G.A.; Adam, L. A miRNA signature associated with human metastatic medullary thyroid carcinoma. Endocr. Relat. Cancer 2013, 20, 809–823. [Google Scholar] [CrossRef]
- Mian, C.; Pennelli, G.; Fassan, M.; Balistreri, M.; Barollo, S.; Cavedon, E.; Galuppini, F.; Pizzi, M.; Vianello, F.; Pellizzo, M.R.; et al. MicroRNA profiles in familial and sporadic medullary thyroid carcinoma: Preliminary relationships with RET status and outcome. Thyroid 2012, 22, 890–896. [Google Scholar] [CrossRef]
- Sharma, S.B.; Ruppert, J.M. MicroRNA-based therapeutic strategies for targeting mutant and wild type RAS in cancer. Drug Dev. Rev. 2015, 76, 328–342. [Google Scholar] [CrossRef]
- Sharma, S.B.; Lin, C.C.; Farrugia, M.K.; McLaughlin, S.L.; Ellis, E.J.; Brundage, K.M.; Salkeni, M.A.; Ruppert, J.M. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol. Cell Biol. 2014, 34, 4143–4164. [Google Scholar] [CrossRef]
- Pennelli, G.; Galuppini, F.; Barollo, S.; Cavedon, E.; Bertazza, L.; Fassan, M.; Guzzardo, V.; Pelizzo, M.R.; Rugge, M.; Mian, C. The PDCD4/miR-21 pathway in medullary thyroid carcinoma. Hum. Pathol. 2015, 46, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Gundara, J.S.; Zhao, J.T.; Gill, A.J.; Clifton-Bligh, R.; Robinson, B.G.; Delbridge, L.; Sidhu, S.B. Nodal metastasis microRNA expression correlates with the primary tumour in MTC. ANZ J. Surg. 2014, 84, 235–239. [Google Scholar] [CrossRef]
- Galuppini, F.; Bertazza, L.; Barollo, S.; Cavedon, E.; Rugge, M.; Guzzardo, V.; Sacchi, D.; Watutantrige-Fernando, S.; Vianello, F.; Mian, C.; et al. MiR-375 and YAP1 expression profiling in medullary thyroid carcinoma and their correlation with clinical-pathological features and outcome. Virchows Arch. 2017, 471, 651–658. [Google Scholar] [CrossRef]
- Hudson, J.; Duncavage, E.; Tamburrino, A.; Salerno, P.; Xi, L.; Raffeld, M.; Moley, J.; Chernock, R.D. Overexpression of miR-10a and miR-375 and downregulation of YAP1 in medullary thyroid carcinoma. Exp. Mol. Pathol. 2013, 95, 62–67. [Google Scholar] [CrossRef]
- Lassalle, S.; Zangari, J.; Popa, A.; Ilie, M.; Hofman, V.; Long, E.; Patey, M.; Tissier, F.; Belleannee, G.; Trouette, H.; et al. MicroRNA-375/SEC23A as biomarkers of the in vitro efficacy of vandetanib. Oncotarget 2016, 7, 30461–30478. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhao, S.M.; Luo, Y.; Zhang, A.W.; Wei, L.H.; Xie, Z.Y.; Li, Y.Y.; Ma, W. MiR-375: A prospective regulator in medullary thyroid cancer based on microarray data and bioinformatics analyses. Pathol. Res. Pract. 2017, 213, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Romeo, P.; Colombo, C.; Granata, R.; Calareso, G.; Gualeni, A.V.; Dugo, M.; De Cecco, L.; Rizzetti, M.G.; Zanframundo, A.; Aiello, A.; et al. Circulating miR-375 as a novel prognostic marker for metastatic medullary thyroid cancer patients. Endocr. Relat. Cancer. 2018, 25, 217–231. [Google Scholar] [CrossRef]
- Lan, S.H.; Wu, S.Y.; Zuchini, R.; Lin, X.Z.; Su, I.J.; Tsai, T.F.; Lin, Y.J.; Wu, C.T.; Liu, H.S. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy 2014, 10, 1687–1689. [Google Scholar] [CrossRef]
- Shen, S.N.; Wang, L.F.; Jia, Y.F.; Hao, Y.Q.; Zhang, L.; Wang, H. Upregulation of microRNA-224 is associated with aggressive progression and poor prognosis in human cervical cancer. Diagn. Pathol. 2013, 8, 1–7. [Google Scholar] [CrossRef]
