The Impact of Preprocessing Methods for a Successful Prostate Cell Lines Discrimination Using Partial Least Squares Regression and Discriminant Analysis Based on Fourier Transform Infrared Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Sample Preparation for Spectroscopic Studies
2.3. FT-IR Measurements
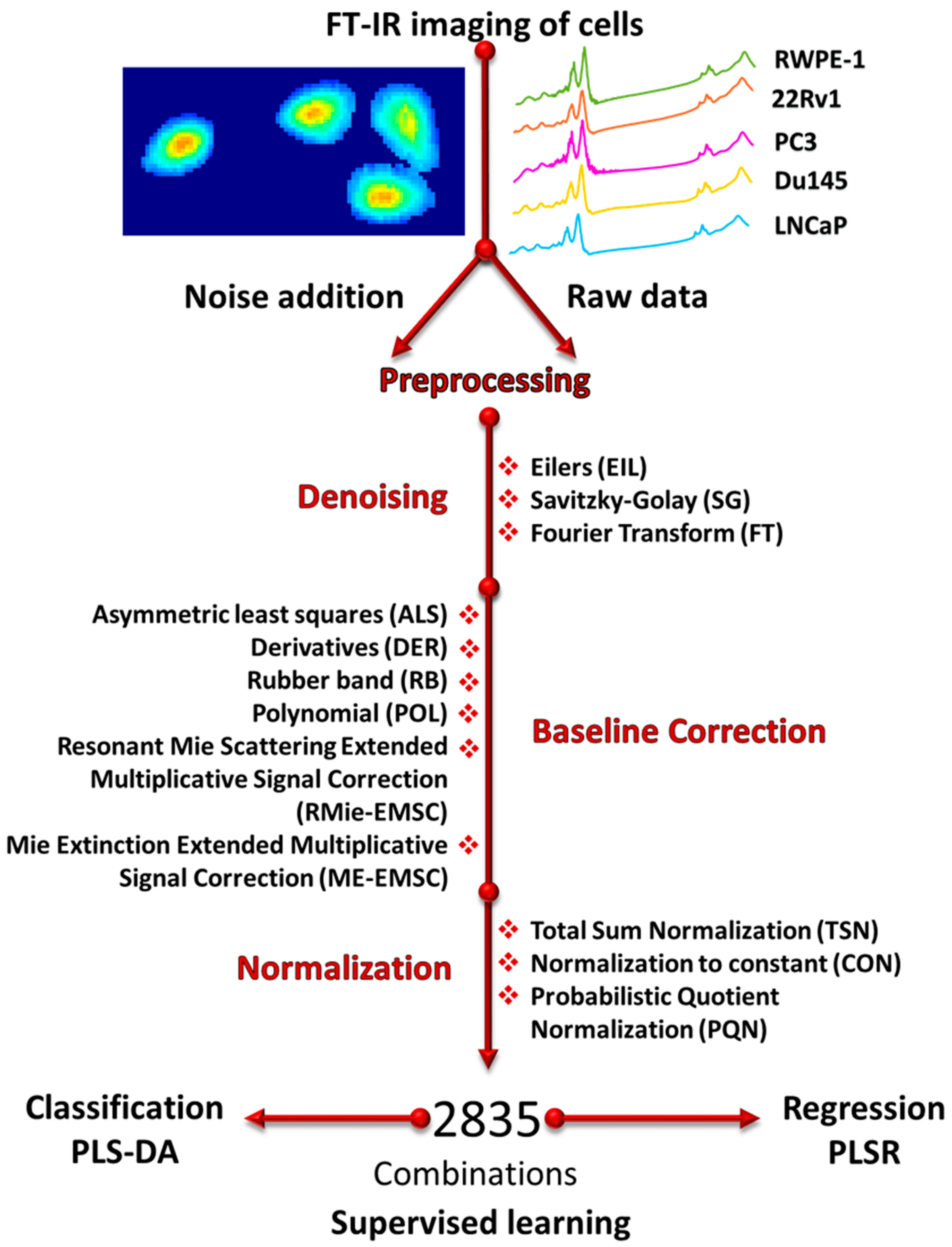
2.4. Noise Addition and Preprocessing
2.5. Regression and Classification
2.6. Model Calibration and Validation
2.7. Description of Methods
2.7.1. Baseline Correction
2.7.2. Normalization
2.7.3. Denoising
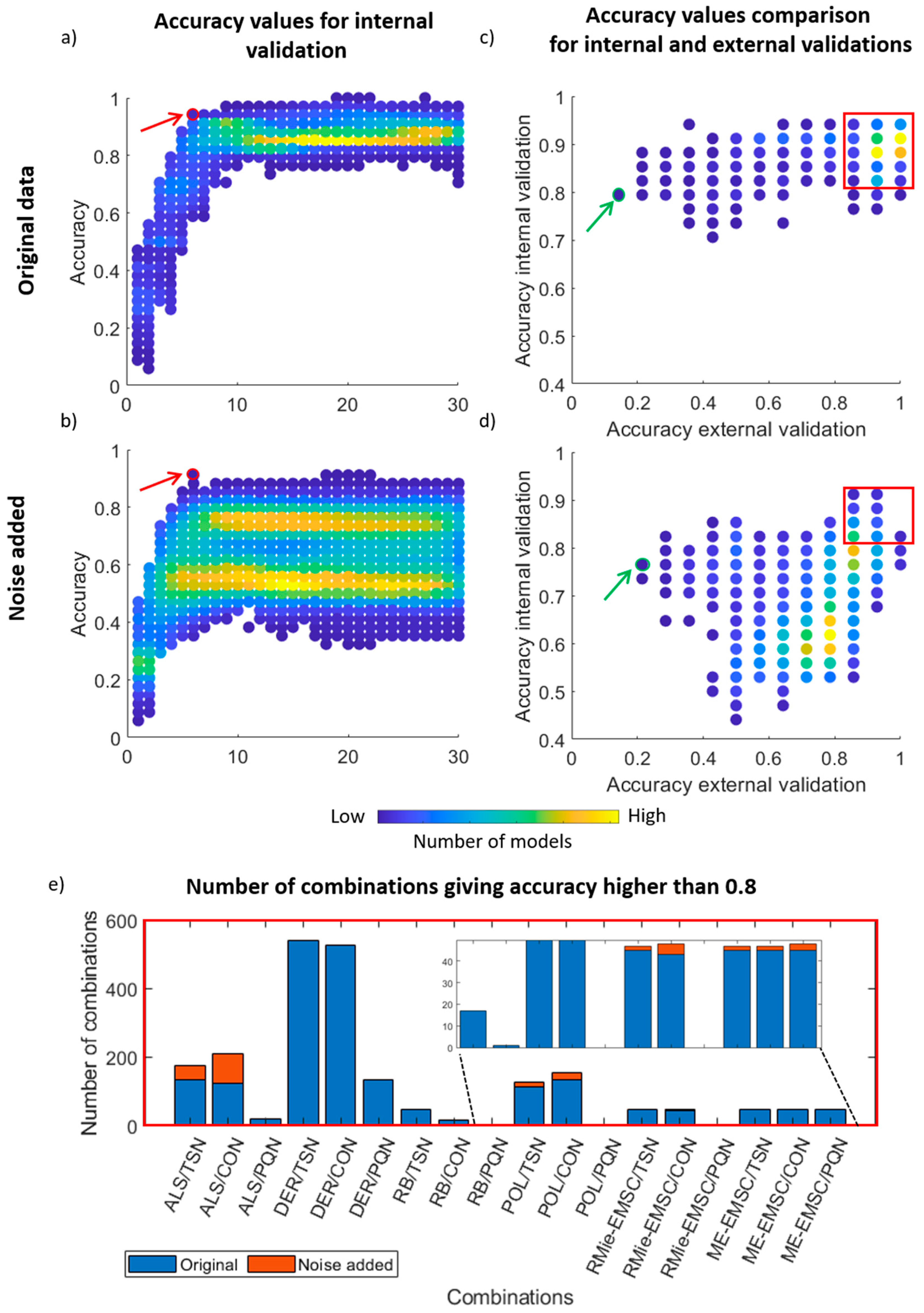
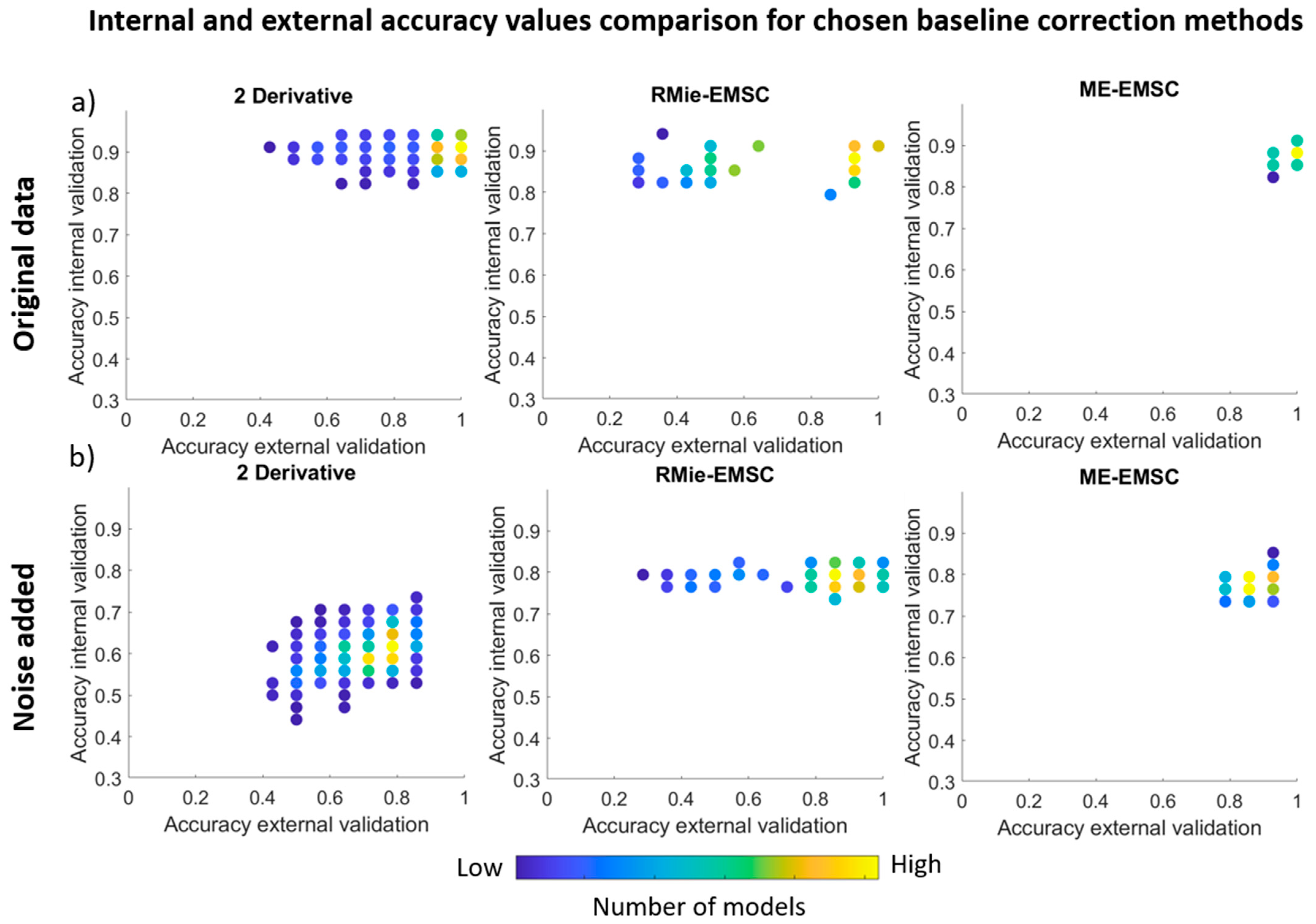
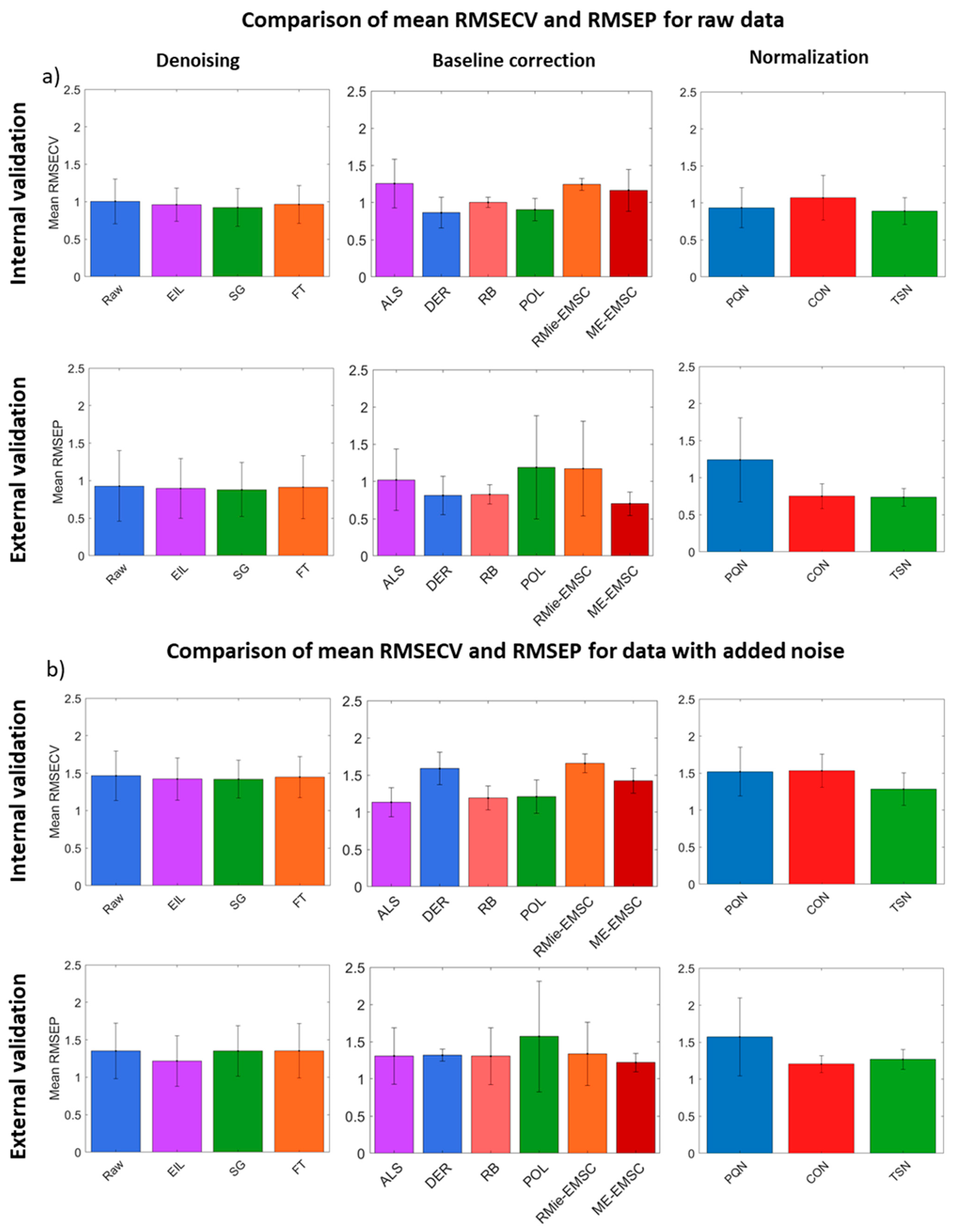
3. Results and Discussion
3.1. Spectral Changes
3.2. PLS Discriminant Analysis
3.3. PLS Regression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin 2016, 66, 7–30. [Google Scholar] [CrossRef]
- Ackerstaff, E.; Pflug, B.R.; Nelson, J.B.; Bhujwalla, Z.M. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res. 2001, 61, 3599–3603. [Google Scholar]
- Augustyniak, K.; Chrabaszcz, K.; Jasztal, A.; Smeda, M.; Quintas, G.; Kuligowski, J.; Marzec, K.M.; Malek, K. High- and Ultra-High definition of IR spectral histopathology gives an insight into chemical environment of lung metastases in breast cancer. J. Biophotonics 2018, e201800345. [Google Scholar] [CrossRef]
- Quaroni, L.; Zlateva, T. Infrared spectromicroscopy of biochemistry in functional single cells. Analyst 2011, 136, 3219–3232. [Google Scholar] [CrossRef]
- Majzner, K.; Kaczor, A.; Kachamakova-Trojanowska, N.; Fedorowicz, A.; Chlopicki, S.; Baranska, M. 3D confocal Raman imaging of endothelial cells and vascular wall: Perspectives in analytical spectroscopy of biomedical research. Analyst 2013, 138, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, T.P.; Mateuszuk, L.; Chlopicki, S.; Malek, K.; Baranska, M. Imaging of lipids in atherosclerotic lesion in aorta from ApoE/LDLR-/ mice by FT-IR spectroscopy and Hierarchical Cluster Analysis. Analyst 2011, 136. [Google Scholar] [CrossRef]