- Mencia, N.; Selga, E.; Noé, V.; Ciudad, C.J. Underexpression of miR-224 in methotrexate-resistant human colon cancer cells. Biochem. Pharmacol. 2011, 82, 1572–1582. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, L.J.; Yang, Y.C.; Wang, Z.X.; Wang, R. MiR-224 promotes the chemoresistance of human lung adenocarcinoma cells to cisplatin via regulating G(1)/S transition and apoptosis by targeting p21(WAF1/CIP1). Br. J. Cancer 2014, 111, 339–354. [Google Scholar] [CrossRef]
- Chen, W.; Fan, X.M.; Mao, L.; Zhang, J.Y.; Li, J.; Wu, J.Z.; Tang, J.H. MicroRNA-224: As a potential target for miR-based therapy of cancer. Tumour Biol. 2015, 36, 6645–6652. [Google Scholar] [CrossRef]
- Cavedon, E.; Barollo, S.; Bertazza, L.; Pennelli, G.; Galuppini, F.; Watutantrige-Fernando, S.; Censi, S.; Iacobone, M.; Benna, C.; Vianello, F.; et al. Prognostic Impact of miR-224 and RAS Mutations in Medullary Thyroid Carcinoma. Int. J. Endocrinol. 2017, 2017, 4915736. [Google Scholar] [CrossRef]
- Ma, D.; Tao, X.; Gao, F.; Fan, C.; Wu, D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol. Lett. 2012, 4, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fu, H.; Xu, C.; Tie, Y.; Xing, R.; Zhu, J.; Qin, Y.; Sun, Z.; Zheng, X. miR-183 inhibits TGF-β1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer 2010, 10, 354. [Google Scholar] [CrossRef] [PubMed]
- Aubert, S.; Berdelou, A.; Gnemmi, V.; Behal, H.; Caiazzo, R.; D’herbomez, M.; Pigny, P.; Wemeau, J.L.; Carnaille, B.; Renaud, F.; et al. Large sporadic thyroid medullary carcinomas: Predictive factors for lymph node involvement. Virchows. Arch. 2018, 472, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Yang, Q.; Yuan, J.; Miao, Q.; Duan, L.; Li, F.; Wang, S. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol. Rep. 2016, 36, 2563–2570. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, L.; Ma, R.; Gong, H.; Xu, P.; Wang, C. MicroRNA-127 is aberrantly downregulated and acted as a functional tumor suppressor in human pancreatic cancer. Tumour Biol. 2016, 37, 14249–14257. [Google Scholar] [CrossRef]
- Zhang, J.; Hou, W.; Chai, M.; Zhao, H.; Jia, J.; Sun, X.; Zhao, B.; Wang, R. MicroRNA-127-3p inhibits proliferation and invasion by targeting SETD8 in human osteosarcoma cells. Biochem. Biophys. Res. Commun. 2016, 469, 1006–1011. [Google Scholar] [CrossRef]
- Jiang, H.; Hua, D.; Zhang, J.; Lan, Q.; Huang, Q.; Yoon, J.G.; Han, X.; Li, L.; Foltz, G.; Zheng, S.; et al. MicroRNA-127-3p promotes glioblastoma cell migration and invasion by targeting the tumorsuppressor gene SEPT7. Oncol. Rep. 2014, 31, 2261–2269. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Jiang, X. miR-127 suppresses gastric cancer cell migration and invasion via targeting Wnt7a. Oncol. Lett. 2019, 17, 3219–3226. [Google Scholar] [CrossRef]
- Wei, S.; LiVolsi, V.A.; Montone, K.T.; Morrissette, J.J.D.; Baloch, Z.W. Detection of molecular alterations in medullary Thyroid Carcinoma using next-generation sequencing: An institutional experience. Endocr. Pathol. 2016, 27, 359–362. [Google Scholar] [CrossRef]
- Joo, L.J.S.; Weiss, J.; Gill, A.J.; Clifton-Bligh, R.; Brahmbhatt, H.; MacDiarmid, J.A.; Gild, M.L.; Robinson, B.G.; Zhao, J.T.; Sidhu, S.B. RET Kinase-Regulated MicroRNA-153-3p Improves Therapeutic Efficacy in Medullary Thyroid Carcinoma. Thyroid 2019, 29, 830–844. [Google Scholar] [CrossRef]