- Wrobel, T.P.; Marzec, K.M.; Chlopicki, S.; Maślak, E.; Jasztal, A.; Franczyk-Zarów, M.; Czyzyńska-Cichoń, I.; Moszkowski, T.; Kostogrys, R.B.; Baranska, M. Effects of Low Carbohydrate High Protein (LCHP) diet on atherosclerotic plaque phenotype in ApoE/LDLR−/− mice: FT-IR and Raman imaging. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Marzec, K.M.; Wrobel, T.P.; Rygula, A.; Maslak, E.; Jasztal, A.; Fedorowicz, A.; Chlopicki, S.; Baranska, M. Visualization of the biochemical markers of atherosclerotic plaque with the use of Raman, IR and AFM. J. Biophotonics 2014, 7, 744–756. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, T.P.; Bhargava, R. Infrared Spectroscopic Imaging Advances as an Analytical Technology for Biomedical Sciences. Anal. Chem. 2018, 90, 1444–1463. [Google Scholar] [CrossRef]
- Wrobel, T.P.; Piergies, N.; Pieta, E.; Kwiatek, W.; Paluszkiewicz, C.; Fornal, M.; Grodzicki, T. Erythrocyte heme-oxygenation status indicated as a risk factor in prehypertension by Raman spectroscopy. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3659–3663. [Google Scholar] [CrossRef] [PubMed]
- Pięta, E.; Petibois, C.; Pogoda, K.; Suchy, K.; Liberda, D.; Wróbel, T.P.; Paluszkiewicz, C.; Kwiatek, W.M. Assessment of cellular response to drug/nanoparticles conjugates treatment through FTIR imaging and PLS regression study. Sens. Actuatorsb Chem. 2020, 313, 1–9. [Google Scholar] [CrossRef]
- Paluszkiewicz, C. SR-FTIR spectroscopic preliminary findings of non-cancerous, cancerous, and hyperplastic human prostate tissues. Vib. Spectrosc. 2007, 43, 237–242. [Google Scholar] [CrossRef]
- Taleb, A.; Diamond, J.; Mcgarvey, J.J.; Beattie, J.R.; Toland, C.; Hamilton, P.W. Raman Microscopy for the Chemometric Analysis of Tumor Cells. J. Phys. Chem. B 2006, 110, 19625–19631. [Google Scholar] [CrossRef]
- Nicholson, J.M.; Lyng, F.M.; Byrne, H.J.; Hart, C.A.; Brown, M.D.; Clarke, N.W.; Gardner, P. An investigation of the RWPE prostate derived family of cell lines using FTIR spectroscopy. Analyst 2010, 135, 887–894. [Google Scholar] [CrossRef]