- Bai, Z.; Sun, J.; Wang, X.; Wang, H.; Pei, H.; Zhang, Z. MicroRNA-153 is a prognostic marker and inhibits cell migration and invasion by targeting SNAI1 in human pancreatic ductal adenocarcinoma. Oncol. Rep. 2015, 34, 595–602. [Google Scholar] [CrossRef]
- Zeng, H.F.; Yan, S.; Wu, S.F. MicroRNA-153-3p suppress cell proliferation and invasion by targeting SNAI1 in melanoma. Biochem. Biophys. Res. Commun. 2017, 487, 140–145. [Google Scholar] [CrossRef]
- Zhou, J.; Xie, M.; Shi, Y.; Luo, B.; Gong, G.; Li, J.; Wang, J.; Zhao, W.; Zi, Y.; Wu, X.; et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol. Rep. 2015, 34, 111–120. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef]
- Chu, Y.H.; Hardin, H.; Schneider, D.F.; Chen, H.; Lloyd, R.V. MicroRNA-21 and long non- coding RNA MALAT1 are overexpressed markers in medullary thyroid carcinoma. Exp. Mol. Pathol. 2017, 103, 229–236. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef]
- Zhang, R.; Hardin, H.; Huang, W.; Chen, J.; Asioli, S.; Righi, A.; Maletta, F.; Sapino, A.; Lloyd, R.V. MALAT1 long non-coding RNA expression in thyroid tissues: Analysis by in situ hybridization and real-time PCR. Endocr. Pathol. 2017, 28, 7–12. [Google Scholar] [CrossRef]
- Laurent-Puig, P.; Grisoni, M.L.; Heinemann, V.; Liebaert, F.; Neureiter, D.; Jung, A.; Montestruc, F.; Gaston-Mathe, Y.; Thiébaut, R.; Stintzing, S. Validation of miR-31-3p expression to predict cetuximab efficacy when used as first-line treatment in RAS wild-type metastatic colorectal cancer. Clin. Cancer Res. 2019, 25, 134–141. [Google Scholar] [CrossRef]
- Zeljic, K.; Jovanovic, I.; Jovanovic, J.; Magic, Z.; Stankovic, A.; Supic, G. MicroRNA meta-signature of oral cancer: Evidence from a meta-analysis. Ups. J. Med. Sci. 2018, 123, 43–49. [Google Scholar] [CrossRef]
- Pedroza-Torres, A.; Fernandez-Retana, J.; Peralta-Zaragoza, O.; Jacobo-Herrera, N.; Cantù de Leon, D.; Cerna-Cortés, J.F.; Lopez-Camarillo, C.L.; Péreez-Plasencia, C. A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol. Oncol. 2016, 142, 557–565. [Google Scholar] [CrossRef]
- Jiang, M.; Shi, X.; Zhu, H.; Wei, W.; Li, J. Two GEO MicroRNA Expression Profile Based High-Throughput Screen to Identify MicroRNA-31-3p Regulating Growth of Medullary Thyroid Carcinoma Cell by Targeting RASA2. Med. Sci. Monit. 2019, 12, 5170–5180. [Google Scholar] [CrossRef] [PubMed]
- Arafeh, R.; Qutob, N.; Emmanuel, R.; Keren-Paz, A.; Madore, J.; Elkahloun, A.; Wilmott, J.S.; Gartner, J.J.; Di Pizio, A.; Winograd-Katz, S.; et al. Recurrent inactivating RASA2 mutations in melanoma. Nat. Genet. 2015, 47, 1408–1410. [Google Scholar] [CrossRef] [PubMed]
- Slabáková, E.; Culig, Z.; Remšík, J.; Souček, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef] [PubMed]
- Fritz, H.K.; Gustafsson, A.; Ljungberg, B.; Ceder, Y.; Axelson, H.; Dahlbäck, B. The Axl-regulating tumor suppressor miR-34a is increased in CCRCC but does not correlate with Axl mRNA or Axl protein levels. PLoS ONE 2015, 10, e0135991. [Google Scholar] [CrossRef]
- Li, Y.; Jia, L.; Ren, D.; Liu, C.; Gong, Y.; Wang, N.; Zhang, X.; Zhao, Y. Axl mediates tumor invasion and chemosensitivity through PI3K/Akt signaling pathway and is transcriptionally regulated by slug in breast carcinoma. IUBMB Life 2014, 66, 507–518. [Google Scholar] [CrossRef]
- Avilla, E.; Guarino, V.; Visciano, C.; Liotti, F.; Svelto, M.; Krishnamoorthy, G.; Franco, R.; Melillo, R.M. Activation of TYRO3/AXL tyrosine kinase receptors in thyroid cancer. Cancer Res. 2011, 71, 1792–1804. [Google Scholar] [CrossRef]
- Xiang, C.; Cui, S.-P.; Ke, Y. MiR-144 inhibits cell proliferation of renal cell carcinoma by targeting MTOR. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2016, 36, 186–192. [Google Scholar] [CrossRef]
- Sun, J.; Shi, R.; Zhao, S.; Li, X.; Lu, S.; Bu, H.; Ma, X.; Su, C. E2F8, a direct target of miR-144, promotes papillary thyroid cancer progression via regulating cell cycle. J. Exp. Clin. Cancer Res. 2017, 36, 40. [Google Scholar] [CrossRef]
- Sun, W.; Lan, X.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, P.; Zhang, H. MicroRNA-144 inhibits proliferation by targeting WW domain-containing transcription regulator protein 1 in papillary thyroid cancer. Oncol. Lett. 2018, 15, 1007–1013. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Zhang, H.; Zhang, J.; Zhang, Y.; Li, S.; Qin, L.; Yang, Z.; Xiong, J. Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol. Ther. 2018, 19, 484–496. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Ho-Fun Lee, V.; Wong, A.M.; Kwong, D.L.; Zhu, Y.H.; Dong, S.S.; Kong, K.L.; Chen, J.; Tsao, S.W.; Guan, X.Y. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis 2013, 34, 454–463. [Google Scholar] [CrossRef]
- Shabani, N.; Razaviyan, J.; Paryan, M.; Tavangar, S.M.; Azizi, F.; Mohammadi-Yeganeh, S.; Hedayati, M. Evaluation of miRNAs expression in medullary thyroid carcinoma tissue samples: miR-34a and miR-144 as promising overexpressed markers in MTC. Hum. Pathol. 2018, 79, 212–221. [Google Scholar] [CrossRef]
- Shabani, N.; Sheikholeslami, S.; Paryan, M.; Zarif Yeganeh, M.; Tavangar, S.M.; Azizi, F.; Mohammadi-Yeganeh, S.; Hedayati, M. An investigation on the expression of miRNAs including miR-144 and miR-34a in plasma samples of RET-positive and RET-negative medullar thyroid carcinoma patients. J. Cell Physiol. 2020, 235, 1366–1373. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbø, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibition of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009, 10, 12. [Google Scholar] [CrossRef]
- Agirre, X.; Jime’nez-Velasco, A.; San Jose’-Ene’riz, E.; Garate, L.; Bandre’s, E.; Cordeu, L.; Aparicio, O.; Saez, B.; Navarro, G.; Vilas-Zornoza, A.; et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34C cells increases USF-mediated cell growth. Mol. Cancer Res. 2008, 6, 1830–1840. [Google Scholar] [CrossRef]
- Garzon, R.; Garofalo, M.; Martelli, M.P.; Briesewitz, R.; Wang, L.; Fernandez-Cymering, C.; Volinia, S.; Liu, C.G.; Schnittger, S.; Haferlach, T.; et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA 2008, 105, 3945–3950. [Google Scholar] [CrossRef]
- Veerla, S.; Lindgren, D.; Kvist, A.; Frigyesi, A.; Staaf, J.; Persson, H.; Liedberg, F.; Chebil, G.; Gudjonsson, S.; Borg, A.; et al. MiRNA expression in urothelial carcinomas: Important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int. J. Cancer 2009, 124, 2236–2242. [Google Scholar] [CrossRef]