- Corsetti, S.; Rabl, T.; Mcgloin, D.; Nabi, G. Raman spectroscopy for accurately characterizing biomolecular changes in androgen-independent prostate cancer cells. J. Biophotonics 2018, 11, 1–8. [Google Scholar] [CrossRef]
- Crow, P.; Barrass, B.; Kendall, C.; Wright, M.; Persad, R.; Stone, N. The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines. Br. J. Cancer 2005, 92, 2166–2170. [Google Scholar] [CrossRef]
- Gazi, E.; Dwyer, J.; Gardner, P.; Wade, A.P.; Miyan, J.; Lockyer, N.P.; Vickerman, J.C.; Clarke, N.W.; Shanks, J.H.; Scott, L.J.; et al. Applications of Fourier transform infrared microspectroscopy in studies of benign prostate and prostate cancer. A pilot study. J. Pathol. 2003, 201, 99–108. [Google Scholar] [CrossRef]
- Henderson, A.; Brown, M.D.; Snook, R.D.; Faria, E.C.; Gardner, P.; Harvey, T.J.; Clarke, N.W.; Ward, A.D.; Gazi, E. Spectral discrimination of live prostate and bladder cancer cell lines using Raman optical tweezers. J. Biomed. Opt. 2008, 13, 064004. [Google Scholar] [CrossRef][Green Version]
- Harvey, T.J.; Gazi, E.; Henderson, A.; Snook, R.D.; Clarke, N.W.; Brown, M.; Gardner, P. Factors influencing the discrimination and classification of prostate cancer cell lines by FTIR microspectroscopy†. Analyst 2009, 134, 1083–1091. [Google Scholar] [CrossRef]
- Harvey, T.J.; Henderson, A.; Gazi, E.; Clarke, N.W.; Brown, M.; Faria, C.; Snook, R.D.; Gardner, P. Discrimination of prostate cancer cells by reflection mode FTIR photoacoustic spectroscopy. Analyst 2007, 132, 292–295. [Google Scholar] [CrossRef]
- He, D.; Guan, Z.; Fan, J.; Cao, P.; Zhang, G.; Wang, J.; Dang, Q.; Wang, X.; Huang, L.; Wang, L.; et al. Raman spectroscopy, a potential tool in diagnosis and prognosis of castration-resistant prostate cancer. J. Biomed. Opt. 2013, 18, 087001. [Google Scholar] [CrossRef][Green Version]
- Pogoda, K.; Pięta, E.; Roman, M.; Piergies, N.; Liberda, D.; Wróbel, T.P.; Janmey, P.A.; Paluszkiewicz, C.; Kwiatek, W.M. In search of the correlation between nanomechanical and biomolecular properties of prostate cancer cells with different metastatic potential. Arch. Biochem. Biophys. 2021, 697. [Google Scholar] [CrossRef]
- Mukherjee, P.; Lim, S.J.; Wrobel, T.P.; Bhargava, R.; Smith, A.M. Measuring and Predicting the Internal Structure of Semiconductor Nanocrystals through Raman Spectroscopy. J. Am. Chem. Soc. 2016, 138. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, T.P.; Kwak, J.T.; Kadjacsy-Balla, A.; Bhargava, R. High-definition Fourier transform infrared spectroscopic imaging of prostate tissue. In Proceedings of the Progress in Biomedical Optics and Imaging—Proceedings of SPIE, San Francisco, CA, USA, 13 February 2016; Volume 9791. [Google Scholar]
- Pérez-Guaita, D.; Kuligowski, J.; Garrigues, S.; Quintás, G.; Wood, B.R. Assessment of the statistical significance of classifications in infrared spectroscopy based diagnostic models. Analyst 2015, 140, 2422–2427. [Google Scholar] [CrossRef]
- Pérez-Guaita, D.; Kuligowski, J.; Lendl, B.; Wood, B.R.; Quintás, G. Assessment of discriminant models in infrared imaging using constrained repeated random sampling—Cross validation. Anal. Chim. Acta 2018, 1–9. [Google Scholar] [CrossRef]
- Koziol, P.; Raczkowska, M.K.; Skibinska, J.; Urbaniak-Wasik, S.; Paluszkiewicz, C.; Kwiatek, W.; Wrobel, T.P. Comparison of spectral and spatial denoising techniques in the context of High Definition FT-IR imaging hyperspectral data. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Koziol, P.; Raczkowska, M.K.; Skibinska, J.; McCollum, N.J.; Urbaniak-Wasik, S.; Paluszkiewicz, C.; Kwiatek, W.M.; Wrobel, T.P. Denoising influence on discrete frequency classification results for quantum cascade laser based infrared microscopy. Anal. Chim. Acta 2018. [Google Scholar] [CrossRef]
- Gorrochategui, E.; Jaumot, J.; Lacorte, S.; Tauler, R. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: Overview and workflow. TRAC Trends Anal. Chem. 2016, 82, 425–442. [Google Scholar] [CrossRef]