- Mancikova, V.; Montero-Conde, C.; Perales-Paton, J.; Fernandez, A.; Santacana, M.; Jodkowska, K.; Inglada-Pérez, L.; Castelblanco, E.; Borrego, S.; Encinas, M.; et al. Multilayer OMIC Data in Medullary Thyroid Carcinoma Identifies the STAT3 Pathway as a Potential Therapeutic Target in RET(M918T) Tumors. Clin. Cancer Res. 2017, 23, 1334–1345. [Google Scholar] [CrossRef]
- Ehyaei, S.; Hedayati, M.; Zarif-Yeganeh, M.; Sheikholeslami, S.; Ahadi, M.; Amini, S.A. Plasma Calcitonin Levels and miRNA323 Expression in Medullary Thyroid Carcinoma Patients with or without RET Mutation. Asian Pac. J. Cancer Prev. 2017, 18, 2179–2184. [Google Scholar] [CrossRef]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.C.; Wolff, R.K.; Samowitz, W.S.; Herrick, J.S. Dysregulated genes and miRNA s in the apoptosis pathway in colorectal cancer patients. Apoptosis 2018, 23, 237–250. [Google Scholar] [CrossRef]
- Yerukala Sathipati, S.; Ho, S.Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci. Rep. 2018, 8, 16138. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhi, Q.; Wang, W.; Zhang, Q.; Fang, T.; Ma, Q. Up-regulation of miR- 592 correlates with tumor progression and poor prognosis in patients with colorectal cancer. Biomed. Pharmacother. 2015, 69, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Du, Y.; Yang, C.; Zhang, D.; Zhang, N.; Liu, X.; Cho, W.C.; Yang, Y. An oncogenic role of miR- 592 in tumorigenesis of human colorectal cancer by targeting forkhead box o3a (FoxO3A). Expert Opin. Ther. Targets 2016, 20, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Rao, P.; Li, W. MiR-592 represses FOXO3 expression and promotes the proliferation of prostate cancer cells. Int. J. Clin. Exp. Med. 2015, 8, 15246–15257. [Google Scholar] [PubMed]
- He, Y.; Ge, Y.; Jiang, M.; Zhou, J.; Luo, D.; Fan, H.; Shi, L.; Lin, L.; Yang, L. MiR- 592 promotes gastric cancer proliferation, migration, and invasion through the Pi3K/Akt and MAPK/ERK signaling pathways by targeting spry2. Cell Physiol. Biochem. 2018, 47, 1465–1481. [Google Scholar] [CrossRef]
- Liu, T.; Meng, J.; Zhang, Y. miR-592 acts as an oncogene and promotes medullary thyroid cancer tumorigenesis by targeting cyclin-dependent kinase 8. Mol. Med. Rep. 2020, 22, 3316–3326. [Google Scholar] [CrossRef]
- Brägelmann, J.; Klümper, N.; Offermann, A.; von Mässenhausen, A.; Böhm, D.; Deng, M.; Queisser, A.; Sanders, C.; Syring, I.; Merseburger, A.S.; et al. Pan-cancer analysis of the mediator complex transcriptome identifies CDK19 and CDK8 as therapeutic targets in advanced prostate cancer. Clin. Cancer Res. 2017, 23, 1829–1840. [Google Scholar] [CrossRef]
- McDermott, M.S.; Chumanevich, A.A.; Lim, C.U.; Liang, J.; Chen, M.; Altilia, S.; Oliver, D.; Rae, J.M.; Shtutman, M.; Kiaris, H.; et al. Inhibition of CDK8 mediator kinase suppresses estrogen dependent transcription and the growth of estrogen receptor positive breast cancer. Oncotarget 2017, 8, 12558–12575. [Google Scholar] [CrossRef]
- Liang, J.; Chen, M.; Broude, E.V.; Roninson, I.B. Role of transcription-regulating kinase CDK8 in colon cancer metastasis. Oncotarget 2019, 10, 622–623. [Google Scholar] [CrossRef]
- Nassa, D.N.; Rosenwald, S.R.; Meiri, E.; Gilad, S.; Tabibian-Keissar, H.; Schlosberg, A.; Kuker, H.; Sion-Vardy, N.; Tobar, A.; Kharenko, O. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009, 19, 375–383. [Google Scholar] [CrossRef]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bi-functional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosc. 2008, 28, 14341–14346. [Google Scholar] [CrossRef]
- Jeon, H.M.; Sohn, Y.W.; Oh, S.Y.; Kim, S.H.; Beck, S.; Kim, S.; Kim, H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011, 71, 3410–3421. [Google Scholar] [CrossRef]
- Ogata, K.; Sumida, K.; Miyata, K.; Kushida, M.; Kuwamura, M.; Yamate, J. Circulating miR-9* and miR-384-5p as potential indicators for trimethyltin-induced neurotoxicity. Toxicol. Pathol. 2015, 43, 198–208. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, S.; Zhao, R.; Zhao, Q.; Zhang, T. Upregulated miR-9-3p Promotes Cell Growth and Inhibits Apoptosis in Medullary Thyroid Carcinoma by Targeting BLCAP. Oncol. Res. 2017, 25, 1215–1222. [Google Scholar] [CrossRef]
- Floros, K.V.; Hellinida, T.; Dimitra, F.; Maroulio, T.; Andreas, S. Alterations in mRNA expression of apoptosis-related genes BCL2, BAX, FAS, caspase-3, and the novel member BCL2L12 after treatment of human leukemic cell line HL60 with the antineoplastic agent etoposide. Ann. N. Y. Acad. Sci. 2006, 1090, 89–97. [Google Scholar] [CrossRef]
- Swanton, E.; Savory, P.; Cosulich, S.; Clarke, P.; Woodman, P. Bcl-2 regulates a caspase-3/caspase-2 apoptotic cascade in cytosolic extracts. Oncogene 1999, 18, 1781–1787. [Google Scholar] [CrossRef]
- Ogawa, R.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitsui, A.; Katada, T.; Harata, K.; Tanaka, T.; Fujii, Y. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med. Mol. Morphol. 2009, 42, 102–109. [Google Scholar] [CrossRef]
- Yu, X.; Song, H.; Xia, T.; Han, S.; Xiao, B.; Luo, L.; Xi, Y.; Guo, J. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene 2013, 532, 87–93. [Google Scholar] [CrossRef]
- Karaayvaz, M.; Zhai, H.; Ju, J. MiR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013, 4, e659. [Google Scholar] [CrossRef]
- Liu, Y.; Hei, Y.; Shu, Q.; Dong, J.; Gao, Y.; Fu, H.; Zheng, X.; Yang, G. VCP/p97, down-regulated by microRNA-129-5p, could regulate the progression of hepatocellular carcinoma. PLoS ONE 2012, 7, e35800. [Google Scholar] [CrossRef]
- Li, M.; Tian, L.; Wang, L.; Yao, H.; Zhang, J.; Lu, J.; Sun, Y.; Gao, X.; Xiao, H.; Liu, M. Down-regulation of miR-129-5p inhibits growth and induces apoptosis in laryngeal squamous cell carcinoma by targeting APC. PLoS ONE 2013, 8, e77829. [Google Scholar] [CrossRef]
- Duan, L.; Hao, X.; Liu, Z.; Zhang, Y.; Zhang, G. MiR-129-5p is down-regulated and involved in the growth, apoptosis and migration of medullary thyroid carcinoma cells through targeting RET. FEBS Lett. 2014, 588, 1644–1651. [Google Scholar] [CrossRef] [PubMed]
- Weeraratne, S.D.; Amani, V.; Teider, N.; Pierre-Francois, J.; Winter, D.; Kye, M.J.; Sengupta, S.; Archer, T.; Remke, M.; Bai, A.H.; et al. Pleiotropic effects of miR-183 ~ 96 ~ 182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012, 123, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Sheng, C.; Huang, L.; Zhang, H.; Cheng, Z.; Zhu, Q. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014, 16, 473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qian, P.; Zhang, X.; Zhang, M.; Wang, H.; Wu, M.; Kong, X.; Tan, S.; Ding, K.; Perry, J.K.; et al. Autocrine/ paracrine human growth hormone-stimulated MicroRNA 96-182-183 cluster promotes epithelial-mesenchymal transition and invasion in breast cancer. J. Biol. Chem. 2015, 290, 13812–13829. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.F.; Hanniford, D.; Menendez, S.; Reavie, L.; Zou, X.; Alvarez-Diaz, S.; Zakrzewski, J.; Blochin, E.; Rose, A.; Bogunovic, D.; et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA 2009, 106, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Shen, J.; Wang, C.; Yang, L.; Zhang, X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer 2012, 12, 227. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Mito, J.K.; Lee, C.L.; Zhang, M.; Li, Z.; Dodd, R.D.; Cason, D.; Luo, L.; Ma, Y.; Van Mater, D.; et al. MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. J. Clin. Invest. 2014, 124, 4305–4319. [Google Scholar] [CrossRef]
- Ning, F.L.; Wang, F.; Li, M.L.; Yu, Z.S.; Hao, Y.Z.; Chen, S.S. MicroRNA-182 modulates chemosensitivity of human non-small cell lung cancer to cisplatin by targeting PDCD4. Diagn. Pathol. 2014, 9, 143. [Google Scholar] [CrossRef]
- Perilli, L.; Vicentini, C.; Agostini, M.; Pizzini, S.; Pizzi, M.; D’Angelo, E.; Bortoluzzi, S.; Mandruzzato, S.; Mammano, E.; Rugge, M.; et al. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget 2014, 5, 6611–6619. [Google Scholar] [CrossRef]
- Spitschak, A.; Meier, C.; Kowtharapu, B.; Engelmann, D.; Pützer, B.M. MiR-182 promotes cancer invasion by linking RET oncogene activated NF-κB to loss of the HES1/Notch1 regulatory circuit. Mol. Cancer 2017, 16, 24. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, C.; Lu, H.; Chen, X.; Ba, Y.; Zhang, C.; Zhang, C.Y. Altered Serum MicroRNA Profile May Serve as an Auxiliary Tool for Discriminating Aggressive Thyroid Carcinoma from Nonaggressive Thyroid Cancer and Benign Thyroid Nodules. Dis. Markers 2019, 2019, 3717683. [Google Scholar] [CrossRef]
- Li, X.; Abdel-Mageed, A.B.; Mondal, D.; Kandil, E. Micro- RNA expression profiles in differentiated thyroid cancer, a review. Int. J. Clin. Exp. Med. 2013, 6, 74–80. [Google Scholar] [PubMed]
- Wang, Y.; Tang, N.; Hui, T.; Wang, S.; Zeng, X.; Li, H.; Ma, J. Identification of endogenous reference genes for RT-qPCR analysis of plasma microRNAs levels in rats with acetaminophen-induced hepatotoxicity. J. Appl. Toxicol. 2013, 33, 1330–1336. [Google Scholar] [CrossRef]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef]
- De Smaele, E.; Ferretti, E.; Gulino, A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010, 1338, 100–111. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Valerio, L.; Pieruzzi, L.; Giani, C.; Agate, L.; Bottici, V.; Lorusso, L.; Cappagli, V.; Puleo, L.; Matrone, A.; Viola, D.; et al. Targeted therapy in thyroid cancer: State of the art. Clin. Oncol. 2017, 29, 316–324. [Google Scholar] [CrossRef]
- Maia, A.L.; Magagnin Wajner, S.; Vaz Ferreira Vargas, C. Advances and controversies in the management of medullary thyroid carcinoma. Curr. Opin. Oncol. 2017, 29, 25–32. [Google Scholar] [CrossRef]
| miRNA | Gene Mechanism | Actions |
|---|---|---|
| miR-21 | Upregulates RAS/ERK pathway (targets RASA1 and SPRED1), inhibits PDCD4 and apoptosis | High levels associated with lymph node metastases, advanced stage and postoperative chronic disease; potential therapeutic target [19] |
| miR-375 | Downregulates the PI3K/Akt pathway (targets YAP1, SLC16a2, SEC23A, PARP, JAK2 and NGFR) | Potential biomarker, plasma levels correlate with tumor burden, distant metastasis and response to vandetinib treatment [25] |