- Singh, R.; Wrobel, T.P.; Mukherjee, P.; Gryka, M.; Kole, M.; Harrison, S. Bulk Protein and Oil Prediction in Soybeans Using Transmission Raman Spectroscopy: A Comparison of Approaches to Optimize Accuracy. Appl. Spectrosc. 2019. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.; Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M.C. Breaking with trends in pre-processing? TRAC Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Zimmermann, B.; Kohler, A. Optimizing savitzky-golay parameters for improving spectral resolution and quantification in infrared spectroscopy. Appl. Spectrosc. 2013, 67, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Filzmoser, P.; Walczak, B. What can go wrong at the data normalization step for identification of biomarkers? J. Chromatogr. A 2014, 1362, 194–205. [Google Scholar] [CrossRef]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New data preprocessing trends based on ensemble of multiple preprocessing techniques. TRAC Trends Anal. Chem. 2020, 132, 116045. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. A contemporary review on Data Preprocessing (DP) practice strategy in ATR-FTIR spectrum. Chemom. Intell. Lab. Syst. 2017, 163, 64–75. [Google Scholar] [CrossRef]
- Oliveri, P.; Malegori, C.; Simonetti, R.; Casale, M. The impact of signal pre-processing on the final interpretation of analytical outcomes—A tutorial. Anal. Chim. Acta 2019, 1058, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Martyna, A.; Menżyk, A.; Damin, A.; Michalska, A.; Martra, G.; Alladio, E.; Zadora, G. Improving discrimination of Raman spectra by optimising preprocessing strategies on the basis of the ability to refine the relationship between variance components. Chemom. Intell. Lab. Syst. 2020, 202. [Google Scholar] [CrossRef]
- Bassan, P.; Kohler, A.; Martens, H.; Lee, J.; Jackson, E.; Lockyer, N.; Dumas, P.; Brown, M.; Clarke, N.; Gardner, P. RMieS-EMSC correction for infrared spectra of biological cells: Extension using full Mie theory and GPU computing. J. Biophotonics 2010, 3, 609–620. [Google Scholar] [CrossRef]
- Solheim, J.; Gunko, E.; Petersen, D.; Großerüschkamp, F.; Gerwert, K.; Kohler, A. An open source code for Mie Extinction EMSC for infrared microscopy spectra of cells and tissues. J. Biophotonics 2019, 10–16. [Google Scholar] [CrossRef]
- Wrobel, T.P.; Liberda, D.; Koziol, P.; Paluszkiewicz, C.; Kwiatek, W.M. Comparison of the new Mie Extinction Extended Multiplicative Scattering Correction and Resonant Mie Extended Multiplicative Scattering Correction in transmission infrared tissue image scattering correction. Infrared Phys. Technol. 2020, 107, 103291. [Google Scholar] [CrossRef]
- Eilers, P.H.C. A Perfect Smoother. Anal. Chem. 2003, 75, 3631–3636. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1 H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.M.; Klein, M.S.; Hochrein, J.; Oefner, P.J.; Spang, R.; Gronwald, W. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics 2012, 8, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Bassan, P.; Kohler, A.; Martens, H.; Lee, J.; Byrne, H.J.; Dumas, P.; Gazi, E.; Brown, M.; Clarke, N.; Gardner, P. Resonant Mie scattering (RMieS) correction of infrared spectra from highly scattering biological samples. Analyst 2010, 135, 268–277. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedure. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Hen, X.I.S.; Iang, L.X.U.; Hubin, S.Y.E.; Ong, R.H.U.; In, L.I.N.G.J.; Anyang, H.X.U.; Iu, W.E.L. Automatic baseline correction method for the open-path Fourier transform infrared spectra by using simple iterative averaging. Opt Express 2018, 26, 609–614. [Google Scholar]
- Eilers, P.H.C. Baseline Correction with Asymmetric Least Squares Smoothing. Anal. Chem. 2005, 75, 3631–3636. [Google Scholar] [CrossRef]
- Bassan, B.P.; Kohler, A.; Byrne, H.J.; Martens, H.; Lee, J.; Bassan, P.; Kohler, A.; Martens, H.; Lee, J.; Byrne, H.J.; et al. Resonant Mie Scattering (RMieS) EMSC correction guide. Analyst 2010. [Google Scholar] [CrossRef]
- Konevskikh, T.; Lukacs, R.; Kohler, A. An improved algorithm for fast resonant Mie scatter correction of infrared spectra of cells and tissues. J. Biophotonics 2017, 1–10. [Google Scholar] [CrossRef]
- Bylesjo, M.; Cloarec, O.; Rantalainen, M. Normalization and Closure. In Comprehensive Chemometrics; Brown, S., Tauler, R., Walczak, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; Volume 2, pp. 109–127. [Google Scholar]