| miR-224 | Dysregulates the PI3K/Akt pathway | Low levels in advanced MTC, high levels and positive prognostic factor in sporadic MTC with RAS mutations [31] |
| miR-183 | Inhibits PDCD4 and apoptosis | High levels associated with lateral cervical lymph node metastases, distant metastases and mortality [14,34] |
| miR-127 | Downregulates BAG5 and SEPT7, upregulates Wnt7a | Low levels in sporadic MTC harboring RET mutation [40] |
| miR-153-3p | Downregulates mTOR pathway (targets RPS6KB1) | Potential therapeutic effects in combination with tyrosine kinase inhibitors [41] |
| Long-non-coding-RNA—MALAT1 | Downregulates B-MYB, p53, upregulates EMT (targets E-cadherin) | High levels in MTC, in vitro inhibition reduces tumor cell proliferation and invasion [46] |
| miR-31-3p | Downregulates RAS pathway (targets RASA2) | Low levels in MTC, reduces in vitro MTC cell proliferation [52] |
| miR-34a; miR-144 | Dysregulates PI3K/Akt/mTOR pathway (targets AXL) | High levels in MTC, proposed as biomarkers but lack sufficient specificity and sensitivity [63,64] |
| miR-10a | Downregulates HOXD4 | High levels in primary MTC but downregulated in metastases [15] |
| miR-200 family | Upregulates TGF-β2 (targets ZEB1 and ZEB2) and EMT (targets E-cadherin) | May correlate with metastatic potential [21] |
| miR-323 | Unknown | High levels in MTC but not related to RET mutational status [70] |
| miR-592 | Downregulates FOXO3a and CDK8 | Potential therapeutic target [77] |
| miR-9-3p | Upregulates p21(WAF1/CIP1) and Bcl-XL/Bcl-2 pathway (targets BLCAP) | Enhances cell growth and inhibits apoptosis [16,85] |
| miR-129-5p | Down-regulates the PI3K/Akt pathway (targets RET) | Low level in MTC, acts as tumor suppressor [93] |
| miR-182 | Downregulates FOXO3, MITF, MTSS1, PDCD4 and HES1/Notch1 pathway | Potential blood biomarker for tumor progression [101,102] |
| miR-222-3p; miR-17-5p | Downregulates p27kip1, p57, PTEN, TIMP3 and c-KIT | Potential blood biomarkers [103,104] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galuppini, F.; Censi, S.; Moro, M.; Carraro, S.; Sbaraglia, M.; Iacobone, M.; Fassan, M.; Mian, C.; Pennelli, G. MicroRNAs in Medullary Thyroid Carcinoma: A State of the Art Review of the Regulatory Mechanisms and Future Perspectives. Cells 2021, 10, 955. https://doi.org/10.3390/cells10040955
Galuppini F, Censi S, Moro M, Carraro S, Sbaraglia M, Iacobone M, Fassan M, Mian C, Pennelli G. MicroRNAs in Medullary Thyroid Carcinoma: A State of the Art Review of the Regulatory Mechanisms and Future Perspectives. Cells. 2021; 10(4):955. https://doi.org/10.3390/cells10040955
Chicago/Turabian StyleGaluppini, Francesca, Simona Censi, Margherita Moro, Stefano Carraro, Marta Sbaraglia, Maurizio Iacobone, Matteo Fassan, Caterina Mian, and Gianmaria Pennelli. 2021. "MicroRNAs in Medullary Thyroid Carcinoma: A State of the Art Review of the Regulatory Mechanisms and Future Perspectives" Cells 10, no. 4: 955. https://doi.org/10.3390/cells10040955
APA StyleGaluppini, F., Censi, S., Moro, M., Carraro, S., Sbaraglia, M., Iacobone, M., Fassan, M., Mian, C., & Pennelli, G. (2021). MicroRNAs in Medullary Thyroid Carcinoma: A State of the Art Review of the Regulatory Mechanisms and Future Perspectives. Cells, 10(4), 955. https://doi.org/10.3390/cells10040955