- Reis, M.S.; Saraiva, P.M.; Bakshi, B.R. Denoising and Signal-to-Noise Ratio Enhancement: Wavelet Transform and Fourier Transform. In Comprehensive Chemometrics; Brown, S., Tauler, R., Walczak, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; Volume 2, pp. 25–55. ISBN 9780444527011. [Google Scholar]
- Wrobel, T.P.; Mukherjee, P.; Bhargava, R. Rapid visualization of macromolecular orientation by discrete frequency mid-infrared spectroscopic imaging. Analyst 2017, 142, 75–79. [Google Scholar] [CrossRef] [PubMed]


| Denoising | Adjusted Parameter | Baseline | Adjusted Parameter | Normalization | Internal Accuracy | External Accuracy | ||
|---|---|---|---|---|---|---|---|---|
| Original Data | ||||||||
| Fourier | frame | 100 | Second derivative | Poly, frame | 2, 27 | CONSTANT | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 2, 29 | CONSTANT | 0.94 | 0.79 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 3, 27 | CONSTANT | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 3, 29 | CONSTANT | 0.94 | 0.79 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 2, 23 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 2, 25 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 2, 27 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 2, 29 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 3, 23 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 3, 25 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 3, 27 | TSN | 0.94 | 0.86 |
| Fourier | frame | 100 | Second derivative | Poly, frame | 3, 29 | TSN | 0.94 | 0.86 |
| Noise Added Data | ||||||||
| Fourier | frame | 140 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.86 |
| Fourier | frame | 220 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| Eilers | λ | 6 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.86 |
| SavitzkyG | Poly, frame | 2, 15 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 2, 17 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.86 |
| SavitzkyG | Poly, frame | 2, 19 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 2, 21 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 2, 23 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 3, 15 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 3, 17 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.86 |
| SavitzkyG | Poly, frame | 3, 19 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 3, 21 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
| SavitzkyG | Poly, frame | 3, 23 | ALS | λ, p | 6, 0.1 | CONSTANT | 0.91 | 0.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liberda, D.; Pięta, E.; Pogoda, K.; Piergies, N.; Roman, M.; Koziol, P.; Wrobel, T.P.; Paluszkiewicz, C.; Kwiatek, W.M. The Impact of Preprocessing Methods for a Successful Prostate Cell Lines Discrimination Using Partial Least Squares Regression and Discriminant Analysis Based on Fourier Transform Infrared Imaging. Cells 2021, 10, 953. https://doi.org/10.3390/cells10040953
Liberda D, Pięta E, Pogoda K, Piergies N, Roman M, Koziol P, Wrobel TP, Paluszkiewicz C, Kwiatek WM. The Impact of Preprocessing Methods for a Successful Prostate Cell Lines Discrimination Using Partial Least Squares Regression and Discriminant Analysis Based on Fourier Transform Infrared Imaging. Cells. 2021; 10(4):953. https://doi.org/10.3390/cells10040953
Chicago/Turabian StyleLiberda, Danuta, Ewa Pięta, Katarzyna Pogoda, Natalia Piergies, Maciej Roman, Paulina Koziol, Tomasz P. Wrobel, Czeslawa Paluszkiewicz, and Wojciech M. Kwiatek. 2021. "The Impact of Preprocessing Methods for a Successful Prostate Cell Lines Discrimination Using Partial Least Squares Regression and Discriminant Analysis Based on Fourier Transform Infrared Imaging" Cells 10, no. 4: 953. https://doi.org/10.3390/cells10040953
APA StyleLiberda, D., Pięta, E., Pogoda, K., Piergies, N., Roman, M., Koziol, P., Wrobel, T. P., Paluszkiewicz, C., & Kwiatek, W. M. (2021). The Impact of Preprocessing Methods for a Successful Prostate Cell Lines Discrimination Using Partial Least Squares Regression and Discriminant Analysis Based on Fourier Transform Infrared Imaging. Cells, 10(4), 953. https://doi.org/10.3390/cells10040953







